Abstract
Wildlife species exposed to habitat fragmentation are often in need of a conservation effort. The African buffalo (Syncerus caffer) is one of the key species in the Serengeti ecosystem as they form a large part of the herbivore biomass, providing ecotourism and valuable trophies. The ecosystem is a part of Tanzanias protected areas and is administrated under different management practices. Among these, we have analysed the genetic structure of buffalo (n = 68) from the Serengeti National Park (SNP), the Ngorongoro conservation area (NCA) and the Maswa game reserve (MGR). Both the sequence variation in a 493 base pair fragment of the mitochondrial D-loop and the allele frequency-distribution in 15 microsatellites suggest genetic structuring of the buffalo populations within the ecosystem. Both the allele frequency-distribution and the amount of genetic variation were high and similar in SNP and MGR, suggesting a high degree of gene flow between these locations. By comparison, the NCA buffaloes had significantly lower genetic variation and were genetically differentiated from SNP and MGR. Approximate Bayesian computation estimates suggest that the observed genetic structure is of a recent origin, indicating that the recent increases in developmental activity in the region may have influenced the genetic structure of the buffalo within the Serengeti ecosystem.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Historically, the vast populations of large wild herbivores in Africa provided a substantial resource, supplying local and regional communities with goods and economic income (Conover 1997; Loibooki et al. 2002; Ogutu 2002). However, during the past few decades, overexploitation, poaching, diseases and habitat loss have reduced many of these large herbivore species to the extent that they are now restricted to protected areas (Hilborn et al. 2006; Chantal et al. 2007). Unfortunately, many protected areas in Africa are currently experiencing size reduction and isolation due to increases in human populations and activities (Newmark 2008), which may disrupt or prevent dispersal and gene flow (Ralph et al. 2006; Heller et al. 2010). As a consequence, small or fragmented populations may experience inbreeding, loss of adaptive genetic variation and an increased risk of extinction (Allendorf 1986; Soulé and Mills 1992; Amos and Balmford 2001; Keller and Waller 2002). Tanzania has reserved more than 25% of its land areas to wildlife conservation and harbours a wide diversity of wild animals (Stuwart et al. 1990; Thirgood et al. 2004). Recently however, protected areas in Tanzania have suffered fragmentation and isolation resulting from human activities. More than two decades ago it was estimated that the country has lost about 43% of its original wildlife habitats (WRI 1989), a situation that requires particular conservation attention (Fynn and Bonyongo 2010).
The Serengeti ecosystem is a geographical region west of the Great Rift Valley in northern Tanzania, often defined as the area encompassed by the wildebeest migration (McNaughton and Campbell 1991). The ecosystem cover areas under different management practices with different degrees of allowed human impact, ranging from strict photographic tourism to trophy hunting and multiple usage of land with livestock keeping, agriculture and human habitation. In some parts of the ecosystem, human population growth, anthropogenic activities and fragmentation of the ecosystem have been rapid (Kurji 1981; Meertens et al. 1995; Kijazi et al. 1997; Estes et al. 2006), posing a serious threat to the long term viability of wildlife populations.
African buffalo (Syncerus caffer) is one of the key species in the Serengeti ecosystem; not only because they form a large part of the herbivore biomass in the area (Van Hooft et al. 2002, 2003; Iain et al. 2004), but also by providing ecotourism and valuable trophy hunting. A number of studies have been conducted on the species’ ecology across the region (George 1975; Sinclair 1977; Runyoro et al. 1995; Estes et al. 2006; Metzger et al. 2010), providing information on its spatial and temporal population fluctuations. Historically, the species had a wide geographical distribution across nearly the whole of sub-Saharan Africa. However, the buffalo has declined in numbers across most of its historical range and its distribution has become highly fragmented (Sidney 1965; Sinclair 1977). In Tanzania the census population amounts to about 400,000 buffalos, but the population is increasingly fragmented and its distribution is becoming more restricted to protected areas (TAWIRI 2008).
