Abstract
Stocking is a widely applied practice for enhancement of fish populations exploited for recreational and commercial uses. In the present study, we investigate the genetic consequences of stocking and river diversion by analyzing 18 microsatellites in 440 brown trout (Salmo trutta L.) from three historical and ten contemporary populations from two river systems in a national park in Norway. Eight sources have been recorded for stocking in this area, but not concurrently. These stocking events took place after historical sampling and river diversion by construction of a barrier and a channel. A complete shift in the genetic structure between historical and contemporary populations was revealed. The genetic differentiation can be explained by stocking with just four non-native sources. The constructed barrier has helped to maintain a fraction of the historical genetic profile. Stocking success is discussed in relation to population exploitation, variation in natural recruitment, and reduced discharge due to river diversion. Our study demonstrates a high vulnerability of natural populations to stocking with non-native fish, of particular importance for fishery management and preservation of native fish.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The conservation of genetic diversity determines the ability of populations to adapt to environmental changes (Amos and Harwood 1998). For wild populations of brown trout (Salmo trutta L.), anthropogenic habitat fragmentation and destruction are important factors responsible for loss of genetic diversity. River barriers often prevent migratory fish from reaching their preferred reproduction habitats (Gosset et al. 2006), and can reduce population sizes. The total genetic variability may be reduced as a result (Taylor et al. 2003; Wofford et al. 2005), thereby increasing the probability of local extinctions (Gilpin and Soulé 1986; Lande 1998). Despite increased extinction risk, small isolated populations may still persist (Letcher et al. 2007; Cook et al. 2010; Whiteley et al. 2010).
Supplement stocking programs using non-native fish is a widespread management practice to support recreational- and commercial fisheries, and to sustain population density. However, such stocking events may lead to hybridization (Hindar et al. 1991; Hansen et al. 2009; Kohout et al. 2012), and near displacement of original stocks (García-Marín et al. 1991; Sønstebø et al. 2008b). Conversely, there are also reports of low introgression levels even after decades of stocking (Hansen 2002; Heggenes et al. 2006; Hesthagen et al. 2010). Despite the negative effects of habitat fragmentation, man-made barriers and natural waterfalls have also been shown to shelter resident salmonid populations from the effects from introgression and interactions with stocked hatchery-reared individuals (e.g. food and habitat competition) (Thompson and Rahel 1998; Yamamoto et al. 2004; Van Houdt et al. 2005).
In Norway, pristine and relatively untouched lakes and rivers in high alpine environments inhabited by allopatric brown trout populations may still be found. Parts of these alpine areas are protected as national parks and landscape protection areas. In 1981, 3,400 km2 of the Hardangervidda mountain plateau in southern Norway was approved as a national park to conserve its flora, fauna, and cultural monuments. Brown trout populations in this area occur in thousands of lakes and rivers. However, many of these populations have been stocked for enhancement of fisheries, and also affected by habitat fragmentation in connection with water diversion that occurred before the establishment of the national park. Stocking has been permitted to continue after the park’s establishment, as well. We studied the combined effects of stocking and watercourse fragmentation in two neighboring watercourses within Hardangervidda National Park by analyzing historical and contemporary brown trout populations. Such utilization of archival historical scale samples has been shown useful for studying population genetic structure in time and space (Nielsen and Hansen 2008). We hypothesize that (i) stocking and river diversion have led to population displacement above a constructed river barrier, and (ii) fragmentation by the barrier has reduced population displacement in former downstream localities.
Materials and methods
Study area and sampling locations
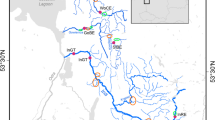
The study area consists of two drainage systems located within the Hardangervidda National Park (Fig. 1). The two systems originate in close vicinity to each other, with the Veig river system running to the west, and the Numedalslågen river system running to the east. Brown trout is the only fish species within the study area. In 1952, the upper part of Veig (called Viersla) was fragmented by construction of a river barrier and opening a channel to the neighbor drainage system, Numedalslågen (Figs. 1, 2). Thereby the discharge from a 12.4 km2 of the drainage area of Veig river system was diverted to the Numedalslågen river system for increasing hydropower production in several power plants located outside the national park. Construction of the barrier took place before known stocking of non-native fish was initiated in the 1960’s (Tysse and Garnås 1996).
