Abstract
Gene flow among small fragmented populations is critical for maintaining genetic diversity, and therefore the evolutionary potential of a species. Concern for two New Zealand endemic subspecies, the Hector’s (Cephalorhynchus hectori hectori) and Maui’s (C. h. maui) dolphins, arises from their low abundance, slow rate of reproduction, and susceptibility to fisheries-related mortality. Our work examined genetic differentiation and migration between the subspecies and among regional and local Hector’s dolphin populations using mitochondrial (mt) DNA and microsatellite genotypes from 438 samples. Results confirmed earlier reports of a single unique mtDNA control region haplotype fixed in the Maui’s dolphin, and provided new evidence of reproductive isolation from Hector’s dolphins (9-locus microsatellite F ST = 0.167, P < 0.001). Independent evolutionary trajectories were also supported for Hector’s dolphin populations of the East Coast, West Coast, Te Waewae Bay and Toetoe Bay. Low asymmetrical migration rates were found among several Hector’s dolphin populations and assignment tests identified five Hector’s dolphins likely to have a migrant father from another regional population. There appears to be sufficient step-wise gene flow to maintain genetic diversity within the East and West Coasts; however, the two local South Coast populations exhibited a high degree of differentiation given their close proximity (~100 km). To maintain the evolutionary potential and long-term survival of both subspecies, genetic monitoring and conservation management must focus on maintaining corridors to preserve gene flow and prevent further population fragmentation and loss of genetic diversity, in addition to maintaining local population abundances.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gene flow among small fragmented populations is critical for the maintenance of genetic diversity, and therefore the evolutionary potential, or ability of a species to adapt to environmental changes. When gene flow is severely restricted, low-abundance population fragments become increasingly vulnerable to characteristics correlated with the risk of extinction, including the loss of genetic variation via genetic drift, accumulation of mildly deleterious mutations, inbreeding depression, and the inability to adapt to change (Frankham 1995). These effects are particularly concerning for endemic island populations, which tend to display higher rates of extinction than non-endemic or mainland species (Frankham 1997, 1998). The Hector’s dolphin (Cephalorhynchus hectori) is the only cetacean endemic to the North and South Islands of New Zealand, and is currently facing threats from both anthropogenic and genetic factors.
Two subspecies of Hector’s dolphin are currently recognized: C. h. hectori, which retains the common name Hector’s dolphin; and C. h. maui, referred to as the Maui’s dolphin (Baker et al. 2002). The Hector’s dolphin has an estimated abundance of 7,270 individuals (95 % CI = 5303–9966; Slooten et al. 2004) distributed discontinuously around the South Island (Fig. 1), and has been listed as ‘Endangered’ by the IUCN since 2000 (Reeves et al. 2008). The most recent estimate for the ‘Critically Endangered’ Maui’s dolphin population is 80 individuals (95 % CI = 48–252) concentrated along the central West Coast of the North Island (Baker et al. In review). These dolphins are generally sighted within a few kilometers of shore (e.g. Dawson and Slooten 1988; Dawson et al. 2004), but have been documented ranging out to 35 km, and exhibit an ‘offshore’ shift during the austral winter (Slooten and Dawson 1988; Bräger et al. 2003; Stone et al. 2005; Rayment et al. 2006). This coastal distribution coincides with areas utilized by commercial and recreational fisheries, whose targets include flatfish, red cod, mullet, butterfish, moki and small sharks (New Zealand Department of Conservation, Ministry of Fisheries 2007). While the quantification of bycatch rates using an observer program has proven challenging (Blezard 2002; Fairfax 2002; Reid 2002), observations made during the 1997–1998 fishing year on the East Coast of the South Island were used to estimate a mean bycatch rate of 0.037 (SE = 0.15) Hector’s dolphins caught per setnet, extrapolated to a count of 18 dolphins in the given area and timeframe (Baird and Bradford 2000). Of the 503 Hector’s and Maui’s dolphin carcasses opportunistically recovered by the New Zealand Department of Conservation between 1921 and 2011, cause of death was able to be determined for 257, of which 185 Hector’s and 5 Maui’s showed direct evidence of fisheries-related mortality (New Zealand Department of Conservation 2011). Incidental gillnet entanglement is considered to be the primary anthropogenic threat to Hector’s and Maui’s dolphins (Dawson 1991; Dawson and Slooten 2005; Slooten 2007), and according to the most recent population viability analysis by Slooten (2007), the low reproductive output of these dolphins is not sufficient to sustain the populations at the estimated levels of anthropogenic mortality.
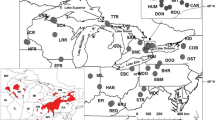
Distribution (shaded coastline) and mitochondrial control region haplotypes (360 bp) for the Maui’s dolphin (NI); regional populations of the Hector’s dolphin: East Coast (EC), West Coast (WC), South Coast (SC); and local populations of the Hector’s dolphin: Cloudy Bay (CB), Kaikoura (KK), Pegasus Bay (PG), Banks Peninsula (BP), Timaru (TM), Westport (WP), Greymouth (GM), Jackson Bay (JB), Te Waewae Bay (TW), Toetoe Bay (TB). EC and WC sample sizes include additional samples collected from unknown local populations within each region
The loss of genetic diversity and restriction of gene flow have also emerged as concerns for the Hector’s and Maui’s dolphins, given their small population sizes and fragmented distribution. A loss of mitochondrial DNA (mtDNA) diversity was documented for the Maui’s dolphin and East Coast Hector’s dolphin populations by a comparison of ‘contemporary’ samples collected 1988–1998 with available historical samples from 1870 to 1987 (Pichler and Baker 2000). The ‘contemporary’ Maui’s dolphin population was found to be fixed for a single unique mtDNA haplotype (‘G’), without detection of two other haplotypes represented by three historical specimens reportedly collected on the North Island. There is, however, doubt about the subspecies origin of these three historical non-‘G’ specimens, which caused them to be excluded from the analyses of Baker et al. (2002) to define the Hector’s and Maui’s dolphin subspecies; their skeletal measurements were consistent with those of Hector’s dolphins; there is doubt about the reported collection location of one in the Bay of Islands; and there is speculation that the other two crossed Cook Strait either pre- or post-mortem. In addition to the lack of gene flow detected between the two subspecies, restrictions in gene flow were also detected between the East, West and South Coast populations of Hector’s dolphins using maternally-inherited mtDNA (Pichler et al. 1998; Pichler 2002). A preliminary analysis of six biparentally-inherited microsatellite loci also supported the differentiation of the East and West Coasts, but did not differentiate the South Coast from either of them (Pichler 2002).