Genetic studies have shown that the African buffalo harbours high levels of genetic diversity but low population differentiation throughout its ecological range (Simonsen et al. 1998; Van Hooft et al. 2000, 2002, 2003). However, to achieve sustainable conservation of the species, and to be able to identify potential management units and barriers to gene flow (De Young and Honeycutt 2005; Heller et al. 2010), it is necessary to attain information on existing genetic structure both between and within protected areas. Besides a study of the genetic differentiation among one introduced and three remnant buffalo populations in South Africa (O’Ryan et al. 1998), little is known about the genetic population structure of buffalo within separate ecosystems. The primary aim of this study was therefore to examine the genetic structure of the African buffalo within the Serengeti ecosystem with special emphasis on the potential differentiation between areas with different management practices.
Materials and methods
Study area
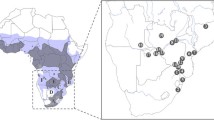
The Serengeti ecosystem is located in the north-east of Tanzania between 34°45′–35°50′ E and 2°–3°20′ S (Fig. 1a). The ecosystem covers different conservation administrations. The main are the Serengeti National Park (SNP), the Ngorongoro conservation area (NCA) and the Maswa game reserve (MGR) (Fig. 1b). In addition there is Ikorongo/Grumeti game reserve (IGGR) and Loliondo game controlled area (LGCA). In 2006, estimates from repeated field counting suggested census population sizes of 1,887 buffaloes in NCA, 70,526 in SNP and 47,668 in MGR (TAWIRI 2008). SNP is a totally protected area where no human activity other than photographic tourism is allowed. The MGR lies along the western boundary of SNP and abuts the south-western corner of NCA (Fig. 1b). Here, photographic tourism and game harvesting including trophy hunting is permitted but human habitation is not. From 1976 to 2003, MGR area has twice been reduced in size due to increasing human activities (Kurji 1985; Meertens et al. 1995; Songorwa 2004). By comparison, the NCA is a multiple land-use area where photographic tourism, human habitation and livestock keeping are allowed but harvesting of wildlife is forbidden. In this area human population growth and development has been rapid (Kurji 1981; Kijazi et al. 1997; Estes et al. 2006). Most buffaloes in NCA are found in the Ngorongoro Crater (Fig. 1b) which is particularly famous for its wildlife and photographic safaris. No human settlement is allowed in the crater, although livestock keepers are allowed to make a seasonal use of the crater floor for grazing, watering and salt licking (Fyumagwa et al. 2007). It has been suggested that human impacts and habitat reductions around the crater have affected buffalo migration to nearby areas (Estes et al. 2006).
Map showing Tanzania and location of Serengeti ecosystem (a), the buffalo distribution in the Tanzanian part of the ecosystem together with geographical location of the different conservation administration areas including Serengeti National Park (SNP), Ngorongoro Conservation Area (NCA), Maswa Game Reserve (MGR), Loliondo Game Controlled Area (LGCA), Ikorongo/Grumeti Game Reserves (IGGR) (b), and the distribution of the human settlement in NCA adjacent to Crater highlands (c)
Samples and DNA extraction
From 2007 to 2009, buffalo blood samples were obtained from within the crater in NCA (n = 19) and SNP (n = 24), while muscle tissue was sampled in MGR (n = 25). In NCA all samples were obtained from the crater floor only, while in SNP and MGR the sampling were more spread out across the respective areas. Whole blood was obtained from immobilized animals during routine disease surveillance. Etorphine hydrochloride (M99® 9.8 mg/ml, Novartis South Africa Ltd) was used for immobilization. Darting was done from a vehicle using a gas powered dart gun (DAN-INJECT® MOD JM, Denmark). Whole blood was collected from any accessible vein in EDTA vacutainers and stored in a cool box for a maximum of 6 h prior to freezing (−20°C). After sampling, immobilized animals were given the antidote Diprenorphine Hydrochloride (M5050® 5 mg/ml, Norvartis South Africa Ltd) and monitored until full recovery and re–joining with the herd. From MGR samples were obtained from hunters and from lion kills. Muscle samples were stored in 5 ml bottles containing absolute ethanol and kept at room temperature. Genomic DNA was isolated from samples using QIAGEN®, DNeasy Blood and Tissue kit.
