Abstract
The alfalfa leafcutting bee, Megachile rotundata (ALCB) is an economically important pollinator necessary for seed production of the critical forage crop alfalfa, Medicago sativa. The pollinator was accidentally introduced to North America from Europe approximately 70 years ago, and it is primarily produced in Canada and shipped to the United States annually en masse for seed field pollination. We investigate how the large-scale commercial movement of this bee affects the genetic structure of populations in the North American seed growing system and compare the genetic diversity and structure of introduced North American bees with two native European populations. Using 16 newly developed microsatellite loci, we describe the North American population structure of this bee. ALCBs collected from alfalfa seed farms have a degree of genetic variability similar to one native European population, but lower than the second. Considering that the species was accidentally introduced into North America, we anticipated more signature of a founder effect. Despite the level of genetic variability, we found little, if any, genetic structuring across North America, other than that the North American populations were distinct from the European populations sampled. While we detected some sub-structure in North American populations using Bayesian methods, the structuring was without geographic pattern, and we propose it is the result of the intense human management and movement of these bees. The trade and movement of these bees by humans has created a nearly panmictic M. rotundata population across the continent, which has implications relevant to the preservation and conservation of other bee pollinators.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
We describe here the population genetic structure of the alfalfa leafcutting bee, Megachile rotundata L., (Hymenoptera: Megachilidae)(ALCB), the second most important, managed pollinator after honey bees (Apis mellifera L., Hymenoptera: Apidae). When an organism is intensively managed for agricultural use, the genetic diversity and structure of the domesticated populations can become altered relative to their wild progenitors (Yukuhiro et al. 2002). When insects are reared en masse, some species lose genetic diversity due to founder effects and inbreeding (e.g., screw worms Bush and Neck 1976), while other species remain quite diverse despite domestication (e.g., silk worms Xia et al. 2009). Honey bee colonies have been managed by humans a few thousand years, probably longer than any other insect (Bloch et al. 2010; Crane 1999). Initially, honey bees were kept primarily for honey production, but in recent history, increased demands for crop pollination have increased the demand for the management and movement of bees; not only honey bees, but other species as well, and this movement has occurred on both local and global scales (James and Pitts-Singer 2008; Pitts-Singer and Cane 2011; Byatt et al. 2016). The realization that crop yields can be improved by bringing bees into the fields for pollination has contributed to the rise of new industries that raise and sell both native and non-native bumble bees (Bombus spp.), mason bees (Osmia spp.), and ALCBs, in addition to honey bees (James and Pitts-Singer 2008). Consequently, human propagation and movement of bees has the potential to change the distribution and the population genetics for all these species, as has widely been documented in honey bees (e.g., Crane 1999; Delaney et al. 2009; Cobey et al. 2012; Harpur et al. 2012).
The genus Megachile is very large and diverse, with an estimated 1525 species worldwide (Ascher and Pickering 2011; Gonzalez et al. 2012). ALCBs are solitary, cavity-nesting bees native to Europe and Africa (Friese 1899; Enkulu 1988), unintentionally introduced into the United States at least as early as the 1940s (Hurd 1954; Pitts-Singer and Cane 2011). ALCBs subsequently became widespread and abundant throughout parts of western North America (Pitts-Singer and Cane 2011), and they became recognized in the 1960s as an effective pollinator of alfalfa (lucerne), Medicago sativa L. (Stephen and Torchio 1961). ALCBs are now propagated and managed on a commercial scale to pollinate alfalfa for seed production systems (for details of bee management and production see Pitts-Singer and James 2005; Pitts-Singer 2008; Pitts-Singer and Cane 2011). This bee greatly enhances alfalfa seed yield, as compared to pollination by honey bees (Richards 1984; Frank 2003). Many commercial suppliers of ALCBs are in Canada (Table S1), where this bee tends to have higher survival rates (Pitts-Singer and Cane 2011), and most of the market for the bees is in the western US alfalfa seed production fields (Pitts-Singer and James 2005).