In many coastal cetaceans gene flow is facilitated by relatively large home ranges, high mobility and the absence of geographic barriers. This is illustrated by the harbor porpoise (Phocoena phocoena), which ranges over 400 km in the Bay of Fundy (Read and Westgate 1997), and the bottlenose dolphins (Tursiops sp.) along the California coast, which can range in excess of 670 km (Wells et al. 1990). The dolphins of the genus Cephalorhynchus, however, exhibit much smaller home ranges, which may hinder gene flow if small population fragments are created with large distances separating them. The average home range observed for the Chilean dolphin (C. eutropia) is 23.1 km (Heinrich 2006), 36.6 km for the Heaviside’s dolphin (C. heavisidii; Elwen et al. 2006), and 31 to 33 km for the Hector’s dolphin (Bräger et al. 2002; Rayment et al. 2009). A kernal density (K 95) analysis for Hector’s dolphins around Banks Peninsula adjusted the observed home range estimate to approximately 50 km (Rayment et al. 2009). Based on the observations of the dolphins around Banks Peninsula by both Bräger et al. (2002) and Rayment et al. (2009), the maximum home range for Hector’s dolphins appears to be just over 100 km, as ‘outlier’ individuals have been sighted across a maximum of 107 km (Rayment et al. 2009), with the next largest distance observed in both studies being 60 km. Results from the satellite-tagging of three individuals around Banks Peninsula were also consistent with a small home range (Stone et al. 2005). During the three to six month observation period, the tagged individuals preferentially occupied areas within a 10–14 km radius from the individual’s mean activity center and the maximum along shore distance over which an individual’s signal was detected was 66 km. These studies all suggest that Hector’s dolphins are not likely to regularly move across distances larger than approximately 60 km, with only rare movements in excess of 100 km. Therefore, we expect to find high levels of gene flow within areas where the distribution of Hector’s dolphins is continuous or separated by distances less than 60 km, and little to no gene flow between populations separated by distances greater than 100 km.
The evolutionary potential of Hector’s and Maui’s dolphins is dependent on maintaining genetic diversity through sustaining local abundances as well as gene flow. To identify natural population boundaries and begin to assess evolutionary potential, we examined the current genetic diversity, gene flow and migration between Hector’s and Maui’s dolphins, as well as among regional and local populations of the Hector’s dolphin. Our work builds upon previous studies (Pichler et al. 1998; Pichler and Baker 2000; Pichler 2002), while offering novel insights that result from larger sample sizes with more representation of living dolphins as compared to primarily beachcast or bycaught specimens; increasing coverage of the distribution and including the Toetoe Bay population; and expanding the preliminary microsatellite analysis from six loci (n = 82) up to 13 loci (n = 266) allowing the examination of biparental gene flow and migration.
Materials and methods
Sample collection, DNA extraction and sex identification
A total of n = 438 tissue samples of Hector’s (n = 342) and Maui’s dolphins (n = 96) were collected from 1988 to 2007 and stored by the University of Auckland Cetacean Tissue Archive. Our sample set was restricted to these years to maintain a current sample of adequate size, while minimizing the potential for generational changeover based on the 20 year maximum lifespan reported for Hector’s dolphins (Slooten and Lad 1991). Samples were obtained from free-swimming dolphins using a minimally invasive biopsy dart (n = 176) as described by Krützen et al. (2002), or using the skin swab (n = 132) method described by Harlin et al. (1999). Samples were also collected during the necropsy of carcasses found beachcast or bycaught (n = 129); and one sample was recovered from the stomach contents of a seven-gill shark during necropsy (C. Thorburn, personal communication). Maui’s dolphin samples spanned the majority of their restricted distribution, and the Hector’s dolphin samples represented ten local populations within three regional populations (East, West, and South Coasts; Fig. 1). All samples were stored in 70 % ethanol at −20 °C prior to tissue digestion with proteinase K followed by total cellular DNA extraction using a standard phenol:chloroform protocol (Sambrook et al. 1989) as modified for small samples by Baker et al. (1994). Sex was identified using a multiplexed PCR protocol to amplify fragments of the sry and ZFX/ZFY genes according to Gilson et al. (1998). The observed sex ratio for each population and each type of sample (i.e. biopsy, swab or beachcast) were compared to an expected 1:1 sex ratio using a two-tailed exact binomial test.
Mitochondrial control region
A fragment of approximately 700 bp from the 5′ end of the maternally inherited mtDNA control region was amplified using the primers M13-Dlp-1.5 (5′-TGTAAAACGACAGCCAGTTCACCCAAAGCTGRARTTCTA) and Dlp-8G (5′-GGAGTACTATGTCCTGTAACCA; Dalebout et al. 2005). Each 10 μl reaction contained 1× PCR II buffer, 2.5 mM MgCl2, 0.4 μM each primer, 0.2 mM dNTP, 0.125 units of thermostable Platinum Taq DNA Polymerase (Invitrogen) and 10–20 ng DNA template. Thermocycling was carried out with an initial denaturation step of 94 °C for 2 min followed by 35 cycles of 94 °C for 30 s, 55 °C for 45 s and 72 °C for 45 s, and concluded with a final extension at 72 °C for 10 min. Excess nucleotides and primers were removed using ExoSap-IT (USB) before cycle sequencing with BigDye version 3.1 terminator chemistry (Applied Biosystems, Inc.). Sequence products were cleaned using CleanSEQ (Agentcourt) and run on an ABI 3730 or 3130XL DNA Analyzer. Sequences were aligned and edited using SEQUENCHER v. 4.7 (Gene Codes Corporation). Haplotypes were assigned based on alignment with 360 bp reference sequences of the 17 haplotypes previously identified for Hector’s and Maui’s dolphins (Pichler et al. 1998; Pichler and Baker 2000; Pichler 2002). Haplotype (h) and nucleotide (π) diversity, as well as a hierarchical AMOVA and pairwise F ST and Φ ST at the subspecies, regional and local levels were calculated in ARLEQUIN v. 3.1 (Excoffier 2006). A parsimony network was created using TCS 1.21 (Clement et al. 2000) to visualize the relationships among haplotypes.