DNA amplification
Samples were genotyped using 15 di-nucleotide microsatellites of bovine and ovine origin (Table 1). The microsatellites were distributed on 13 different chromosomal locations on the cattle genome. The PCR was carried out on approximately 20–40 ng of genomic DNA, 2 pmol of each primer, 50 mM KCl, 1.5 mM MgCl2, 10 mM Tris–HCl, 1 μl dNTP and 0.5 U Taq polymerase (Amplicon®), in a total volume of 10 μl. Amplification was done using Gene Amp® PCR system 9700 (Applied Biosystems). Thermocycling parameters after denaturation at 95°C in 2 min were: 95°C for 30 s, annealing for 30 s and extension at 72°C for 45 s followed by 10 min at 72°C. The number of PCR cycles and annealing temperatures for the different loci are given in Table 1. Forward primers were fluorescently labeled for electrophoresis on an ABI3100 automated sequencer (Applied Biosystems). Commercially prepared size standards (ROX GENESCAN® 400HD) were run with every sample. Alleles were scored using GeneMapper 3.7 (ABI 3100, Applied Biosystems), and new PCR’s were performed for samples where genotypes were unclear. Furthermore, 10% of all the samples were selected at random and new PCR and genotyping were performed.
All samples were also amplified and sequenced for a 493 bp region of the mitochondrial control region adjacent to the tRNA pro gene, using the primers 5′-AATAGCCCCACTATCAGCACCC-3′ (Flagstad et al. 2000) paired with 5′-GTGAGATGGCCCTGAAGAAA-3′. Amplification was done in Gene Amp® PCR system 9700 (Applied Biosystems) using an initial denaturation at 94°C for 4 min followed by 35 cycles of 40 s at 94°C, 40 s at 60°C and 40 s at 72°C and ended by an additional 7 min extension step at 72°C. Amplifications were performed in 25 μl volumes containing 1.35 mM MgCl2, 200 μM of each dNTP, 5 pmol of each primer and 0.5 units of AmpliTaq DNA polymerase (Applied Biosystems). PCR amplicons were purified with ExoSap-IT (GE Healthcare) and sequenced using BigDye terminator chemistry version 1.1 on an ABI3100 automated sequencer (Applied Biosystems) following the manufactures’ protocol. Sequences were aligned manually in Proseq 2.9 (Filatov 2002). Sequences were deposited in the GenBank database under accession numbers JN849157-JN849224.
Statistical analysis
For the mitochondrial data Arlequein 3.11 (Excoffier et al. 2005) was used to estimate nucleotide and gene diversity and to obtain pair-wise F ST estimates, taking haplotype frequencies as well as nucleotide variation among haplotypes into account. Statistical significance was evaluated by 1,000 permutations.
The microsatellite data were checked for null alleles, stutter errors or short allele dominance by use of the Micro-Checker 2.2.3 (van Oosterhout et al. 2004). Genetic diversity was estimated from the mean number of alleles per locus, allele frequencies, allelic richness (El Mousadik and Petit 1996), observed and expected heterozygosity using both FSTAT 2.9.3 (Goudet 2001) and CERVUS 3.0.3 (Marshall et al. 1998). Deviations from Hardy–Weinberg equilibrium (HWE) were assessed and Fisher’s exact tests of pairwise genetic differentiation among the sampled individuals performed, using default settings in GENEPOP 4.0.7 (Rousset 2008). The significance level was sequentially Bonferroni adjusted for repeated tests (Rice 1989). Population genetic differentiation was further addressed through traditional F ST statistics (Weir and Cockerham 1984), analysed in GENEPOP and through the more recently proposed estimator D est (Jost 2008), using the web based application SMOGD (Crawford 2010). F ST has been suggested to be well suited when both sample size and the numbers of applied loci are relatively low (Gaggiotti et al. 1999), while D est appears to more accurately account for differences in allelic diversity, especially for highly polymorphic markers such as microsatellites (Jost 2008).
Genetic structuring was also assessed through Bayesian clustering of the microsatellite genotypes as implemented in STRUCTURE 2.3 (Pritchard et al. 2000; Hubisz et al. 2009). The log likelihood of our data was estimated given different numbers of genetic clusters (ln Pr (X |K), K ∈ [1, 6]), using an admixture model with correlated allele frequencies and ten runs of each K. Each run consisted of 500,000 MCMC repetitions and 100,000 burnin cycles. For increasing values of K, the variation between runs of each K value, was used to assess the main genetic structure of the data set (Evanno et al. 2005). The STRUCTURE analyses were done both with and without prior knowledge of sampling locations (with and without LOCPRIOR) (Pritchard et al. 2000; Hubisz et al. 2009). The amount of information provided by the sampling location is expressed by the value r, calculated across all runs. Values equal to or greater than 1 imply that information on the sampling location of individuals is uninformative about ancestry, while smaller r values imply the opposite. To assess first-generation dispersal, STRUCTURE was run in a second analysis using POPINFO at default setting.