(a) Map of Southern Norway, showing the approximate location of the study area on the Hardangervidda mountain plateau. (b) Geographical positions of the sampling locations, HEP Søre Heisandtjørn, HER Herrevatn, OLA Olavsdalselva, SAM Samkomehølen, MGR Midtre Grøndalsvatn, VIR Vierslaelva, VIP “Vierslapytten”, VIV Vierslatjørn, NOR Nordmannslågen, and NDV Nordvatn. Closed arrowheads indicate position of the constructed barrier and the channel. Arrows in rivers indicate water flow direction, and dotted bars waterfalls preventing upstream migration. (c) Simplified illustrations of the hydrologic conditions before 1952, with dotted line indicating the separation of the two river systems, and (d) after 1952, with solid line indicating the diversion of the catchment (VIP, VIR, VIV) to Nordmannslågen (NOR). Arrowheads indicate water flow and waterfalls preventing upstream migration. Asterisk after population abbreviations indicate locations that have been stocked
The barrier (a) and the channel (b) constructed in 1952 to redirect a 12.4 km2 drainage area away from the Veig river system to the Numedalslågen river system. For exact location of the constructed barrier and channel see Fig. 1. (Photos: Reidar Borgstrøm)
About half of the Hardangervidda National Park area is private land, while the remaining part is governmental area, divided into different crown lands, each with local mountain boards. To secure local acceptance for the establishment of the park, land use activities such as sheep summer grazing, outdoor life, hunting, and fishing had to be allowed to continue as before. Brown trout have most likely been present on the mountain plateau for thousands of years, as documented by about 6,500 year old brown trout bones found in kitchen middens in Stone Age camps not far from the study area (Indrelid 1985). The historical use of the natural resources in our study area is well documented by pit–fall traps and connected guiding stone fences used for reindeer hunting (Bakke 1984). In addition to hunting, the brown trout fishery was of high importance, as documented in lawsuits from the 16th century regarding the fishery in the lake Nordmannslågen (Qvenild 2004).
A total of 62 historical samples were retrieved from scale archives of brown trout sampled in 1933 from NOR (NOR33), from the river Olavsdalselva (OLA) in 1945 (OLA45), and from the lake Midtre Grøndalsvatn (MGR) in 1967 (MGR67). The MGR67 and NOR33 samples were also included in a previous analysis by Sønstebø et al. (2008b). Between 2009 and 2011, an additional 378 brown trout were collected from ten contemporary locations representing both sides of the artificial barrier in the upper part of OLA, and thereby also both drainage systems (Veig and Numedalslågen) (Fig. 1). From the river system below the barrier, sampling was performed in the tributary lakes Søre Heisandtjørn (HEP, located 1,245 meter above sea level (m ASL)) and Herrevatn (HER, 1,368 m ASL), in the river itself between two waterfalls preventing upstream migration (OLA, 1,245–1,145 m ASL), in the pool Samkomehølen (SAM, 1,145 m ASL), positioned at the confluence of OLA and the river from MGR (1,270 m ASL), as well as in MGR (Fig. 1). From above the barrier, sampling was performed in the river Vierslaelva (VIR, 1,245–1,310 m ASL), in the small lake “Vierslapytten” (VIP, 1,247 m ASL), the lakes Vierslatjørn (VIV, 1,245 m ASL), Nordmannslågen (NOR, 1,244 m ASL), and Nordvatn (NDV, 1,258 m ASL). The contemporary samples were collected by means of gillnetting, electrofishing and rod fishing. The fish were aged by otoliths and in a few cases by scales. All contemporary samples consisted of several year classes, including both mature and immature individuals of both sexes.
Stocking information
Stocking of brown trout on the Hardangervidda mountain plateau has been and still is common practice for enhancement of both sport and gill net fisheries, often without precise record keeping. For the stocking in the study area, several donor populations have been used at different time periods from 1968 to 2005 (Table 1). From 1972 until 2005, stocking material has originated from an unknown locality as well as from lakes both within the national park (Bjornesfjorden, 1,223 m ASL) and from outside (Tesse, 854 m ASL, and Tunhovdfjorden, 734 m ASL) (Table 1). The stocking of NOR and NDV has more or less been continuous since at least 1972, with fish of non-local origin until 1996, and thereafter based on supportive breeding (Table 1). Eggs and milt for the supportive breeding have been collected from fish on spawning run in a tributary to NOR. In 1971, HEP was expected to be fishless and therefore stocked in 1973 and 1974 (Table 1). Since 1973, HER has been stocked multiple times with fish from at least three stocking sources (Table 1). VIV is the only Viersla locality that has been stocked (Table 1). The presence of brown trout in MGR was first identified in 1910 (Huitfeldt-Kaas 1911), with first registered stocking in 1919, with fish originating from SAM (Sekse 1971) (Fig. 1). MGR was later restocked in 1974 and 1975 (Table 1).
Microsatellite analysis
DNA was extracted with the DNeasy® Tissue kit (Qiagen) from the collected brown trout tissue samples (fin clips) preserved in 96 % ethanol or dried scales from contemporary and historical samples, respectively, according to the manufacturer’s specifications. DNA extractions from tissue and scale samples were done separately to minimize risk of cross contamination, as were all additional laboratory analyses. The three historical samples were analyzed independently from one another. DNA concentration was estimated with an Epoch Microplate Spectrophotometer and a Take3 Micro-Volume Plate (BioTek®), and adjusted to 65 ng/μL. In total, 18 microsatellites were selected from the brown trout linkage map (Gharbi et al. 2006), each representing individual linkage groups, with the exception of loci Ssa54NVH and Str15INRA, which both represent linkage group 12 (Table 2). The two loci are not positioned closely to one another (Gharbi et al. 2006). Six loci (Str15INRA, Str60INRA, Str73INRA, Ssa85, Ssa197 and Strutta58) overlap with Sønstebø et al. (2008b). All loci were amplified singly or in multiplex reactions, using 700 and 800 IRDye labeled forward primers and analyzed on a LI-COR 4300 DNA analyzer (LI-COR Biosciences) after polymerase chain reaction (PCR) amplification. The 6.5 μL PCR cocktail contained 65–130 ng genomic DNA, 0.2 mM of both forward and reverse primers, 1× QIAGEN Multiplex PCR Master MIX (QIAGEN) (3 mM MgCl2) and RNase-free water. Thermo-cycling parameters were initiated with heat-activation at 95 °C for 15 min followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C (Hansen et al. 2009) for 90 s and extension at 72 °C for 60 s. The PCR was terminated after 30 min of final extension at 60 °C. PCR conditions were the same for scale samples, except for 0.4 mM of each primer without multiplexing and extension to 37 cycles. Each 6.5 % KB Plus polyacrylamide gel (LI-COR Biosciences) included positive controls to ensure consistency of allele scoring between runs. Allele scoring was performed using the SAGAMX Software (LI-COR Biosciences). Additionally, 10 % of the samples were rerun to ensure correct scoring of alleles. Evidence of genotyping errors due to stuttering (i.e. amplification errors during PCR read as true alleles), large allele drop-out (i.e. dominance of small alleles during amplification) and / or null alleles (i.e. alleles not amplified because of mutations at primer sites) were searched for, using MICRO-CHECKER v2.2.3 (Van Oosterhout et al. 2004).