Microsatellites
Hector’s dolphin samples were genotyped for 13 loci and Maui’s dolphins for 15 loci using published cetacean primers (Schlotterer et al. 1991; Buchanan et al. 1996; Valsecchi and Amos 1996; Hoelzel et al. 1998a, b; Rosel et al. 1999; Bérubé et al. 2000; Krützen et al. 2001; Caldwell et al. 2002; Online Resource 1). As the datasets for the two subspecies were originally generated for independent objectives, nine loci have genotypes for both subspecies datasets and were used for subspecies level comparisons. Each 10 μl PCR reaction contained 1× PCR II buffer, 2.5 mM MgCl2, 0.4 μM each primer, 0.2 mM dNTP, 0.125 units Platinum Taq (Invitrogen) and 10–20 ng DNA template. Thermocycling conditions differed among loci and are described in Online Resource 1. Products were run on an ABI 3730 or 3130XL DNA Analyzer and allele peaks were binned and visually verified using GENEMAPPER v.3.7 (Applied Biosystems). To minimize genotyping error, each amplification and sizing run included a negative control to detect contamination and 4 internal control samples to ensure comparable allele sizing across all runs. Additionally, 4 blind replicates were independently run for 13 loci; an additional 4 replicates for 9 loci; and 51 replicates for 4 loci that were reported by Pichler (2002). Independent visual verification of automated allele binning was also repeated for 95 samples by researchers experienced with genotyping (R. M. Hamner and A. Alexander). Genotyping error rates were calculated by dividing the number of incongruent allele calls by the total number of alleles compared (Bonin et al. 2004).
Replicate samples of the same individual were identified by comparing genotypes in CERVUS v. 3.0 (Kalinowski et al. 2007) and the overall probability of identity (P ID) and probability of identity for siblings (P IDsib) were calculated in GenAlEx v. 6.1 (Peakall and Smouse 2006). To avoid false exclusion, initial matching allowed for up to five mismatching loci, which were examined for potential allelic dropout or processing error. Sex and mtDNA haplotypes were subsequently compared to support our confidence in correctly identifying re-samples. After review and revision, samples with matching genotypes for at least 10 loci (P ID ≤ 1.59 × 10−5) were accepted as re-samples of the same individual and were only represented once in subsequent analyses.
Micro-Checker v. 2.2.3 (Van Oosterhout et al. 2004) was used to assess the presence of null alleles, and the independence of microsatellite loci was tested using the linkage disequilibrium analysis in GENEPOP (Raymond and Rousset 1995). ARLEQUIN v. 3.1 (Excoffier 2006) was used to calculate the number of alleles and the observed and expected heterozygosity per locus, assess departures from Hardy–Weinberg equilibrium, run a locus by locus hierarchical AMOVA, and calculate pairwise F ST values for the regional and local populations.
Isolation by distance
A correlation between genetic differentiation and geographic distance was evaluated by Mantel tests implemented in GenAlEx v. 6.1 (Peakall and Smouse 2006). Independent tests of mtDNA and microsatellite data were conducted using corrected pairwise F ST values (F ST/1−F ST) as a representation of genetic distance. Geographic distance was measured along the coastline between the mid-points of each local population using Google Earth 5.1. Two representations of geographic distance were analyzed: (1) the shortest coastal distance connecting each pair of populations and (2) a circum-linear pathway proposed by Pichler (2002) connecting Timaru and Toetoe Bay (extended from the original Te Waewae Bay endpoint) by taking the long way around the South Island via the West Coast (Fig. 2).
Mantel tests comparing corrected a microsatellite F ST and b mtDNA F ST with circumlinear geographic distance connecting Timaru (TM) and Toetoe Bay (TB) via the West Coast, as shown by the thick black line around the South Island, with double dashed segments indicating areas of low gene flow between regions. The single dashed line representing the shortest coastal distance between TM and TB, indicates very low gene flow, likely to be male-biased
Sex-biased dispersal
The apparently greater differentiation between mtDNA and microsatellites (see Results) prompted tests for male-biased dispersal to determine if this pattern was due to differences in the effective population size of the two markers or an indication of male-biased dispersal. Therefore, sex-biased dispersal around the South Island was examined by comparing sex-specific F ST, inbreeding coefficient (F IS), mean corrected assignment index (mAIc) and variance of mean corrected assignment index (vAIc) based on microsatellite genotypes using two-tailed tests and 1,000 permutations of the resampling procedure in FSTAT v. 2.9.3 (Goudet 2001). Additionally, sex-specific F ST values were calculated based on mtDNA data (whereby individuals were coded as homozygotes) and compared using the resampling procedure in FSTAT (see Oremus et al. 2007). As the pattern of genetic differentiation suggested that gene flow occurs more commonly between local populations within regions, the data for each region was analyzed independently, as well as all together.
Migration rates and identification of migrants
The direction and magnitude of recent migration rates (m) between the Maui’s dolphin and three Hector’s dolphin regional populations were estimated using the 9-locus microsatellite dataset and the Bayesian method implemented in BayesAss v3.0 (Wilson and Rannala 2003). Following a burn-in of 106, we performed 3 × 106 iterations with a sampling frequency of 2000, and deltas set to 0.30, 0.40, and 0.15 for allele frequency, inbreeding coefficient and migration rate, respectively, to achieve acceptance rates between 20 and 40 %, as recommended by Rannala (2011). To confirm convergence, the analysis was repeated three times with different seed numbers and the trace files were examined for consistent oscillations.
After confirming the ability of Structure v 2.3.2 (Pritchard et al. 2000, 2007) to identify the Maui’s dolphin and three regional Hector’s dolphin populations without a priori location information (Online Resource 2), individual assignment and identification of migrants were assessed at the subspecies and regional population levels using this program. The “Use PopInfo” option (G = 0) was applied to run 106 Markov chain Monte Carlo (MCMC) replicates following a burn-in of 105 for K = 4 (9-locus Maui’s and Hector’s dolphin dataset) and K = 3 (13-locus Hector’s dolphin dataset). Individuals with a membership coefficient <0.6 for the population from which they were sampled were considered as potential migrants or having migrant ancestry. Additionally, the Bayesian method of Rannala and Mountain (1997) was implemented in GeneClass2 v. 2.2.2 (Piry et al. 2004), with an alpha of 0.01 and 10,000 repetitions of Paetkau et al.’s (2004) MCMC re-sampling algorithm. Individuals with −log(L home/L max) ≥ 2 were identified as potential migrants or having migrant ancestry.