The microsatellite data were further analysed in BOTTLENECK 1.2 (Cornuet and Luikart 1996) to test the probability of recent bottlenecks. We applied a two-phase model of stepwise mutation (SMM; Kimura and Ohta 1978) with 20 and 10% variation from the infinite allele model (Kimura and Crow 1964), as this fits most microsatellites better than a strict one-step model (Di Rienzo et al. 1994).
The time of divergence for observed microsatellite differentiation was estimated through the coalescent-based approximate Bayesian computation (ABC) algorithm of DIY ABC (Cornuet et al. 2008). Alternative historic scenarios’ of divergence and admixture that may explain today’s observed populations were specified and explored with and without demographic events (Fig. 3, Table S1 (supplementary)), some involving un-sampled populations (outside the study area). Conditions were set for 1) the sequence order of historic events and 2) for demographic events (VarNe) according to historic records. From each scenario 500,000 data sets were simulated by drawing from a specified set of prior distributions of the parameters. The 5,000 of the simulated data sets most similar to the observed data were identified through logistic regression and a set of four within and six among populations default summary statistics (means across loci for; the number of alleles, gene diversity (Nei 1987), allele size variance, M-ratio (Garza and Williamson 2001) per sample and between each pair of samples, the F ST (Weir and Cockerham 1984), (δμ)2 (Goldstein et al. 1995), number of alleles, gene diversity, allele size variance and shared allele distance (Chakraborty and Jin 1993)). These 5,000 simulated data sets were then used to estimate posterior probabilities and distributions, for each alternative scenario, respectively. The generalized stepwise model of mutation was applied with default values (GSM; Fu and Chakraborty 1998; Estoup et al. 2002). The 2006 field counting (TAWIRI 2008) was used as basis for the census population sizes. SNP and MGR are geographically continuous and, as our analysis indicate, genetically non-differentiated, and were in the analysis combined to 118,194 individuals. Generally the effective size of populations, Ne, is much smaller than the census size because of age structure, uneven sex-ratios, and variation in family size and temporal fluctuations in population size (Grant and Grant 1992; Frankham 1995). In Buffalo, Ne has been reported to be from 10 to 30% of the census size (O’Ryan et al. 1998; Van Hooft et al. 2003). Priors with normal distributions and means of 10 and 30% of the census size were therefore used for present effective sizes. Priors for ancient effective sizes were set large and wide (10,000–100,000).
Results
Among the 493 sequenced mtDNA base pairs (bp), insertions and deletions were identified at four sites which were omitted in further analysis. Among the remaining 489 bp, a total number of 28 haplotypes were identified through substitutions at 75 positions. At only two positions more than two different nucleotides were observed. The level of genetic variability showed a similar gene and nucleotide diversity among the three sampling areas (Table 2), while the number of haplotypes appeared to be lower in NCA. Pair-wise comparisons of F ST (Table 3) indicated low and non-significant differences between the SNP and MGR but significant differences between NCA and each of the two other populations (both P < 0.001). The haplotype sharing was generally low. Among the 39 haplotype observed, eight were shared between SNP and MGR while only one and two haplotype were shared between NCA and each of SNP and MGR, respectively. Most haplotype were rare, and as much as 18 haplotype occurred in only one or two copies while only two haplotype occurred in five or more copies. With such a high level of variation, haplotype frequency estimates become very inaccurate and are thus of limited value in analysing population structure.
All microsatellites were polymorphic in the investigated areas, with allele numbers ranging from four to 14 (Table 4). Micro-Checker gave no evidence of scoring errors, large allele dropout or null alleles in any locus/population combinations except for BM4208 in MGR where a homozygote excess was indicated. Neither of the areas showed any deviations from HWE after sequential Bonferroni corrections. Overall, the level of genetic variation was high in all sampled areas with the mean number of alleles across loci ranging from 6.0 in NCA to 7.9 in SNP and MGR (Table 2). The mean expected heterozygosity (H E) ranged from 0.68 in NCA to 0.71 and 0.73 in SNP and MGR, respectively.