Genetic diversity analysis
Allelic richness (No) and sample size corrected (rarefaction method) allelic richness (NA) were calculated with HP-RARE (Kalinowski 2005). ARLEQUIN v3.5 (Excoffier et al. 2005) was used to calculate observed and expected heterozygosity and GENEPOP v4.0 (Raymond and Rousset 1995; Rousset 2008) was used to test for departures from Hardy–Weinberg equilibrium (HWE) and for linkage disequilibrium (LD). Genetic distances between population pairs was estimated by computing Cavalli-Sforza and Edwards (1967) genetic chord distances (D CE ), and bootstrapping 2,000 times (Hedges 1992) with MSA v4.05 (Dieringer and Schlotterer 2003). Principal coordinate analysis (PCoA) of D CE was conducted in GENALEX v6.2 (Peakall and Smouse 2006). This software was also used to perform an analysis of molecular variance (AMOVA). A Neighbor-Joining (NJ) tree (Takezaki and Nei 1996) based on D CE was generated with the PHYLIP software package (Felsenstein 2004) and visualized in SPLITSTREE v4.0 (Huson and Bryant 2006). Overall genetic differentiation was estimated by calculating F ST (θ ST, Weir and Cockerham 1984), with statistical significance being tested by 10,000 permutations of individuals between samples using MSA v4.05. All tests of statistical significance were adjusted for multiple tests by the false discovery rate (FDR) correction (Benjamini and Yekutieli 2001). Number of populations (K) represented by sampled individuals was estimated using the Bayesian clustering algorithm implemented in STRUCTURE v2.3.3 (Pritchard et al. 2000, 2007; Hubisz et al. 2009) assuming an admixture model and correlated allele frequencies (Falush et al. 2003), thereby allowing individual genomes to represent a mixture of different populations. K was selected from ten independent runs, each consisting of 5 × 104 burn-in iterations and 3 × 105 MCMC repeats, varying K between one and ten. The average log-likelihood of the posterior probability of the data [P(D)] between the ten independent replicate runs was plotted, and K determined as the point on the curve where P(D) reaches a plateau (Falush et al. 2003). After selection of the most probable number of genetic clusters represented in the data, the proportions of each cluster within each of the sampled localities were calculated based on individual membership coefficients (Q) and averaged between all ten independent replicate runs. Only values above 10 % were evaluated as true genetic components. Individual Q values were plotted for visualization in DISTRUCT v1.1 (Rosenberg 2004).
Results
Statistical validity of the data
All loci amplified successfully in the contemporary samples, and missing data was low (0.1 %). In the historical samples, the loci Ssa408UOS and Ssa54NVH failed to amplify reliably in NOR33 and OLA45, the latter also in MGR67, and were therefore treated as missing data in all further analyses. Excluding these, missing data in the historical samples occurred at a modest rate (4 %). A total of 286 alleles were recorded from the 440 contemporary and historical samples (Table 3). The highest total number of scored alleles within a sample was found in NOR (188 alleles), while the lowest of the contemporary samples was found in HEP (102 alleles). The number of genotyped alleles per loci in the contemporary samples ranged from three (Str60INRA and Str73INRA) to 32 (Ssa41NVH and Strutta11) (Table 3). The NOR sample had the highest allele count at a single locus, with 21 alleles from loci Ssa41NVH and Ssa94NVH (Table 3). None of the historical samples were genotyped for more than 47 alleles, and eight of the 18 loci that were successfully amplified in the historical samples were fixed for one allele (Table 3). Six of the eight monomorphic loci were also found monomorphic by Sønstebø et al. (2008b). One locus (Str60INRA) was found monomorphic in one of the contemporary samples (HEP), and fixed for the same allele as detected in the historical samples. MICRO-CHECKER identified the possibility of null alleles due to an excess of homozygotes in Str73INRA in the HER sample, but this constellation was not found to be out of HWE (Table 3). Careful inspection of gel electrophoresis revealed no apparent explanation. Significant deviations from HWE were found in 20 of 234 tests, and the 12 that remained significant after correction for multiple tests (P < FDR corrected α = 0.01431) were predominantly present in the historical samples (Table 3). Linkage disequilibrium was significant in 103 comparisons after FDR correction (P < FDR corrected α = 0.00891). No case of LD was detected systematically for the same pair of loci across different populations.