The biology of the ALCB is fairly typical of cavity nesting solitary bees (Johansen and Eves 1973). These bees nest in small cavities, constructing cells from cut leaf pieces in the summer. In managed systems, the bees are provided communal nesting sites of cavities in large wood or polystyrene blocks (Pitts-Singer and Cane 2011). The female ALCB provisions each cell with a pollen and nectar mass, then the eggs hatch and larvae feed on the pollen provision and develop over the summer. Mature fifth instar larvae (pre-pupae) spin a cocoon and diapause within their natal cells, spending the winter in this stage, although some portion of the population is often multivoltine (Krunic 1972; Johansen and Eves 1973; Hobbs and Richards 1976; Parker and Tepedino 1982; Rank and Rank 1990; Yocum et al. 2015).
ALCBs have been in North America for at least 70 years and are used agriculturally throughout the western United States and Canada, resulting in an introduced range that includes several latitudes, altitudes, and climate types. The purpose of this study is to describe the current population genetics of ALCBs in agricultural fields of North America. Despite the economic importance of this bee for pollination, there are only two studies that describe its genetic variability, and both were based on small, localized populations (Blanchetot 1992; Lu and Rank 1996). As such, relatively little is known about the genetic structure of this species, neither in its native range nor in North America, thus we would benefit by understanding the population genetics of this important species.
Methods
Sample collection
We obtained 282 female pre-pupae as loose cells from the United States and Canada and 51 samples of adult females from Europe (Table 1). North American bees were obtained as overwintering pre-pupae (as is standard for the industry; see Pitts-Singer 2008) from ALCB producers in the United States and Canada in 2012 to the USDA-ARS-Pollinating Insect Research Unit in Logan, UT, where subsequent analyses were performed. We randomly selected ten individual bees from each producer for the molecular analyses and after an initial screening, removed the males from that number, resulting in a final sample size of 282 bees from North America. We obtained additional European samples of adult bees from Copenhagen (Denmark) and Barcelona (Spain), which had been freeze-killed and shipped dry in petri dishes. We confirmed species identity, through assessment of morphological characters.
Sample preparation
We removed pre-pupae from their respective cells and placed them into individual 2.0 ml screw top tubes. To each, we then added 1 ml DNA extraction buffer (100 mM Tris–HCL, pH 8.0; 2% CTAB; 20 mM EDTA) and crushed the larvae with a plastic pestle. We incubated each preparation at 65 °C for 30 min, then added 0.8 ml of phenol:chloroform (1:1) before centrifuging the tubes for 20 min at 13,500 rpm. Following centrifugation, we transferred the supernatant to another tube, washed it with 0.7 ml of chloroform, and centrifuged again for 3 min at 13,500 rpm. We then transferred the supernatant to another tube, added 0.8 ml of cold isopropanol (−20 °C), and incubated at −20 °C for at least 30 min, after which we centrifuged the tubes at 13,500 for 30 min, poured off the isopropanol, washed the pellet with 0.5 ml of 70% (v/v) ethanol, and dried it in a fume hood for 10 min. We re-suspended the pellet in 200 µl of TE (10 mM Tris–HCl; 1 mM EDTA, pH 8.0) and aliquoted 100 µl into two microcentrifuge tubes to store at −20 °C until PCR amplification.
We extracted DNA from a single leg of each adult bee from Europe and the congeneric bees that were tested for cross-species amplification using a Chelex® extraction protocol. We removed a single leg (generally a mid-leg) and placed it in an individual well of a 96 well PCR plate. We submerged the legs in 150 µl of a 5% Chelex® solution and crushed with forceps. We then added Proteinase K (5 µl of 10 mg/ml) to each well and incubated the samples for 1 h at 55 °C, 15 min at 99 °C, 1 min at 37 °C and 15 min at 99 °C (modified from Walsh et al. 1991). We stored the extracted DNA at −20 °C until PCR amplification.