The number of private alleles ranged from four in NCA to 14 in SNP and 17 in MGR. No single individual had a pronounced number of private alleles (three at most), nor did any markers (Table 4). Among a total of 142 observed alleles, 78 were observed in all three samples and 82 alleles had frequencies >0.05 when pooled across the three samples. Both the average number of alleles, allelic richness, and heterozygosity were similar in SNP and MGR but relatively lower in NCA (Table 2). A Student t-test revealed that the mean number of alleles was significantly lower in NCA than in MGR (t = 2.160, P = 0.040). A similar trend was also seen when NCA was compared with SNP, although the difference was not significant (t = 1.63, P = 0.114). Furthermore, the mean number of alleles and allelic richness were both lower in NCA when compared with SNP and MGR pooled (t = 2.917, P = 0.007; t = 3.335, P = 0.002). Similarly, a χ2-test of the total numbers of heterozygote’s and homozygote’s across all loci revealed a significant reduction of genetic diversity in NCA compared with the pooled SNP and MGR (χ2 = 4.496, P = 0.034).
Buffaloes from the three conservation areas were genetically differentiated according to Fisher’s exact tests (P < 0.0001), with ten out of 15 loci showing significant differentiation. Analysis of each sample pair across loci revealed that NCA was significantly different from both SNP and MGR (P < 0.001), while the differences between SNP and MGR were insignificant (P = 0.197). The lower level of genetic differentiation between SNP and MGR was further illustrated by their substantially lower pair-wise F ST value (θ = 0.005) compared to that between NCA and each of the two others (SNP: θ = 0.039; MGR θ = 0.035) (Table 3). A similar trend was detected for D est, with values of 0.069 and 0.042 for NCA-SNP and NCA-MGR compared to a value of 0.004 between SNP and MGR. No signals of recent bottlenecks were detected in any of the populations with the use of neither 20 nor 10% variation from the infinity model.
The STRUCTURE analyses provided support for the genetic differentiation indicated by the Fisher exact tests. With prior information on sampling locations (LOCPRIOR) there was a significant increase in mean likelihood when the number of genetic clusters was increased from one to two, after which no further subdivision was indicated (Fig. 2a). The Bayesian assignment revealed for all runs with K = 2 that all NCA individuals assign to one cluster while all SNP and MGR individuals assigned to the other, suggesting a low level of admixture (Fig. 2b). An r value of 0.1 suggests that information about sampling locations contributed substantially in inferring the dichotomy population structure. Without using the sample location information the STRUCTURE analyses did not uncover any significant genetic structure. Use of POPINFO with K = 2 indicated that no first-generation dispersers were among the sampled individuals, (q > 0.8 for all individuals, data not shown).
Bayesian assignment of African buffaloes in the Serengeti Ecosystem. a gives mean posterior probability (Ln Pr(X|K)) and standard deviation (error bars) over 10 runs for each of the K genetic clusters (K ∈ [1, 6]). b gives individual probability of assignment to each of two clusters (K = 2, different colors) among 68 buffalo sampled in the MGR, SNP and NCA
The most likely scenario according to the DIY ABC posterior probabilities was a demographic model with previous population reduction (tm2), subsequent expansion (td1) and a very recent split between the NCA and the pooled SNP/MGR (ta, scenario 4, Fig. 3, Table S1). In this scenario the divergence and founding of NCA was as recent as around 30 generations ago, while the posterior estimate of previous population reduction and subsequent expansion were around 6,400 and 1,600 generations old, respectively. The results were quite similar regardless of using a Ne of 30 or 10% of census population size (Fig. 3 and Fig. S1 (supplementary), respectively). The other less likely scenarios involved either a relatively recent divergence time between NCA and SNP/MGR (scenario 1, Fig. 3) or an origin of the NCA from an even more recent admixture event with an unsampled population (scenario 2, Fig. S1).