Given the strong differentiation of maternally-inherited mtDNA between regions, this marker was used a posteriori to evaluate the individuals identified by both programs as being actual migrants or having migrant ancestry. If a potential migrant’s mtDNA haplotype was characteristic of the population to which it was assigned, then its identification as a migrant was supported. Alternatively, if a potential migrant’s mtDNA haplotype was characteristic of the region from which it was sampled, and not the region to which it was assigned, then it is not likely to be an actual migrant itself, but more likely the offspring of a female in her natal region and a migrant male (e.g., an F1 migrant).
Results
Genetic sex identification
Amplification of the sry and ZFX/ZFY fragments allowed for identification of 114 females and 149 males, excluding replicate samples of the same individual (see Microsatellite diversity for identification of individuals). The sex of the remaining 126 individuals (n = 125 swab samples; n = 1 beachcast sample) was not identified due to low quality and quantities of DNA, which were consistent characteristics of the swab samples analyzed in this study. Significant deviations from the expected 1:1 sex ratio (P < 0.05) were found only for beachcast Maui’s and Hector’s dolphins, with an excess of female Maui’s dolphins and an excess of male Hector’s dolphins (Table 1). The overall male bias in Hector’s dolphin samples stems from an excess of male beachcast samples from the East Coast of the South Island. No significant sex bias was observed for biopsy samples collected from any area (Table 1).
Mitochondrial DNA diversity
Mitochondrial DNA control region sequence of approximately 650 bp was obtained for 388 individuals, excluding replicate samples from the same individual (see Microsatellite diversity for identification of individuals). Sixteen of the 17 previously described haplotypes (Pichler et al. 1998; Pichler and Baker 2000; Pichler 2002) were found, as well as five additional haplotypes (S, T, U, V and W; GenBank: JQ890071-JQ890075). Only the historical haplotype ‘N’, previously observed in one individual sampled on the North Island and one from Kaikoura on the East Coast of the South Island (Pichler and Baker 2000), was not found in the current sample. Overall haplotype diversity (h) was 0.846 ± 0.008 and nucleotide diversity (π) was 0.787 ± 0.460 %. The increased sample size available for the Maui’s dolphin (n = 70) confirmed earlier reports (Pichler et al. 1998; Pichler and Baker 2000; Pichler 2002) that this subspecies is fixed for a single mtDNA haplotype (‘G’) not found among the 20 haplotypes currently detected in the South Island Hector’s dolphin populations (Figs. 1, 3). Each regional Hector’s dolphin population was characterized by one or two predominant haplotypes, along with additional low frequency haplotypes (Fig. 1). The haplotype network reflects the phylogeographic structure presented by Pichler et al. (1998), however with our increased sample size, novel Hector’s dolphin haplotypes were detected and the most common haplotypes were found to be shared with at least one other region in low frequencies (Fig. 3). Most local Hector’s dolphin populations exhibited multiple haplotypes, with h ranging from 0.410 to 0.721 and π ranging from 0.239 to 0.982 % (Table 2). The exception was the Timaru population, for which the current sample (n = 14) was fixed for the most common East Coast haplotype (‘C’).
Parsimony network of 21 Hector’s (n = 318) and one Maui’s (n = 70) dolphin mtDNA haplotypes (360 bp) created using TCS 1.21. The circles representing each haplotype are proportional to sample size (n) and are shaded according to the geographic region from which they were sampled. Changes in base pairs between haplotypes and their relative positions in the sequence are mapped onto the lines connecting them
Microsatellite diversity
Microsatellite genotypes were obtained for 266 samples, of which 49 were excluded based on genotype matches (i.e. resamples of the same individual or processing error; P ID = 6.7 × 10−14; P IDsib = 6.8 × 10−6). Therefore, 217 individuals were included in the microsatellite-based analyses. Each locus was genotyped for an average of 191 individuals, producing 2 to 29 alleles per locus with no consistent significant deviations from Hardy–Weinberg equilibrium across populations (Table 3) and no evidence of null alleles. The Maui’s dolphin showed less allelic diversity than the Hector’s dolphin populations at eight of the nine loci with data for both subspecies, even when compared to the South Coast population (n = 31), which was represented by almost half as many samples as the Maui’s dolphin (n = 58). A genotyping error rate of 2.2 % was calculated from the blind replicates and 1.1 % from the independent allele binning. The majority of error from the blind replicates appeared to be allelic dropout that occurred in the earlier runs of EV1 and EV14, and is likely to be the result of advances in genotyping technology (i.e. the move to capillary systems) since the generation of the previous dataset by Pichler (2002).
Subspecies isolation and regional differentiation
In addition to the fixed difference in mtDNA (Φ ST = 0.563; P < 0.001), the Hector’s and Maui’s dolphins also showed a high degree of differentiation based on biparentally-inherited genotypes from the nine microsatellite loci (F ST = 0.167, P < 0.001). All individuals were assigned to the expected subspecies by the methods of both GeneClass2 and Structure (Fig. 4).
Assignment of individuals as Hector’s or Maui’s dolphin subspecies based on the Structure v.2.3.2 analysis of 9-locus microsatellite genotypes. Each vertical bar represents an individual and is shaded according to its coefficient of membership to the Maui’s dolphin (orange) and Hector’s dolphin (yellow) subspecies
Within the Hector’s dolphin, hierarchical analyses of both mtDNA and 13-locus microsatellite data confirmed strong genetic differentiation among the East, West and South Coast regions (mtDNA F ST = 0.321, P < 0.001; Φ ST = 0.395; microsatellite F ST = 0.058, P < 0.001), with large differences among regions, but some difference among local populations within regions (Table 4). All pairwise regional mtDNA FST and Φ ST and microsatellite F ST values were significant (P < 0.001; Table 5), however not all individuals assigned clearly to the region from which they were sampled in the GeneClass2 (Table 6) and Structure (Fig. 5) analyses, suggesting that a very low level of gene flow is occurring between the regions (see Migration Rates and Identification of Migrants).