We amplified all 333 female ALCBs and visualized fragments on an ABI® 3730xl sequencer. We performed four 10 µl multiplex reactions each containing: 1 µl extracted DNA, 1x Promega (Madison, WI) reaction buffer, 0.6 mM dNTP mixture, 0.1–0.4 μM primer, 0.001 mg BSA, 0.4 units Taq polymerase (Promega, Madison, WI), and the MgCl2 concentration was adjusted to 1.4 mM. The four reactions contained the following primers (Table S2): (A) Mrot0001, Mrot0005, Mrot0006, and Mrot0012 and Mrot0017; (B) Mrot0009, Mrot0015, Mrot0016, and Mrot0018; (C) Mrot0013, Mrot0019, Mrot0020, Mrot0026, and Mrot0035; (D) Mrot0023, Mrot0027, Mrot0029, and Mrot0031. The PCR conditions for multiplex reactions were: one 4 min cycle at 95 °C, 30 cycles of 95 °C for 30 s, and annealing at 54 °C for 75 s, then 72 °C for 45 s. The cycles were followed by a final extension period of 15 min at 72 °C. We separated the DNA amplifications on an Applied Biosystems 3730xl automatic sequencer (Life Technologies), and we scored allele sizes using GeneMapper™ v4.0 Software (Applied Biosystems). We removed two loci that were either monomorphic (Mrot0020), exhibited significant linkage disequilibrium, or deviated from HWE (Mrot0013), resulting in 16 loci for the population genetics analyses (see Supplemental Material for details on marker development and selection).
Population genetics and spatial analysis
Because of differences in sample sizes across regions, we used the program HP-Rare (Kalinowski 2005) to calculate allelic richness and private allelic richness. We used FSTAT (Goudet 2001) to determine the fixation index F ST for each pair of groups. F ST, in this case, is a useful measure for examining population substructure to assess genetic divergence among population pairs and to test for isolation by distance. Heterozygosity and population pairwise comparisons were calculated using Arlequin v.3.5.1.3.
To assess geographic structuring in North American populations and determine the appropriate geographic level for conducting our analyses, we first performed a test for isolation by distance. For all North American farms sampled that had six or more bees (n = 18 farms), we calculated pairwise genetic (F ST ) and linear geographic distances (km) then conducted an “Isolation by Distance” (IBD) analysis (Genepop v.4.2, Isolation By Distance) (Raymond and Rousset 1995; Rousset 2008) with 1000 permutations for the Mantel test and the minimum distance for regression set at 100 km, so that adjacent farms which might show inbreeding would not bias a distance effect. We used a log transformation of linear distances and converted F-statistics to F/(1−F) for the analysis. We detected no significant farm-by-farm F ST values and so chose to pool samples by state or province for subsequent analyses, in an effort to elucidate differences in genetic structure geographically.
Population structure
We used the program STRUCTURE (Pritchard et al. 2000) to infer population structure, employing a Bayesian method to determine the probability of assigning each individual to K hypothetical clusters, when K was initially unknown (admixture model, correlated allele frequencies). We separated runs for each value of K from 2 to 13, with 50,000 burn-ins and 100,000 replications after burn-in. We estimated the optimal K value using the ΔK method (Evanno et al. 2005) in STRUCTURE HARVESTER (Earl and vonHoldt 2012). Neither this method nor the IBD analysis revealed a fine structure (i.e., farm-by-farm or region-by-region) in populations, thus the state-by-state analyses that follow are appropriate.
Bottleneck and migration
Using the program BOTTLENECK (Cornuet and Luikart 1997), we tested all populations for heterozygote excess under the infinite alleles model and the stepwise mutation model as described in Cornuet and Luikart (1997) to detect a significant mode shift in allele frequency distributions. This enabled us to assess the possibilities of a population bottleneck in North American ALCBs by employing a simulation to test variation in the mutation-drift equilibrium that would result from a recent population bottleneck (Maruyama and Fuerst 1985; Cornuet and Luikart 1997). To detect migrants in individual populations, we chose to use population information in our populations under the ancestory model tab in the STRUCTURE analyses. We set the parameters at the default (GENSBACK = 2; MIGRPRIOR = 0.05) using the admixture model.
Results
Population genetic diversity
The average allelic richness (±SEM) across all loci for each sampling location varied from 3.99 ± 0.54 (Spain) to 2.50 ± 0.52 (Utah), and the private allelic richness across all loci for each sampling location varied from 1.44 ± 0.72 (Spain) to 0.08 ± 0.14 (WA) (Table 1). The samples of ALCBs from Spain had a markedly higher average number of private alleles per locus than ALCBs from any of the other sample locations (Table 1). Specifically, ALCBs from Spain had more private alleles at 15 of the 16 loci.