Scenarios explored with DIY ABC to explain observed genetic structure in the Serengeti buffalo population. Pop 1 consist of SNP & MGA (n = 49) and pop 2 is NCA (n = 19). Applied present effective population sizes are 30% of estimated census sizes. Posterior probabilities of each scenario (PP (95% quartile)) after logistic regression on the 1% simulated data most similar to the observed data, and median (95% CI) estimated time in numbers of generations since divergence between NCA and SNP/MGA (t1), and admixture (ta) and divergence (t2) with an unsampled population outside the study area
Discussion
The buffalo has been reported to harbour high levels of nucleotide diversity compared to the other species of large mammals in Africa (Simonsen et al. 1998). Accordingly, both our mtDNA and microsatellite data suggest a high level of within population variation similar to that reported for buffaloes across even larger areas in Africa (Simonsen et al. 1998; Van Hooft et al. 2002; Heller et al. 2008). However, to obtain accurate estimates of the allele frequencies of highly variable markers may be challenging with regard to sample size, particularly within protected areas where all handling of wildlife initially is forbidden. This is exemplified by the 28 observed haplotypes of mtDNA which are distributed across only 68 samples in this study, limiting the accuracy of estimates of frequencies and genetic differentiation. The relatively high numbers of private microsatellite alleles within the sampled populations probably reflect a similar effect. However, most microsatellite alleles were present in all three samples and had frequencies higher than 0.05 across the material, making the present analyses of genetic structure reasonably appropriate and reliable.
In this study, both the applied mtDNA and microsatellites demonstrate genetic differentiation and structure between buffalo in the different management areas of the Serengeti ecosystem, with the NCA being significantly differentiated from the SNP and MGR. The observed structure, particularly for the microsatellites, is not very strong, as illustrated by the absence of any clear structure in the STRUCTURE analyses without priors on sampling locations, although this may have been influenced by the relatively low sample sizes and the highly variable markers. However, the lower level of genetic variation in NCA, the F ST statistics, Dest estimator and the STRUCTURE analyses with priors on sampling locations suggested the same dichotomy in genetic structure between buffalo in the Serengeti. The differentiation between the NCA and the two others sampling areas appear substantial higher for mtDNA (F ST = 0.152) than for microsatellites (F ST = 0.039). On one hand, mtDNA is haploid and maternally inherited, it is thus effectively a quarter of the population size compared to diploid nuclear DNA, and consequently more sensitive to demographic events like bottlenecks and genetic drift. However, if dispersal has been male biased, the reduced gene flow we report may also have contributed to the relatively higher mtDNA differentiation.
We did not detect any signals of recent bottlenecks despite records of catastrophic droughts and disease epidemics that resulted in population crashes of the African buffalo in many areas in the late 1800s (Sinclair 1977; O’Ryan et al. 1998; Simonsen et al. 1998; Wenink et al. 1998; Van Hooft et al. 2000). Similar results have been reported for the buffalo in both the nearby Masai Mara population (Kenya) and the Queen Elizabeth population (Uganda) in a microsatellite study by Heller et al. (2008), who suggested that the late 1800s Rinderpest plague had little effect on the genetic variation and structure of African buffalo. Overall, many species in this region display absence of genetic signatures from recent population bottlenecks, as in the black rhinoceros, Diceros bicornis (Harley et al. 2005), waterbuck, Kobus ellipsiprymnus (Lorenzen et al. 2006a), impala, Aepycerus melampus (Lorenzen et al. 2006b) and elephant, Loxodonta africana (Okello et al. 2008). Generally, a reduction in effective population size following a bottleneck is correlated with a decrease in allelic diversity and later on heterozygosity (Wright 1931), a phenomenon we did not observe in our study. Rather, the general level of genetic variation in the Serengeti buffalo was high, which may indicate that the population has been large historically. However, the African buffalo is suggested to be capable of maintaining non-critical population sizes, high growth rates and good dispersal capabilities, enabling re-colonisation of available habitat and exchange of genetic material across large distances following adverse conditions (Heller et al. 2010). This could explain why no signals of previous bottlenecks or any old genetic structure have been detected.