Assignment of individuals to the East, West or South Coast regional populations of Hector’s dolphins based on the Structure v.2.3.2 analysis of 13-locus microsatellite genotypes. Each vertical bar represents an individual and is shaded according to its coefficient of membership to the East (red), West (blue) and South Coast (green) regional populations. Arrows indicate potential migrants
Hector’s dolphin local differentiation
Pairwise comparisons of the local Hector’s dolphin populations within the East and West Coasts reflected the differentiation among the regions, however relatively little significant differentiation was detected among most local populations within the same region (Table 7). Notable exceptions were found between local populations at the extremes of the East Coast (Timaru and both Cloudy Bay and Kaikoura) and West Coast (Westport and Jackson Bay; Fig. 1), suggesting that a stepping-stone pattern of gene flow is occurring within these regions (see Hector’s dolphin isolation by distance). Unlike the East and West Coasts, however, the two local populations within the South Coast, Te Waewae and Toetoe Bays, displayed a notable restriction in gene flow based on both mtDNA (F ST = 0.136; P = 0.03) and microsatellites (F ST = 0.043; P = 0.005) over the small distance of only ~100 km that separates them.
Hector’s dolphin isolation by distance
Mantel tests supported a one-dimensional stepping stone pattern of gene flow following the long way around the South Island from Timaru to Toetoe Bay, with an apparent barrier to gene flow between them for both mtDNA (r 2 = 0.59, P = 0.001) and microsatellites (r 2 = 0.51, P = 0.001; Fig. 2). The tests using the shortest possible coastal distance connecting each population pair also produced a significant correlation between genetic differentiation and distance, but showed much lower correlation coefficients: mtDNA (r 2 = 0.12, P = 0.013) and microsatellite (r 2 = 0.33, P = 0.001). Interestingly, among the East Coast–South Coast comparisons the Timaru–Toetoe Bay comparison produced the highest pairwise F ST for the mtDNA data, but the lowest pairwise F ST for the microsatellite data. This suggests the likely influence of a low level of male-biased gene flow between these populations, despite a likely historical female gene flow connection between the South and West Coasts.
Hector’s dolphin sex-biased dispersal
No prevalent pattern of sex-biased dispersal was found among Hector’s dolphins. Within the West Coast, males exhibited significantly higher F IS than females (P = 0.048), and approached significantly lower mAIc (P = 0.077), suggesting male-biased dispersal among the local populations within this region (Table 8). On the other hand, males exhibited a higher microsatellite F ST (P = 0.012) within the South Coast, suggesting female-biased dispersal between the two South Coast populations. The overall sample exhibited a higher mtDNA F ST (P = 0.001) for males, also suggesting female-biased dispersal among the three coastal regions; however this may be an artifact of a strong overall bias toward male samples (Table 8).
Migration rates and identification of migrants
Migration rates estimated between the Maui’s dolphin and each of the Hector’s dolphin regional populations were very low with large standard errors (Table 9), providing no evidence for migration between the two subspecies. Furthermore, we did not detect any migrants or individuals with migrant ancestry between the Hector’s and Maui’s dolphin subspecies (Fig. 4).
Although most migration rates estimated among the regional populations of Hector’s dolphins were also low with large standard errors, several showed a directional bias in migration that correlates with the prevailing currents around the South Island (Table 9). The high migration rate of 0.189 (SE = 0.078) from the West Coast to the East Coast corresponds to the D’Urville Current, which flows east through Cook Strait (Brodie 1960). The next largest migration rate was 0.066 (SE = 0.36) from the South Coast to the East Coast, corresponding to the Southland Current, which flows from the West Coast of the South Island east through Foveaux Strait before it hooks north along the East Coast (Brodie 1960).
Although the majority of the Hector’s dolphins showed very strong assignment to the region from which they were sampled, five individuals were identified as potential migrants between regions by the methods of both Structure and GeneClass2 (Fig. 5; Table 6). An a posteriori review of mtDNA showed that the haplotypes of these individuals were characteristic of the region from which they were sampled, and were only detected in very low frequencies or not at all in the region to which they were assigned. This suggests that these five individuals are more likely F1 migrants produced by resident females and migrant males than actual migrants themselves.
Discussion
Our findings confirmed the subspecies classification of the Maui’s dolphin (Cephalorhynchus hectori maui) and the Hector’s dolphin (C. h. hectori) based on a larger sample of mtDNA and more extensive survey of microsatellites (Pichler et al. 1998; Pichler and Baker 2000; Pichler 2002). The two subspecies appear to be entirely isolated with no detectable levels of current female or male gene flow or migration. While these findings would qualify the Hector’s and Maui’s dolphins for elevation to species status under some species concepts, we feel that evidence of diagnostic nuclear DNA differences and irreversible divergence is necessary before such a proposal is suggested (Reeves et al. 2004).
In addition to the previously described differentiation between the East and West Coast populations of Hector’s dolphins (Pichler et al. 1998; Pichler and Baker 2000; Pichler 2002), significant differentiation for two South Coast populations (Te Waewae Bay and Toetoe Bay) was also supported by both our mtDNA and microsatellite analyses. Therefore, independent management plans should be considered for each of these populations. Although Toetoe Bay is represented by a small sample size (eight individuals) and would benefit from further sampling, the genetic differentiation detected by this first investigation of the genetic diversity in this small population warrants attention. Almost half of the sampled individuals represent a newly detected haplotype, ‘S’, which has never been sampled in any other population. Furthermore, it lacks haplotype ‘M’, which characterizes almost half of the closest neighboring population in Te Waewae Bay and would likely be detected if female gene flow were occurring regularly between these two populations. Within the larger East and West Coast populations, there appears to be a sufficient level of step-wise gene flow to maintain genetic diversity. However among the four Hector’s dolphin populations, only very rare dispersal events are facilitating gene flow across the approximate 100–370 km gaps separating them. The cause of the gaps separating these four genetically differentiated populations of Hector’s dolphins is likely environmental as well as behavioral. An avoidance of deep water seems to be a primary factor limiting the distribution and density of these dolphins, as most sightings occur in depths less than 39 m (Bräger et al. 2003). This is consistent with the rarity of Hector’s dolphin in the Fiordland area between the West and South Coasts, where depths are in excess of 300 m (Dawson and Slooten 1988; Slooten et al. 2004). It is also consistent with the apparent reluctance of these dolphins to cross the deep water of Cook Strait, which divides the North and South Islands (Dawson and Slooten 1988), and may have spurred the divergence of the two subspecies when it opened near the end of the last ice age 15–16,000 years ago (Lewis et al. 1994; Pichler 2001). The North Island–South Island separation is a common biogeographic pattern seen among both terrestrial and marine organisms of New Zealand (Wallis and Trewick 2009; Trewick and Bland 2011), so it is plausible that a host of ecological and environmental factors played a role in the divergence of Hector’s and Maui’s dolphins and continue to maintain their differentiation.