Based on the pairwise multi-locus F ST estimates, we determined that the ALCB samples from Denmark were significantly, genetically distinct from ALCBs from California, Utah, and Spain (for correlation coefficients and statistical tests, see Table 2). Significant population differentiation was also observed between Spain and all other locations. The ALCB samples sizes from Washington were not large enough to calculate a reliable F ST estimate, so results from that population should be viewed with caution; however, they are consistent with the other North American samples.
Spatial analysis
No significant signature of IBD was found among the 18 farms with more than six bees that we tested (n = 122 pairwise comparisons; r 2 = 0.028; df = 121; P = 0.142). The correlation of genetic distance to geographic distance was very weak and not statistically significant. We observed some evidence of high genetic similarity at sites less than 100 km apart (evidenced by F ST values not significantly different from zero among nearby farms), which is perhaps only a sign that neighboring farms are likely to purchase bees from the same supplier.
Population structure
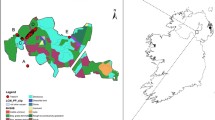
For samples from North America only (data not shown), STRUCTURE and STRUCTURE HARVESTER both identified two distinct population clusters (K = 2). This conclusion is supported by three separate estimations of K: ΔK, rate of change of the likelihood distribution, and the mean of ln(p(X/K)). As such, there is little, if any, geographic population structuring across the entire North American continent for this species. Both putative populations in the North American ALCB were represented in each region and generally were found among the bees of each sampled farm (Fig. 1 ).
Map of North America indicating the collection locations for Megachile rotundata. Individual farms are represented by white dots. Population genetic identity was pooled by province or state, and these pooled populations are represented by pie charts that show the proportion of genetic assignment within the political entity, using K = 4 populations averaged across all individuals with that political region
When the European samples were included in a full analysis, STRUCTURE and STRUCTURE HARVESTER both identified four distinct population clusters (K = 4; Fig. 2). This conclusion is supported by three separate estimations of K: ΔK, rate of change of the likelihood distribution, and the mean of ln(p(X/K)) (see Figure S1). The clusters were geographically associated in Europe (Denmark and Spain), but not in North America where the weakly differentiated populations showed no relation to geographic origin.
STRUCTURE diagrams of the individuals within sampled regions for different genetic clusters (K): a K = 2; b K = 3; and c K = 4 and K = 7 genetic populations. The K = 4 and K = 7 graphs were chosen because these two estimations had the highest support in the population structure analyses (K = 4, ΔK = 14.024; K = 7, ΔK = 14.025). For each individual on the x-axis, the percentage of genetic assignment is given
Population bottleneck and migration
We found no clear evidence of population bottlenecks in the North American ALCB populations as evidenced by a mode shift from a normal L-shaped distribution of the allele frequencies. However, in some populations we did find heterozygote deficiencies and excesses, but only in one population was the mode of the distribution shifted significantly, and indication of a recent bottleneck (Table S4). It was among the Washington State samples that allele frequency distribution deviated from the normal L-shaped distribution, and this deviation was statistically significant under the stepwise mutation model (p = 0.02), but was not significant in the infinite alleles model (p = 0.52). This population is the only one that showed any sign of a recent bottleneck; however, it also had the smallest sample size (n = 3), and thus should be viewed with caution.
Only three of the 333 individuals (0.9%) tested were found to be potential migrants in our analyses. One individual from California and two individual bees from Spain were identified as potential migrants in the analysis. The bee from California assigned primarily to the Denmark population with California being the secondary assignment; whereas the two bees from Spain each had weak association with several potential areas of origin and the strongest association with the Spanish population. Given the low level of population differentiation in North America, as evidenced by F ST values, the power to detect migration among regions using this method may be limited.
Discussion
We found that managed ALCB populations in North America had high genetic diversity, little to no geographic genetic structuring, and no evidence of genetic bottlenecks, except perhaps at one sampling site. Based on the results from our European samples, the ALCB appears to have a high genetic diversity in its natural range, a characteristic that could make this bee adaptable to many different environments (De Palma et al. 2015), providing at least some explanation as to why it has been such a successful invader.