Our DIY ABC analyses thus suggests that the most likely scenario to explain the genetic structure of Buffalo in the Serengeti Ecosystem is a demographic model with population decline and subsequent expansion before a very recent divergence of the NCA from SNP/MGA (scenario 4 in Fig 3). The recent estimates of divergence or admixture events explaining the origin of the NCA in the other explored scenarios supports this, suggesting that the lower level of genetic variation in the NCA population stems from a founder event rather than an old bottleneck event. The estimated onsets of a 90% decline around 6,400 generations ago and a subsequent tenfold expansion at about 1,600 generations ago (Fig. 3), are in agreement with the previous results of Heller et al. (2008), suggesting a 75–98% reduction 3–7,000 years ago during mid Holocene. However, our estimates could be biased by sample size. Moreover, the DIY ABC algorithm assumes no migration between the events of scenarios (Cornuet et al. 2008). Even though the STRUCTURE analyses with POPINFO support such an assumption, the sample sizes may be too low to detect first-generation dispersers if dispersal rates are low. A reduction of genetic structure through gene flow and introduction of new gene copies may thus have introduced a bias on the divergence estimates, potentially involving estimates that are closer to present than actual divergence time. Nonetheless, the genetic differentiation of NCA is real and most probably of a recent origin. Considering the lack of genetic signatures also in many of the other regional species (Harley et al. 2005; Lorenzen et al. 2006a, b; Okello et al. 2008), this may suggest that it is not until now that fragmentation and isolation have become grave enough to have a genetic impact in the region. This highlights how rapidly fragmentation can lead to isolation and genetic differentiation, even in such a highly genetically variable and numerous key species as the buffalo, illustrating the real risk of fragmentation in the region.
Much has changed over the last decades in NCA, including large increases in the human populations of both Maasai pastoralists and other human immigrants with interests in agriculture (Kijazi et al. 1997; Galvin et al. 2004; Boone et al. 2006). Diseases and drought, among other factors, have resulted in decreasing livestock, which are the sole reliant source of food and income to pastoralists (NCAA 1999; Lynn 2000; Fyumagwa et al. 2007). Therefore, land-use has become intensified in recent years as the Maasai pastoralists are forced to become more sedentary and practice agriculture in response to food scarcity (McCabe et al. 1997b; Neumann 1998; Boone et al. 2006). Unfortunately, much of the present agriculture is taking place on fertile areas that otherwise would have provided a good habitat for buffaloes and that could have represented a corridor important for genetic exchange between the NCA and other Serengeti management areas (Fig. 1c). The relatively recent separation of the buffaloes in NCA from SNP and MGR could therefore be attributed to human settlements, infrastructure and agriculture around the Crater highlands potentially blocking natural dispersal corridors. Besides anthropogenic factors, Estes et al. (2006) outlined that also the recent climatic changes may have influenced wildlife in the area, and Van Hooft et al. (2000) stated that migration among buffalo populations in East Africa could be limited by the distribution of dry habitats. The progressively drier environment to the eastern terminus of the Serengeti plains may thus have made this area unsuitable for large wild ungulates like the buffaloes and could be acting as a barrier against their dispersal.
In small populations, harvesting may cause significant genetic drift as age structures are altered, sex-ratios skewed and effective sizes decreased, but to assess these effects requires a correct assessment of population genetic structure, which acts as an enforcing factor (Frankham 1996; Coltman 2008). Harvest may in MGR have involved genetic drift and differentiation, as compared to the protected SNP. Absence of any genetic change in MGR, despite its harvesting regime probably reflects a large effective population size. Moreover, the hunting regime in the area is trophy based and targets mainly reproductively senescent individuals that already have undergone most of their successful reproduction with subsequently small effects on effective population sizes. There are no distinct physical barriers between MGR and SNP, except for a small seasonal river in the southern part of the borderline. This, together with the high ability of African buffalo to migrate or switch between herds and populations (Estes 1991; Prins 1996; Halley et al. 2002; Cross et al. 2005; Korte 2009) may explain why MGR and SNP appear as one large population in which genetic effects of hunting are of minor importance. The present study points therefore towards the importance of maintaining buffaloes in large populations or in populations with extensive gene flow to maintain historical levels of genetic diversity, a conservation effort which may become important also in other African savannah ungulates.