Population structure
There was significant genetic differentiation between samples (F ST = 0.13, P < 0.001). Exclusion of the historical samples resulted in a lowering of the overall genetic differentiation (F ST = 0.086, P < 0.001). The AMOVA indicated that most of the genetic variability occurred within populations (74 %) rather than among populations (26 %). Exclusion of historical samples changed these proportions to 84 and 16 %, respectively. The most likely number of genetic populations identified by the replicate estimation of the probability of the data using STRUCTURE 2.3.3 showed that P(D) reached a plateau for K = 5 (Fig. 3). The admixture proportions of the 440 analyzed fish at K = 5 are presented in Fig. 4. Five individuals from contemporary samples below the constructed barrier were identified as showing >50 % admixture proportions to the historical samples (Fig. 4). The average admixture proportions for each of the sampled locations are given in Fig. 5 and Table S1. With five genetic clusters representing the data, the admixture proportions indicate that the most pronounced separation was found between the historical (Cluster 1) and contemporary samples (Fig. 5). Thereafter the geographic separation of samples on each side of the barrier became apparent, with cluster two and three representing all samples above the constructed barrier (NDV, NOR, VIV, VIR, and VIP). However, clusters were not represented equally among populations, and cluster three was not detected at a notable level in VIR and VIP (Fig. 5). Cluster four represents samples below the constructed barrier (HEP, OLA, and SAM), while cluster five corresponds to MGR (Fig. 5). Clusters two, three, and four were all represented in HER (Fig. 5). The genetic relationship between the 13 analyzed samples by NJ show separation into five groups (Fig. 5). Historical samples group (cluster 1, red) away from the contemporary samples with complete bootstrap support. The Viersla samples (i.e. VIV, VIR, and VIP) separate away as a consequence of almost complete representation by cluster two (purple), whereas the NOR and NDV samples group away due to the addition of cluster three (orange, Fig. 5). The remaining samples (i.e. those below the barrier), except HER, were grouped together via admixture from HEP (cluster four, green) into OLA and SAM, and from MGR (cluster five, blue) into SAM. The HER sample was separated from all other samples, as clusters two, three and four are all represented (Fig. 5). Thus, the groupings by the NJ analysis correspond to admixture proportions identified by the STRUCTURE analysis (Fig. 5). The identified genetic relationships were furthermore in general agreement with the PCoA (Fig. 6). PCoA based on D CE clustered the samples into three groups along the first axis (Dim. 1), (i) the historical, (ii) populations above the constructed barrier, and (iii) populations below the barrier, and explains 47.6 % of the variation (Fig. 6). The HER sample was positioned between the two clusters from either side of the constructed barrier. Dim. 2, explaining 15.6 % of the total variation, showed sub-clustering in the samples above the barrier, between VIV, VIR and VIP in one cluster, and NOR and NDV in another (Fig. 6). Dim. 2 also separated HEP from the remaining samples below the constructed barrier. Dim. 3, explaining 11.9 % of the variation, separated MGR from the remaining samples below the barrier, and also revealed the relative intermediate positioning of SAM between OLA and MGR.
Neighbor-Joining tree based on D CE genetic distance, showing the genetic relationship between the 13 analyzed brown trout populations from the Hardangervidda mountain plateau. Bootstrap support (2,000 replicates) of 100 % is shown as line breaks. Stacked bar plots with percentages indicate the >10 % average population assignment to the five genetic clusters identified by STRUCTURE. Colors correspond to clusters in Fig. 4
Discussion
The significant genetic differentiation found in the studied populations indicate a change in the spatial genetic structuring between the historical and contemporary samples, with only a limited genetic signature from historical samples still persisting. Nearly all of the contemporary samples show some level of admixture from two or more of the suggested genetic clusters. Despite this, the overall genetic differentiation between the ten contemporary samples indicates a strong degree of population subdivision, despite partial gene flow. Similar genetic differentiation levels are reported from brown trout in other watercourses (Lehtonen et al. 2009; Swatdipong et al. 2010; Kohout et al. 2012), and assumed to evolve naturally over time (Carlsson and Nilsson 2000, 2001; Carlsson et al. 1999). The genetic differentiation was not homogeneous across the sampling area, revealing the presence of divergent populations across the constructed barrier and natural waterfalls, as also observed in other fragmented river systems with dam constructions (Heggenes and Røed 2006) or natural barriers (Griffiths et al. 2009). Comparison of the admixture proportions of each individual from each of the sampled locations may provide information regarding the minor inconsistencies between results from the different analyses applied in our study. Nearly all of the contemporary samples show some level of admixture from two or more of the suggested genetic clusters. Despite this, the overall genetic differentiation between the ten contemporary samples indicates a strong and significant degree of population subdivision, despite partial gene flow.