While the sample size from the Old World is relatively small, the ALCBs we sampled from North American alfalfa seed farms had a high degree of genetic variability that was comparable to the Spanish population. It would be interesting to see if the allelic richness and degree of structure that we observed in the Spanish sample is reflective of broader patterns of European ALCB populations, or if the lower level of diversity observed in Denmark is more the norm within the native range of this bee. Considering that ALCBs were accidentally introduced into North America, and presumably had very few introduction events (Hurd 1954), we expected it to exhibit some degree of a founder effect there. We propose several possible reasons for this lack of a genetic bottleneck in the invaded region. The first possibility is that the North American populations have already reached migration-drift equilibrium. Alternatively, although it has been assumed the founding population came from a small migration event (Hurd 1954), multiple introductions might actually have occurred. The first introductions were estimated to have occurred in the 1940s (Hurd 1954), but it is plausible that multiple introductions occurred over time from bees nesting in wooden shipping crates from Europe. Furthermore, contemporary gene flow may still be occurring as recent invasions into North America have been documented for other Megachilidae bees (Paiero and Buck 2003; Strange et al. 2011). Multiple introduction events can create a novel and diverse gene pool, founded from multiple European populations that are being homogenized by the subsequent human-mediated mass movement of ALCBs within North America, similar to what is observed in other domesticated organisms (Yukuhiro et al. 2002; Delaney et al. 2009). A third possibility is that the initial founder population in North America was very large, providing the relatively diverse population we now observe.
Although we found the population structure to lack any geographic pattern in North America, these populations were markedly different from those sampled in Europe, especially Spain. Thus, there has either been a lack of localized adaptation in North America, a frequent and thorough mixing of the gene pool, or both. In other words, despite having high genetic variability and occurring across a very large region that includes a large range in latitudes, climate zones, and elevations, the ALCB has not differentiated into unique populations. This phenomenon is likely caused by human intervention, because alfalfa seed growers import bees from Canada every year. The Canadian producers tend to produce healthier bees, and nearly half of all the bees used in the United States are imported from Canada annually, on average (Table S1). Thus, a large proportion of the bees are reared in Canada and then moved south to the United States. In addition, a few of the U.S. farmers raise and sell their own bees, though never to Canada. Canada does not allow the bees to be imported (Canada Food Inspection Agency 2013). It is likely that some movement of bees is also occurring within Canada, thus preventing local population structure from developing in the northern portions of the range. In contrast, the ALCB is not typically managed in Europe, and where it is, it is managed locally and not transported across regions as in North America (Pitts-Singer and Cane 2011).
The degree of allelic diversity we observed within the North American ALCB samples, coupled with the weak structuring found among widely distributed North American sample sites, is in contrast to what we observed in the endemic range of the species in Europe, where high levels of diversity were coupled with strong structuring of populations. The lack of structure in ALCB in North America is similar to that observed with honey bees in North America, another introduced, managed bee (Harpur et al. 2012, 2015). In the areas where the bees are non-native, the high rates of human-mediated migration have not reduce the genetic diversity, but rather they have created a geographically widespread, panmictic population with high levels of diversity (Delaney et al. 2009). In contrast, movement of bees in Europe risks mixing distinct ecotypes that are well adapted to local environments within the native range (Strange et al. 2008). What is yet unclear from this current study is whether the maintenance of genetic diversity is sufficient in these panmictic populations to adapt to local environmental variation or climate changes. Genetic diversity also has implications for bee conservation efforts that reintroduce or translocate species from areas of abundance to areas of the native range where the species has been extirpated. The implications for such movement goes beyond genetic considerations (Lozier et al. 2015), but using this endemic bee as an example, perhaps moving other bee species in re-population efforts is not as harmful to population-wide genetic diversity as previously assumed.