Regardless of the causes, gaps in Hector’s dolphin distribution larger than the average 30–50 km home range (maximum reported ~107 km; Bräger et al. 2002; Rayment et al. 2009) seem to be presenting barriers to dispersal that are acting as a force of isolation. This is consistent with the high degree of genetic differentiation observed between the regions, and over the approximate 100 km distance between the Te Waewae Bay and Toetoe Bay populations. Furthermore, Rayment et al. (2009) identified ‘hotspots’ of Hector’s dolphin density around Banks Peninsula that were spaced approximately 30 km apart, and suggested that dolphins with home ranges centered around hotspots would have little interaction with dolphins of adjacent hotspots. Therefore, within the East and West Coasts, each local population is likely to serve as a critical link for maintaining the stepping-stone chain of gene flow. The loss of a local population could interrupt the regional connectivity, resulting in smaller isolated subpopulations facing increased susceptibility to genetic drift and extinction. This may already be the case for Te Waewae and Toetoe Bays.
The protection of discrete local populations alone, however, is not sufficient to preserve the evolutionary potential of the Hector’s dolphin. Consideration must be given to protecting corridors for the individuals dispersing between local populations, and the even more rare dispersers between regions. It is this occasional gene flow that enhances the evolutionary potential of small populations. Dispersal events are particularly important for the local populations at the extremes of the East Coast and West Coast regions, as the genetic diversity of these extreme populations is primarily maintained by gene flow with a single adjacent intra-regional population. These populations also border the gaps between regions, and therefore, are likely to be critical in facilitating the rare dispersal responsible for maintaining the connectivity of the subspecies as a whole. If these ‘peripheral’ local populations are extirpated, the regions will be separated by larger geographic gaps, further reducing the frequency of dispersal. The two South Coast populations illustrate the relatively small scale over which differentiation can occur. The approximate 100 km coastal distance that separates the eastern edge of Te Waewae Bay and western edge of Toetoe Bay sampling areas is sufficient to restrict local gene flow, and the approximate 160 km to the nearest regional population (East Coast, Timaru) appears to be sufficient for the complete isolation of maternal lineages.
The East and South Coasts, however, appear to be connected by a low level of male-mediated gene flow showing a directionally-biased migration rate favoring movement from the South Coast to the East Coast. This directional bias is consistent with the detection of two F1 migrants sampled in the East Coast inferred to have fathers from the South Coast, compared to a single F1 migrant for which the opposite was found. Interestingly, the father of the one F1 migrant detected between the East and West Coasts would have traveled in opposition to the prevailing direction of migration and D’Urville Current, suggesting that more complex factors than just currents influence dispersal direction.
We did not find evidence of a strong sex-bias in dispersal among Hector’s dolphin populations. In general, the microsatellite-based tests implemented have low power to detect sex-biases unless the rate of dispersal is high and the sex-bias is large (Goudet et al. 2002). It is possible that the significant biases detected could be artifacts of the small samples sizes or variability in sample sizes that results when the samples representing each population are divided by sex. Conversely, a low rate of male-biased gene flow, consistent with the F IS test within the West Coast and the F1 migrants resulting from migrant fathers that were detected, could be masked by an isolation by distance effect. If a low number of males were more likely to disperse over longer distances, the signal could be over-shadowed by that of both sexes engaging in the step-wise pattern of shorter-range dispersal and gene flow between adjacent local populations (Goudet et al. 2002). Given the disagreement in the direction of bias for the few significant results, we cannot conclude that dispersal in Hector’s dolphins is strongly skewed toward either sex.
Conservation approaches should, however, consider the implications of the differences in sex biases observed in beachcast samples from Hector’s and Maui’s dolphins. In the Hector’s dolphin, the male bias represented in beachcast specimens suggests that the protection of corridors may be particularly vital for male Hector’s dolphins. A pattern of male-biased dispersal would be consistent with the suggestion that males are likely to encounter more nets while dispersing between areas, thereby increasing the likelihood of entanglement (Pichler 2002). In the Maui’s dolphin, the observed female bias in beachcast samples could be an indication of deleterious inbreeding effects. Given the isolation and small size of this remnant population, some degree of inbreeding is inevitable. The discovery of beachcast pregnant females and neonates is consistent with lethal pregnancy-related complications that have been associated with inbreeding depression in captive populations (Adamec et al. 2006; Strom Holst and Frossling 2009).
The continued genetic monitoring (Schwartz et al. 2007) of these populations is critical to assessing the effectiveness of current management strategies for Hector’s and Maui’s dolphins and the potential need for more active conservation interventions in the future. The opportunity for monitoring across the relatively short lifespan of these dolphins, compared to other cetaceans, will also provide broader insight into the demographic and genetic forces influencing the survival, or extinction, of small populations.