The stocking history and genetic structure of the studied populations seem to be clearly linked. However, the shift in the genetic structure between the contemporary and historical samples is surprising, since post-release survival of hatchery-reared non-native salmonids (Weiss and Schmutz 1999; Hesthagen et al. 2010), as well as native fish bred in captivity (Kostow 2004; Christie et al. 2012), have been found to be low compared to progeny of wild fish in natural waters. Unexpectedly, the historical brown trout in the study area were almost monomorphic and nearly identical among the two drainage systems, as also found by Sønstebø et al. (2008b), indicating a historical connectivity between the two drainage systems. It is, however, not possible to determine whether the genetic connection between the historical brown trout in these two neighboring systems (NOR and OLA) is caused by early translocation of fish, or by natural immigration during flood periods with overflow between the watercourses, as observed before the channel was constructed (Nils Runar Sporan, pers. comm.). The genetic monomorphism and low diversity of the historical samples could be the consequence of population establishment by a small number of initial individuals thousands of years ago. Rapid population expansion after initial introduction would most likely prevent extinction (Reznick and Ghalambor 2001). After introduction, even at a relative short time frame, selection could tend to give fixation of specific alleles (Koskinen et al. 2002), resulting in low genetic diversity in the established population. This may be especially pertinent if diversity was initially low as a consequence of founder events and bottlenecks (Dlugosch and Parker 2008; Barson et al. 2009). Even when considering founder effects, genetic differentiation between the historical samples would still be expected if the populations independently had experienced bottlenecks that could lead to random fixation of different alleles, as long as noticeable levels of gene flow and population expansion do not exist (Lacy 1987). The existence of monomorphic populations is rare in salmonid populations, although not unique (Hynes et al. 1996; Prodöhl et al. 1997; Whiteley et al. 2010). Since persistence of monomorphic populations of brown trout has existed for more than 100 years in Scotland (Prodöhl et al. 1997), it is likely that the historical genetic structure of brown trout on Hardangervidda would have persisted as well, if not being impacted by the stocking events. Consequently, anthropogenic interferences have most likely led to the genetic displacement of the monomorphic, historical brown trout populations, but at the same time resulted in a threefold increase in the genetic diversity of current populations, based upon the number of genotyped alleles. This does not imply reduced fitness in historical brown trout populations, since they likely survived during periods with extensive changes in climate during the last ca. 6,000 years, as described by Nesje et al. (2008, 2012). The historical change in the genetic structure, with an increase in allelic richness, may nonetheless provide a good basis for future local adaptation, and enable long term population persistence of the current brown trout populations in the study area.
The stocking success documented in the present study is contradictory to unsuccessful stocking events observed in other studies (García-Marín et al. 1998; Heggenes et al. 2002; Hesthagen et al. 2010), where reduced survival of hatchery-reared brown trout (García-Marín et al. 1998) and large indigenous populations (Glover et al. 2012) may have prevented stocking success. As shown for Atlantic salmon (Salmo salar L.), the degree of introgression from immigrated non-native, escaped farmed fish appears to be strongly dependent of native population size (Glover et al. 2012). Thus, the success of stocked non-native brown trout on the Hardangervidda mountain plateau (Sønstebø et al. 2008b, and our study), may similarly be connected to small native populations, as a combined result of heavy exploitation and periodic low natural recruitment (Borgstrøm and Museth 2005).
Preservation of the original genetic profile by construction of a barrier, as also observed elswhere (Thompson and Rahel 1998; Yamamoto et al. 2004; Van Houdt et al. 2005), was not obvious from our data. Indeed, none of the contemporary brown trout analyzed had a genetic profile identical to the historical samples. However, since some individuals from below the constructed barrier showed more than 50 % genetic identity to the historical samples, according to the Bayesian clustering analysis, this may indicate that at least some preservation of the historical genetic profile has occurred. The genetic shift observed below the barrier may be related to stocking of the tributary lakes (HEP, HER and MGR), with downstream immigrations to the river sections OLA and SAM. The difference in genetic components between these sections are most likely explained by the presence of waterfalls preventing upstream migration, constituting natural barriers for gene flow (Heggenes and Røed 2006; Griffiths et al. 2009), and indirect stocking by downstream migration from tributary lakes (Sønstebø et al. 2008a). Heavy reduction of the river discharge, as a consequence of water diversion, may have increased the risk of bottom freezing during especially cold winters, leading periodically to reduced recruitment (Borgstrøm and Museth 2005). This may explain the success of downstream immigrants from the tributary lakes.
Contrary to the findings below the barrier, the stocking events between 1972 and 1974, seems not to have been as successful in the locations above the barrier. Here the populations have most likely changed their genetic identity as a consequence of stocking by especially the two latest and most extensive non-native sources. Despite opening of the channel between the Viersla area and NOR, leading to an increase in available spawning habitat for the brown trout in NOR, the Viersla populations (VIV, VIR, and VIP) still distinguish themselves by showing limited admixture and introgression from the latest non-native stocking source only used in NOR and NDV. The enhancement stocking of NOR, with direct releases into the lake (Gunnar Elnan pers. comm.) may have restricted the spawning runs to streams and rivers (O’Grady 1984; Hesthagen et al. 1999). Thus, this stocking practice may have restricted the introgression and admixture from NOR into the Viersla area, and potentially explain the genetic structure between these populations. Furthermore, with several available spawning sites in NOR, both in tributaries, outlet river, and the lake itself, as suggested by Qvenild (2004), it is possible that only a small proportion of the stocked brown trout released in NOR have migrated through the opened channel. Stocking sources may show varying introgression success, as shown for one of the stocking sources when used in another lake, possibly as a consequence of variations in preferences for different spawning habitats (Heggenes et al. 2006). However, without tagging experiments, it is not possible to identify how the mature fish distributes among the many spawning sites, and thereby estimate their potential effects on the genetic structure of the populations.