References
Ascher JS, Pickering J (2011) Bee species guide (Hymenoptera:Apoidea: Anthophila). http://www.discoverlife.org/mp/20q?guide=Apoidea_species. Accessed Mar 2016
Blanchetot A (1992) DNA fingerprinting analysis in the solitary bee Megachile rotundata: variability and nest mate genetic relationships. Genome 35:681–688
Bloch G, Francoy TM, Wachtel I, Panitz-Cohen N, Fuchs S, Mazar A. (2010) Industrial apiculture in the Jordan valley during Biblical times with Anatolian honeybees. Proc Nat Acad Sci USA 107:11240–11244
Bush GL, Neck RW (1976) Ecological genetics of screwworm fly, Cochliomyia hominivorax (Diptera-Calliphoridae) and its bearing on quality-control of mass-reared insects. Environ Entomol 5:821–826
Byatt MA, Chapman NC, Latty T, Oldroyd BP (2016) The genetic consequences of the anthropogenic movement of social bees. Insectes Soc 63:15–24
Canadian Food Inspection Agency (2013) Alfalfa leaf cutting bee producers guide to the national bee farm-level biosecurity standard. Section1: Bee health management. http://www.inspection.gc.ca/. Accessed Jan 2017
Cobey SW, Sheppard WS, Tarpy DR (2012) Status of breeding practices and genetic diversity in domestic us honey bees. In: Sammataro D, Yoder JA (eds) Honey bee colony health: challenges and sustainable solutions. CRC Press, Boca Raton, pp. 25–36
Cornuet JM, Luikart G (1997) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Crane E (1999) Beekeeping and honey hunting. Routledge, New York
De Palma A, Kuhlmann M, Roberts SPM, Potts SG, Börger L, Hudson LN, Lysenko I, Newbold T, Purvis A (2015) Ecological traits affect the sensitivity of bees to land-use pressures in European agricultural landscapes. J Appl Ecol 52:1567–1577. doi:10.1111/1365-2664.12524
Delaney DA, Meixner MD, Schiff NM, Sheppard WS (2009) Genetic characterization of commercial honey bee (Hymenoptera: Apidae) populations in the United States by using mitochondrial and microsatellite markers. Ann Entomol Soc Am 102:666–673
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Enkulu L (1988) Les Megachiles (Hymenoptera, Apoidea) D’Europe et D’Afrique. Faculte´ Des Sciences Agronomiques de L’Etat.
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Frank G (2003) Alfalfa seed and leafcutter bee productions and marketing manual. Association ASP, Brooks
Friese H (1899) Die bienen Europa’s (Apidae eropaeae). Theil V. Solitaire Apiden: Genus Lithurgus Genus Megachile (Chalkicodoma). Insbruck.
Gonzalez VH, Griswold T, Praz CJ, Danforth BN (2012) Phylogeny of the bee family Megachilidae (Hymenoptera: Apoidea) based on adult morphology. Syst Entomol 37:261–286
Goudet J (2001) FSTAT Version 2.9.3: a program to estimate and test gene diversities and fixation indices. Institute of Ecology, University of Lausanne.
Harpur BA, Minaei S, Kent CF, Zayed A (2012) Management increases genetic diversity of honey bees via admixture. Mol Ecol 21:4414–4421
Harpur BA, Chapman NC, Krimus L, Maciukiewicz P, Sandhu V, Sood K, Lim J, Rinderer TE, Allsopp MH, Oldroyd BP, Zayed A (2015) Assessing patterns of admixture and ancestry in Canadian honey bees. Insectes Soc 62:479–480
Hobbs GA, Richards KW (1976) Selection for a univoltine strain of Megachile (Eutricharaea) pacifica (Hymenoptera-Megachilidae). Can Entomol 108:165–167
Hurd PD (1954) Distributional notes on Eutricharea, a Palearctic subgenus of Megachile, which has become established in the United States (Hymenoptera: Megachilidae.). Ent News 65:93–95
James RR, Pitts-Singer TL (2008) Bee pollination in agricultural ecosystems. Oxford University Press, Oxford
Johansen CA, Eves JD (1973) Effects of chilling humidity and seasonal conditions on emergence of the alfalfa leaf-cutting bee. Environ Entomol 2:23–26
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Krunic MD (1972) Voltinism in Megachile rotundata (Megachilidae-Hymenoptera) in southern Alberta. Can Entomol 104:185–188
Lozier JD, Cameron SA, Duennes MA, Strange JP, Williams PH, Goulson D, Brown MJF, Morales C, Jepsen S (2015) Relocation risky for bumblebee colonies. Science 350:286–287
Lu R, Rank GH (1996) Use of RAPD analyses to estimate population genetic parameters in alfalfa leaf-cutting bee, Megachile rotundata. Genome 39:655–663