References
Adamec V, Cassell BG, Smith EP, Pearson RE (2006) Effects of inbreeding in the dam on dystocia and stillbirths in US holsteins. J Dairy Sci 89:307–314
Baird SJ, Bradford E (2000) Estimation of Hector’s dolphin bycatch from inshore fisheries, 1997/98 fishing year. Published Client Report on Contract 3024, Department of Conservation, Wellington
Baker CS, Hamner RM, Cooke J, Heimeier D, Vant M, Steel D, Constantine R (In review) Abundance and probable decline of the Maui’s dolphin (Cephalorhynchus hectori maui) estimated from capture–recapture of microsatellite genotypes, 2001–2006
Baker CS, Slade RW, Bannister JL, Abernethy RB, Weinrich MT, Lien J, Urban RJ, Corkeron P, Calambokidis J, Vasquez O, Palumbi SR (1994) Hierarchical structure of mitochondrial DNA gene flow among humpback whales, Megaptera novaeangliae, world-wide. Mol Ecol 3:313–327
Baker AN, Smith ANH, Pichler FB (2002) Geographical variation in Hector’s dolphin: recognition of new subspecies of Cephalorhynchus hectori. J R Soc NZ 32:713–727
Bérubé M, Jørgensen H, McEwing R, Palsbøll PJ (2000) Polymorphic di-nucleotide microsatellite loci isolated from the humpback whale, Megaptera novaeangliae. Mol Ecol 9:2181–2183
Blezard RH (2002) Observations of set-net and inshore trawl fishing operations in the South Canterbury Bight, 2001. DOC Sci Intern Ser 85, Department of Conservation, Wellington, 19 p
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Bräger S, Dawson SM, Slooten E, Smith S, Stone GS, Yoshinaga A (2002) Site fidelity and along-shore range in Hector’s dolphin, an endangered marine dolphin from New Zealand. Biol Conserv 108:281–287
Bräger S, Harraway JA, Manly BFJ (2003) Habitat selection in a coastal dolphin species (Cephalorhynchus hectori). Mar Biol 143:233–244
Brodie JW (1960) Coastal surface currents around New Zealand. NZ J Geol Geophys 3:235–252
Buchanan FC, Friesen MK, Littlejohn RP, Clayton JW (1996) Microsatellites from the beluga whale Delphinapterus leucas. Mol Ecol 5:571–575
Caldwell M, Gaines MS, Hughes CR (2002) Eight polymorphic microsatellite loci for bottlenose dolphin and other cetacean species. Mol Ecol Notes 2:393–395
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Dalebout ML, Robertson KM, Frantzis A, Engelhaupt D, Mignucci-Giannoni AA, Rosario-Delestre RJ, Baker CS (2005) Worldwide structure of mtDNA diversity among Cuvier’s beaked whales (Ziphius cavirostris): implications for threatened populations. Mol Ecol 14:3353–3371
Dawson SM (1991) Incidental catch of Hector’s dolphin in inshore gillnets. Marine Mammal Sci 7:283–295
Dawson SM, Slooten E (1988) Hector’s dolphin, Cephalorhynchus hectori: distribution and abundance. In: Brownell RL Jr, Donovan GP (eds) Biology of the genus Cephalorhynchus. Reports of the International Whaling Commission, special issue 9. International Whaling Commission, Cambridge, pp 315–324
Dawson SM, Slooten E (2005) Management of gillnet bycatch of cetaceans in New Zealand. J Cetacean Res Manag 7:59–64
Dawson S, Slooten E, DuFresne S, Wade P, Clement D (2004) Small-boat surveys for coastal dolphins: line-transect surveys for Hector’s dolphins (Cephalorhynchus hectori). Fish Bull 201:441–451
Elwen S, Meyer MA, Best PB, Kotze PGH, Thornton M, Swanson S (2006) Range and movements of female Heaviside’s dolphins (Cephalorhynchus heavisidii), as determined by satellite-linked telemetry. J Mammal 87:866–877
Excoffier L (2006) ARLEQUIN ver 3.1: an integrated software package for population genetics data analysis. http://cmpg.unibe.ch/software/arlequin3
Fairfax D (2002) Observations of inshore trawl fishing operations in Pegasus Bay and the Canterbury Bight, 2002. DOC Sci Intern Ser 86, Department of Conservation, Wellington, 11 p
Frankham R (1995) Conservation genetics. Annu Rev Genet 29:305–327
Frankham R (1997) Do island populations have less genetic variation than mainland populations? Heredity 78:311–327
Frankham R (1998) Inbreeding and extinction: island populations. Conserv Biol 12:665–675
Gilson A, Syvanen M, Levine K, Banks J (1998) Deer gender determination by polymerase chain reaction: validation study and application to tissues, bloodstains, and hair forensic samples from California. Calif Fish Game 84:159–169
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). Available from http://www.unil.ch/izea/softwares/fstat.html
Goudet J, Perrin N, Waser P (2002) Tests for sex-biased dispersal using bi-parentally inherited genetic markers. Mol Ecol 11:1103–1114
Harlin AD, Wursig B, Baker CS, Markowitz TM (1999) Skin swabbing for genetic analysis: application to dusky dolphins (Lagenorhynchus obscurus). Marine Mammal Sci 15:409–425
Heinrich S (2006) Ecology of Chilean dolphins and Peale’s dolphins at Isla Chiloé, southern Chile. Ph.D. Thesis, University of St. Andrews, St. Andrews
Hoelzel AR, Dahlheim M, Stern SJ (1998a) Low genetic variation among killer whales (Orcinus orca) in the eastern North Pacific and genetic differentiation between foraging specialists. J Hered 89:121–128
Hoelzel AR, Potter CW, Best PB (1998b) Genetic differentiation between parapatric ‘nearshore’ and ‘offshore’ populations of the bottlenose dolphin. Proc R Soc Lond B 265:1177–1183
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1099–1106
Krützen M, Valsecchi E, Connor RC, Sherwin WB (2001) Characterization of microsatellite loci in Tursiops aduncus. Mol Ecol Notes 1:170–172
Krützen M, Barre LM, Moller LM, Heithaus MR, Simms C, Sherwin WB (2002) A biopsy system for small cetaceans: darting success and wound healing in Tursiops spp. Marine Mammal Sci 18:863–878
Lewis KB, Carter L, Davey FJ (1994) The opening of Cook Strait: interglacial tidal scour and aligning basins at a subduction to transform plate edge. Mar Geol 116:293–312
New Zealand Department of Conservation (2011) Hector’s dolphin incident database. http://www.doc.govt.nz/conservation/native-animals/marine-mammals/dolphins/hectors-dolphin/docs-work/hectors-dolphin-incident-database/
New Zealand Department of Conservation, Ministry of Fisheries (2007) Hector's and Maui's dolphin threat management plan draft for public consultation. 5 March 2009. http://www.fish.govt.nz/NR/rdonlyres/2088EFD2008C-E2207-4798-9315-C2000AF2002FC2006FFB2002/2000/DRAFTTMPFINAL.pdf
Oremus M, Poole MM, Steel D, Baker CS (2007) Isolation and interchange among insular spinner dolphin communities in the South Pacific revealed by individual identification and genetic diversity. Mar Ecol Prog Ser 336:275–289