In recent years, the local mountain board has stopped stocking of non-native brown trout, and shifted to supportive breeding, which may greatly benefit the preservation of the original fish as long as it is possible to reliably identify such individuals (Hansen et al. 2006; Iwamoto et al. 2012). An extensive genetic survey would be needed in order to identify these for initiation of supportive breeding based on fish with historical genetic profile. Alternatively, graphical genotyping (Young and Tanksley 1989) with selection in progenies of fish with partially historical marker composition could be used to re-create “original” fish. Given the low diversity mentioned above, it is questionable whether such a DNA marker-based breeding effort would be ecologically and evolutionary meaningful.
In conclusion, the extensive shift in the genetic structure of brown trout in the two analyzed high alpine river systems within the Hardangervidda National Park results from large-scale stocking over a long time period (1968–2005) in combination with low population densities due to fluctuations in natural recruitment and heavy exploitation. Four stocking sources of non-native fish facilitated the genetic differentiation between the contemporary populations. The habitat fragmentation by construction of a barrier may have helped to maintain a tiny fraction of the historical genetic profile, and stocking should therefore not be re-initiated here. The management of the Hardangervidda National Park aims at combining nature conservation and historical landowner rights. Our study demonstrates that research based management is needed to retain a minimum of the original natural diversity.
References
Amos W, Harwood J (1998) Factors affecting levels of genetic diversity in natural populations. Philos Trans Roy Soc B 353:177–186
Bakke Ø 1984. Dyregravene på Hardangervidda. En skisse av deres forekomst og bakgrunn. In: Norsk skogbruksmuseum årbok nr. 10, Elverum, pp 129–198 (in Norwegian)
Barson NJ, Haugen TO, Vøllestad LA, Primmer CR (2009) Contemporary isolation-by-distance, but not isolation-by-time, among demes of european grayling (Thymallus thymallus, Linnaeus) with recent common ancestors. Evolution 63:549–556
Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188
Borgstrøm R, Museth J (2005) Accumulated snow and summer temperature: critical factors for recruitment to high mountain populations of brown trout (Salmo trutta L.). Ecol Freshw Fish 14:375–384
Cairney M, Taggart JB, Høyheim B (2000) Characterization of microsatellite and minisatellite loci in Atlantic salmon (Salmo salar L.) and cross-species amplification in other salmonids. Mol Ecol 9:2175–2178
Carlsson J, Nilsson J (2000) Population genetic structure of brown trout (Salmo Trutta L.) within a northern boreal forest stream. Hereditas 132:173–181
Carlsson J, Nilsson J (2001) Effects of geomorphological structures on genetic differentiation among brown trout populations in a northern boreal river drainage. Trans Am Fish Soc 130:36–45
Carlsson J, Olsen KH, Nilsson J, Overli O, Stabell OB (1999) Microsatellites reveal fine-scale genetic structure in stream-living brown trout. J Fish Biol 55:1290–1303
Cavalli-Sforza Ll, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Evolution 21:550–570
Christie MR, Marine ML, French RA, Blouin MS (2012) Genetic adaptation to captivity can occur in a single generation. PNAS 103:238–242
Cook N, Rahel FJ, Hubert WA (2010) Persistence of Colorado River cutthroat trout populations in isolated headwater streams of Wyoming. Trans Am Fish Soc 139:1500–1510
Dieringer D, Schlotterer C (2003) Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Estoup A, Presa P, Krieg F, Vaiman D, Guyomard R (1993) CT)n and (GT)n microsatellites: a new class of genetic markers for Salmo trutta L. (brown trout. Heredity 71:488–496
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Felsenstein J (2004) PHYLIP (Phylogeny Inference Package), version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle, Washington
García-Marín JL, Jorde PE, Ryman N, Utter F, Pla C (1991) Management implications of genetic differentiation between native and hatchery populations of brown trout (Salmo trutta) in Spain. Aquaculture 95:235–249
García-Marín JL, Sanz N, Pla C (1998) Proportions of native and introduced brown trout in adjacent fished and unfished Spanish rivers. Conserv Biol 12:313–319
Gharbi K, Gautier A, Danzmann RG, Gharbi S, Sakamoto T, Høyheim B, Taggart JB, Cairney M, Powell R, Krieg F, Okamoto N, Ferguson MM, Holm LE, Guyomard R (2006) A linkage map for brown trout (Salmo trutta): chromosome homeologies and comparative genome organization with other salmonid fish. Genetics 172:2405–2419
Gilpin ME, Soulé ME (1986) Minimum viable populations: processes of species extinction. In: Soulé ME (ed) Conservation biology: the science of scarcity and diversity. Sinauer Associates Sunderland, pp 19–34
Glover KA, Quintela M, Wennevik V, Besnier F, Sørvik AGE, Skaala Ø (2012) Three decades of farmed escapees in the wild: a spatio-temporal analysis of Atlantic salmon population genetic structure throughout Norway. PLoS One 7:e43129