Maruyama T, Fuerst PA (1985) Population bottlenecks and non equilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
Paiero SM, Buck M (2003) The giant resin bee,Megachile sculpturalisSmith, and other newly introduced and newly recorded native Megachilidae and Andrenidae (Apoidea) from Ontario. J Entomol Soc Ont 134:141–143
Parker FD, Tepedino VJ (1982) Maternal influence on diapause in the alfalfa leafcutting bee Megachile, rotundata (F.) (Hymenoptera, Megachilidae). Ann Entomol Soc Am 75:407–410
Pitts-Singer TL (2008) Past and present management of alfalfa bees. In: Pitts-Singer TL, James RR (eds) Bee Pollination in Agricultural Ecosystems. Oxford University Press, New York, pp 105–123
Pitts-Singer TL, Cane JH (2011) The alfalfa leafcutting bee,Megachile rotundata : the world’s most intensively managed solitary bee. Annu Rev Entomol 56:221–237
Pitts-Singer TL, James RR (2005) Emergence success and sex ratio of commercial alfalfa leafcutting bees from the United States and Canada. J Econ Entomol 98:1785–1790
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rank GH, Rank FP (1990) Preharvest diurnal temperature-variations increase diapause development in the alfalfa leafcutting bee Megachile rotundata (Fab) (Hym Megachilidae). J Appl Entomol 110:313–317
Raymond M, Rousset F (1995) GENEPOP (Version-1.2)—Population-genetics software for exact tests and ecumenicism. J Hered 86:248–249
Richards KW (1984) Alfalfa leafcutter bee management in western Canada. Agriculture Canada Publication, Canada, pp. 1–53
Rousset F (2008) Genepop’007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
Stephen WP, Torchio PF (1961) Biological notes on the leaf-cutter bee, Megachile (Eutricharaea) rotundata (Fabricius). (Hymenoptera : Megachilidae). Pan Pacific Ent 37:85–93
Strange JP, Garnery L, Sheppard WS (2008) Morphological and molecular characterization of the Landes honey bee (Apis mellifera L.) ecotype for genetic conservation. J Insect Conserv 12:527–537
Strange JP, Koch JB, Gonzalez VH, Nemelka L, Griswold T (2011) Global invasion byAnthidium manicatum(Linnaeus)(Hymenoptera: Megachilidae): assessing potential distribution in North America and beyond. Biol Invasions 13:2115–2133
Walsh PS, Metzger DA, Higuchi R (1991) Chelex® 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513
Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, Cheng T, Jiang T, Becquet C, Xu X, Liu C, Zha X, Fan W, Lin Y, Shen Y, Jiang L, Jensen J, Hellmann I, Tang S, Zhao P, Xu H, Yu C, Zhang G, Li J, Cao J, Liu S, He N, Zhou Y, Liu H, Zhao J, Ye C, Du Z, Pan G, Zhao A, Shao H, Zeng W, Wu P, Li C, Pan M, Li J, Yin X, Li D, Wang J, Zheng H, Wang W, Zhang X, Li S, Yang H, Lu C, Nielsen R, Zhou Z, Wang J, Xiang Z, Wang J (2009) Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326:433–436
Yocum GD, Rinehart JP, Horvath DP, Kemp WP, Bosch J, Alroobi R, Salem S (2015) Key molecular processes of the diapause to post-diapause quiescence transition in the alfalfa leafcutting bee Megachile rotundata identified by comparative transcriptome analysis. Physiol Entomol 40:103–112
Yukuhiro K, Sezutsu H, Itoh M, Shimizu K, Banno Y (2002) Significant levels of sequence divergence and gene rearrangements have occurred between the mitochondrial genomes of the wild mulberry silkmoth,Bombyx mandarina, and its close relative, the domesticated silkmoth,Bombyx mori. Mol Biol Evol 19:1385–1389
Acknowledgements
We thank Jordi Bosch and Anja Wynns for providing samples from Europe. We thank Craig Huntzinger for organizing and collecting bees from the North American farmers, and for providing most of the DNA extractions. We also thank Joyce Knoblett and Marjorie Gurganus for technical assistance with extracting DNA from specimens and for conducting PCR. We thank Terry Griswold for verifying specimen identity, helping to select a diversity of subgenera within the Megachile, and providing museum specimens to complete our study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Strange, J.P., Delaney, D.A., Tarpy, D.R. et al. Novel microsatellite loci reveal high genetic diversity yet low population structure for alfalfa leafcutting bees in North America. Conserv Genet 18, 679–687 (2017). https://doi.org/10.1007/s10592-017-0943-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-017-0943-9