Paetkau D, Slade R, Burden M, Estoup A (2004) Genetic assignment methods for the direct, real-time estimation of migration rate: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Peakall R, Smouse PE (2006) GenAlEx 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pichler FB (2001) Population structure and genetic variation in Hector’s dolphin (Cephalorhynchus hectori). PhD Thesis, University of Auckland, Auckland
Pichler FB (2002) Genetic assessment of population boundaries and gene exchange in Hector’s dolphin. DOC Sci Intern Ser 44, Department of Conservation, Wellington, 37 p
Pichler FB, Baker CS (2000) Loss of genetic diversity in the endemic Hector’s dolphin due to fisheries-related mortality. Proc R Soc Lond B 267:97–102
Pichler FB, Dawson SM, Slooten E, Baker CS (1998) Geographic isolation of Hector’s dolphin populations described by mitochondrial DNA sequences. Conserv Biol 12:676–682
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GENECLASS2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pritchard JK, Wen X, Falush D (2007) Documentation for structure software: version 2.2. Available from http://pritch.bsd.uchicago.edu/software
Rannala B (2011) BayesAss edition 3.0 user’s manual. Downloaded from http://www.rannala.org/?page_id=245. Accessed 5 Jan 2012
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci 94:9197–9201
Rayment W, Dawson S, Slooten L, Childerhouse S (2006) Offshore distribution of Hector’s dolphin at Banks Peninsula. DOC Res Dev Ser 232, Department of Conservation, Wellington, 23 p
Rayment W, Dawson S, Slooten E, Braeger S, Fresne SD, Webster T (2009) Kernel density estimates of alongshore home range of Hector’s dolphins at Banks Peninsula, New Zealand. Marine Mammal Sci 25:537–556
Raymond M, Rousset F (1995) GENEPOP. Available at http://genepop.curtin.edu.au/
Read AJ, Westgate AJ (1997) Monitoring the movements of harbour porpoises (Phocoena phocoena) with satellite telemetry. Mar Biol 130:315–322
Reeves RR, Perrin WF, Taylor BL, Baker CS, Mesnick SL (2004) Report of the workshop on shortcomings of cetacean taxonomy in relation to needs of conservation and management. NOAA Technical Memorandum NOAA-NMFS-SWFSC-363, National Oceanic and Atmospheric Administration, La Jolla
Reeves RR, Dawson SM, Jefferson TA, Karczmarski L, Laidre K, O’Corry-Crowe G, Rojas-Bracho L, Secchi ER, Slooten E, Smith BD, Wang JY, Zhou K (2008) Cephalorhynchus hectori maui. In: IUCN 2011. IUCN Red List of Threatened Species. Version 2011.1. http://www.iucnredlist.org
Reid PRJ (2002) Report on an inshore fishery observer programme in Pegasus Bay and Canterbury Bight, 1999/2000. Unpublished report. Fisheries Audit Services (NZ), Nelson
Rosel PE, France SC, Wang JY, Kocher TD (1999) Genetic structure of harbour porpoise Phocoena phocoena populations in the northwest Atlantic based on mitochondrial and nuclear markers. Mol Ecol 8:S41–S54
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Schlotterer C, Amos B, Tautz D (1991) Conservation of polymorphic simple sequence loci in cetacean species. Nature 354:63–65
Schwartz MK, Luikart G, Waples RS (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33
Slooten E (2007) Conservation management in the face of uncertainty: effectiveness of four options for managing Hector’s dolphin bycatch. Endanger Species Res 3:169–179
Slooten E, Dawson SM (1988) Studies on Hector’s dolphin, Cephalorhynchus hectori: a progress report. In: Brownell RL Jr, Donovan GP (eds) Reports of the International Whaling Commission, special issue 9. International Whaling Commission, Cambridge, pp 325–338
Slooten E, Lad F (1991) Population biology and conservation of Hector’s dolphin. Can J Zool 69:1701–1707
Slooten E, Dawson SM, Rayment WJ (2004) Aerial surveys for coastal dolphins: abundance of Hector’s dolphins off the South Island west coast, New Zealand. Marine Mammal Sci 20:477–490
Stone G, Hutt A, P Duignan, Teilmann J, Cooper R, Geschke K, Yoshinaga A, Russell K, Baker A, Suisted R, Baker S, Brown J, Jones G, Higgins D (2005) Hector’s dolphin (Cephalorhynchus hectori hectori) satellite tagging, health and genetic assessment project. Unpublished report submitted to the New Zealand Department of Conservation. Auckland Conservancy, Auckland
Strom Holst B, Frossling J (2009) The Swedish breeding cat: population description, infectious diseases and reproductive performance evaluated by a questionnaire. J Feline Med Surg 11:793–802
Trewick SA, Bland KJ (2011) Fire and slice: palaeogeography for biogeography at New Zealand’s North Island/South Island juncture. J R Soc NZ, iFirst. doi:10.1080/03036758.2010.549493
Valsecchi E, Amos W (1996) Microsatellite markers for the study of cetacean populations. Mol Ecol 5:151–156
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wallis GP, Trewick SA (2009) New Zealand phylogeography: evolution on a small continent. Mol Ecol 18:3548–3580
Wells RS, Hansen LJ, Baldridge A, Dohl TP, Kelly DL, Defran RH (1990) Northward extension of the range of bottlenose dolphins along the California coast. In: Leatherwood S, Reeves RR (eds) The bottlenose dolphin. Academic Press, San Diego, pp 421–431
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Acknowledgments
This work was made possible by access to the University of Auckland Cetacean Tissue Archive, maintained and curated by CSB and RC. Biopsy samples were collected under permit to CSB from the New Zealand Department of Conservation and animal ethics protocols AEC/02/2002/R9 and AEC/02/2005/R334 from the University of Auckland. We thank everyone involved in biopsy sampling and the collection of samples from beachcast specimens, including Ros Cole, Padraig Duignan, Al Hutt, Don Neale, Wendi Roe, Kirsty Russell, Greg Stone and the numerous field staff of the New Zealand Department of Conservation. We are grateful for assistance and discussions from Murdoch Vant, Alana Alexander, Shane Lavery, Marc Oremus and Debbie Steel. Funding for this work was provided by grants or contracts to CSB from the New Zealand Department of Conservation, the U.S. Marine Mammal Commission and the Marsden Fund, and support to RC from the Performance Based Research Fund of the University of Auckland. RMH was supported in part by a Fulbright US Graduate Fellowship to New Zealand. Publication of this paper was supported, in part, by the Thomas G. Scott Publication Fund.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hamner, R.M., Pichler, F.B., Heimeier, D. et al. Genetic differentiation and limited gene flow among fragmented populations of New Zealand endemic Hector’s and Maui’s dolphins. Conserv Genet 13, 987–1002 (2012). https://doi.org/10.1007/s10592-012-0347-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0347-9