Gosset C, Rives J, Labonne J (2006) Effect of habitat fragmentation on spawning migration of brown trout (Salmo trutta L.). Ecol Freshw Fish 15:247–254
Griffiths AM, Koizumi I, Bright D, Stevens JR (2009) A case of isolation by distance and short-term temporal stability of population structure in brown trout (Salmo trutta) within the River Dart, southwest England. Evol Appl 2:537–554
Hansen MM (2002) Estimating the long-term effects of stocking domesticated trout into wild brown trout (Salmo trutta) populations: an approach using microsatellite DNA analysis of historical and contemporary samples. Mol Ecol 11:1003–1015
Hansen MM, Bekkevold D, Jensen LF, Mensberg K-LD, Nielsen EE (2006) Genetic restoration of a stocked brown trout Salmo trutta population using microsatellite DNA analysis of historical and contemporary samples. J Appl Ecol 43:669–679
Hansen MM, Fraser DJ, Meier K, Mensberg K-LD (2009) Sixty years of anthropogenic pressure: a spatio-temporal genetic analysis of brown trout populations subject to stocking and population declines. Mol Ecol 18:2549–2562
Hedges SB (1992) The number of replications needed for accurate estimation of the bootstrap p value in phylogenetic studies. Mol Biol Evol 9:366–369
Heggenes J, Røed KH (2006) Do dams increase genetic diversity in brown trout (Salmo trutta)? Microgeographic differentiation in a fragmented river. Ecol Freshw Fish 15:366–375
Heggenes J, Røed KH, Høyheim B, Rosef L (2002) Microsatellite diversity assessment of brown trout (Salmo trutta) population structure indicate limited genetic impact of stocking in a Norwegian alpine lake. Ecol Freshw Fish 11:93–100
Heggenes J, Skaala O, Borgstrøm R, Igland OT (2006) Minimal gene flow from introduced brown trout (Salmo trutta L.) after 30 years of stocking. J Appl Ichthyol 22:119–124
Hesthagen T, Floystad L, Hegge O, Staurnes M, Skurdal J (1999) Comparative life-history characteristics of native and hatchery-reared brown trout, Salmo trutta L., in a sub-Alpine reservoir. Fish Manag Ecol 6:47–61
Hesthagen T, Johnsen SI, Gran R (2010) Effect of supplementary stocking of juvenile brown trout, Salmo trutta, on yield in a Norwegian mountain reservoir. Fish Manag Ecol 17:186–191
Hindar K, Ryman N, Utter F (1991) Genetic effects of cultured fish on natural fish populations. Can J Fish Aquat Sci 48:945–957
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9:1322–1332
Huitfeldt-Kaas HH (1911) Indberetning om fiskeriforholdene på Hardangervidden. In: Indstilling fra Fjeldbeitekomiteen om Hardangerviddens utnyttelse (Landbruksdepartementet, ed). Centraltrykkeriet, Kristiania, pp 81–98 (in Norwegian)
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267
Hynes RA, Ferguson A, McCann MA (1996) Variation in mitochondrial DNA and post-glacial colonization of north western Europe by brown trout. J Fish Biol 48:54–67
Indrelid S (1985) De første bosetterne. In: Barth EK (ed) Hardangervidda. Luter Forlag, Oslo, pp 97–111 (in Norwegian)
Iwamoto EM, Myers JM, Gustafson RG (2012) Resurrecting an extinct salmon evolutionarily significant unit: archived scales, historical DNA and implications for restoration. Mol Ecol 21:1567–1582
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Kohout J, Jašková I, Papoušek I, Šedivá A, Šlechta V (2012) Effects of stocking on the genetic structure of brown trout, Salmo trutta, in Central Europe inferred from mitochondrial and nuclear DNA markers. Fish Manag Ecol 19:252–263
Koskinen MT, Haugen TO, Primmer CR (2002) Contemporary fisherian life-history evolution in small salmonid populations. Nature 419:826–830
Kostow KE (2004) Differences in juvenile phenotypes and survival between hatchery stocks and a natural population provide evidence for modified selection due to captive breeding. Can J Fish Aquat Sci 61:577–589
Lacy RC (1987) Loss of genetic diversity from managed populations: interacting effects of drift, mutation, immigration, selection, and population subdivision. Conserv Biol 1:143–158
Lande R (1998) Anthropogenic, ecological and genetic factors in extinction and conservation. Res Popul Ecol 40:259–269
Lehtonen PK, Tonteri A, Sendek D, Titov S, Primmer CR (2009) Spatio-temporal genetic structuring of brown trout (Salmo trutta L.) populations within the River Luga, northwest Russia. Conserv Genet 10:281–289
Letcher BH, Nislow KH, Coombs JA, O’Donnell MJ, Dubreuil TL (2007) Population response to habitat fragmentation in a stream-dwelling brook trout population. PLoS One 2:e1139
Nesje A, Bakke J, Dahl SO, Lie O, Matthews JA (2008) Norwegian mountain glaciers in the past, present and future. Glob Planet Change 60:10–27
Nesje A, Pilø LH, Finstad E, Solli B, Wangen V, Ødegård RS, Isaksen K, Støren EN, Bakke DI, Andreassen LM (2012) The climatic significance of artefacts related to prehistoric reindeer hunting exposed at melting ice patches in southern Norway. Holocene 22:485–496
Nielsen EE, Hansen MM (2008) Waking the dead: the value of population genetic analyses of historical samples. Fish Fish 9:450–461
O’Grady MF (1984) Observations on the contribution of planted brown trout (Salmo trutta L.) to spawning stocks in four Irish lakes. Aquac Res 15:117–122
O’Reilly PT, Hamilton LC, McConnell SK, Wright JM (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci 53:2292–2298
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Eco Notes 6:288–295
Poteaux C, Bonhomme F, Berrebi P (1999) Microsatellite polymorphism and genetic impact of restocking in Mediterranean brown trout (Salmo trutta L.). Heredity 82:645–653
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: version 2.2. University of Chicago, Chicago, p 36
Prodöhl PA, Walker AF, Hynes R, Taggart JB, Ferguson A (1997) Genetically monomorphic brown trout (Salmo trutta L) populations, as revealed by mitochondrial DNA, multilocus and single-locus minisatellite (VNTR) analyses. Heredity 79:208–213
Qvenild T (2004) Hardangervidda: fiske og fjelliv. Naturforlaget, Oslo (in Norwegian)
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rexroad CE, Coleman RL, Hershberger WK, Killefer J (2002) Rapid communication: thirty-eight polymorphic microsatellite markers for mapping in rainbow trout. J Anim Sci 80:541–542
Reznick DN, Ghalambor CK (2001) The population ecology of contemporary adaptations: what empirical studies reveal about the conditions that promote adaptive evolution. Genetica 112–113:183–198
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) GENEPOP ‘007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Sekse T (1971) Jakt og fiske på Hardangerviddi. In: Kolltveit O (ed) Odda, Ullensvang og Kinsarvik i gamal og ny tid. Boktrykk L/L, Bergen, pp 153–163 (in Norwegian)
Sønstebø JH, Borgstrøm R, Heun M (2008a) Genetic structure in alpine brown trout Salmo trutta L. shows that indirect stocking affects native lake populations. J Fish Biol 72:1990–2001
Sønstebø JH, Borgstrøm R, Heun M (2008b) High genetic introgression in alpine brown trout (Salmo trutta L.) populations from Hardangervidda Norway. Ecol Freshw Fish 17:174–183
Swatdipong A, Vasemägi A, Niva T, Koljonen ML, Primmer CR (2010) High level of population genetic structuring in lake-run brown trout, Salmo trutta, of the Inari Basin, northern Finland. J Fish Biol 77:2048–2071
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Taylor EB, Stamford MD, Baxter JS (2003) Population subdivision in westslope cutthroat trout (Oncorhynchus clarki lewisi) at the northern periphery of its range: evolutionary inferences and conservation implications. Mol Ecol 12:2609–2622
Thompson PD, Rahel FJ (1998) Evaluation of artificial barriers in small Rocky Mountain streams for preventing the upstream movement of brook trout. North Am J Fish Manag 18:206–210
Tysse Å, Garnås E (1996) Status og strategi for kultivering av ferskvassfisk i Buskerud. Fylkesmannen i Buskerud, Miljøavdelingen. Report nr. 5, Drammen (in Norwegian)
Van Houdt JKJ, Pinceel J, Flamand MC, Briquet M, Dupont E, Volckaert FAM, Baret PV (2005) Migration barriers protect indigenous brown trout (Salmo trutta) populations from introgression with stocked hatchery fish. Conserv Genet 6:175–191
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Weiss S, Schmutz S (1999) Performance of hatchery-reared brown trout and their effects on wild fish in two small Austrian streams. Trans Am Fish Soc 128:302–316
Whiteley A, Hastings K, Wenburg J, Frissell C, Martin J, Allendorf F (2010) Genetic variation and effective population size in isolated populations of coastal cutthroat trout. Conserv Genet 11:1929–1943
Wofford JEB, Gresswell RE, Banks MA (2005) Influence of barriers to movement on within-watershed genetic variation of coastal cutthroat trout. Ecol Appl 15:628–637
Yamamoto S, Morita K, Koizumi I, Maekawa K (2004) Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: spatial–temporal changes in gene frequencies. Conserv Genet 5:529–538
Young ND, Tanksley SD (1989) Restriction fragment length polymorphism maps and the concept of graphical genotypes. Theor Appl Genet 77:95–101
Acknowledgments
John Gunner Dokk, Markus Sydenham, and Carsten Thaulow Pedersen assisted during field sampling. We thank Arne Bu and Olaf Bu for supplying samples from Nordvatn, Sylfest Laingen for samples from Nordmannslågen, and Odd Terje Sandlund for providing the historical scale samples. We furthermore acknowledge Gunnar Elnan in Eidfjord mountain board, Nils Runar Sporan and Henning Syvertsen in Statkraft, Sverre Tveiten, former at Norwegian directorate for nature management, for information regarding the stocking history, Thrond Haugen for comments on an earlier version of this manuscript, and Marie Davey for improving the English. We are also grateful to two anonymous reviewers whose comments considerably improved the manuscript. Financial support was given by Department of Ecology and Natural Resource Management (INA), Norwegian University of Life Science (UMB) and Statkraft. An unchangeable file containing all data and analyses is stored at UMB.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Thaulow, J., Borgstrøm, R. & Heun, M. Brown trout population structure highly affected by multiple stocking and river diversion in a high mountain national park. Conserv Genet 14, 145–158 (2013). https://doi.org/10.1007/s10592-012-0438-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0438-7