Conclusions and conservation implications
We have shown that within the Serengeti ecosystem, buffalo in the NCA are genetically different from the buffalo in the SNP and MGR management areas, and that the differentiation probably is of recent origin. This may indicate that recently onset factors in this area are restricting the gene flow between NCA and the other parts of the Serengeti Ecosystem. It also demonstrates and highlights the importance of genetic studies in actual conservation planning and management. Although African buffalo are highly capable of maintaining non-critical population sizes, dispersal and re-colonization of different habitats, we believe that this may not be the case when anthropogenic developments reach the extent of blocking and depleting potential dispersal corridors and habitats between protected areas. We therefore acknowledge the need for, and encourage, the facilitation of gene flow between protected areas to prevent further genetic differentiation.
In essence, both humans and buffaloes compete for the same land. While humans need land for agriculture and other developments, buffaloes are an indigenous part of the original habitat. However, Tanzania as well as other countries in the region is facing a rapid human population growth and with the anticipated associated increases in anthropogenic activities, wildlife habitat exploitation and fragmentation, both the buffalo and other wildlife may soon be threatened in the region. The potential effects are illustrated by this paper, and as further fragmentation may be critical, this problem will obviously pose a conservation challenge in the future.
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Amos W, Balmford A (2001) When does conservation genetics matter? Heredity 87:257–265
Bishop MD, Kappes SM, Keele JW, Stone RT, Sunden SLF, Hawkins GA, Toldo SS, Fries R, Grosz MD, Yoo L, Beattie CW (1994) A genetic linkage map for cattle. Genetics 136:619–639
Boone RB, Galvin KA, Thornton PK, Swift DM, Coughenour MB (2006) Cultivation and conservation in Ngorongoro conservation area, Tanzania. Hum Ecol 34:809–828
Buchanan FC, Galloway SM, Crawford AM (1993) Ovine microsatellites at the OarFCB11, OarFCB128, OarFCB193, OarFCB266 and OarFCB304 loci. Anim Genet 25:60
Chakraborty R, Jin L (1993) A unified approach to study hypervariable polymorphisms; statistical considerations of determining relatedness and population distance. EXS 67:153–175
Chantal S, Tim C, Simon M, Charles M, George S, Markus B (2007) Assessment of effectiveness of protection strategies in Tanzania based on a decade of survey data for large herbivores. Conserv Biol 21:635–646
Coltman DW (2008) Molecular ecological approaches to studying the evolutionary impact of selective harvesting in wildlife. Mol Ecol 17:221–235
Conover MR (1997) Monetary and intangible valuation of deer in the United States. Wildl Soc Bull 25:298–305
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cornuet JM, Santos F, Beaumont MA et al (2008) Inferring population history with DIY ABC: a user-friendly approach to approximate Bayesian computation. Bioinformatics 24:2713–2719
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557
Crawford AM, Dodds KG, Ede AJ, Pierson CA, Montgomery GW, Garmonsway HG, Beattie AE, Davies K, Maddox JF, Kappes SM, Stone RT, Nguyen TC, Penty JM, Lord EA, Broom JE, Buitkamp J, Schwaiger W, Epplen JT, Matthew P, Matthews ME, Hulme DJ, Beh KJ, McGraw RA, Beattie CW (1995) An autosomal genetic linkage map of the sheep genome. Genetics 140:703–724
Cross PCL, Lloyd JI, Getz WM (2005) Disentangling association patterns in fission-fusion societies using African buffalo as an example. Anim Behav 69:499–506
DeYoung RW, Honeycutt RL (2005) The molecular toolbox: genetic techniques in wildlife ecology and management. J Wildl Manag 69:1362–1384
Di Rienzo A, Peterson AC, Garza JC (1994) Mutational processes of simple-sequence repeat loci in human population. Proc Natl Acad Sci USA 91:3166–3170
El Mousadik A, Petit RJ (1996) High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L. Skeels)] endemic to Morocco. Theor Appl Genet 92:832–839
Estes RD (1991) The behaviour guide to African mammals. The University of California Press, Berkeley
Estes RD, Atwood JL, Estes AB (2006) Downward trends in Ngorongoro Crater ungulate populations 1986–2005: conservation concerns and the need for ecological research. Biol Conserv 131:106–120
Estoup A, Jarne PJ, Cornuet JM (2002) Homoplasy and mutation model at microsatellite loci and their consequences for population genetic analysis. Mol Ecol 11:1591–1604
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Laval G, Schneider S (2005) Arlequin 3.01: an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Filatov DA (2002) Proseq: a Software for preparation and evolutionary analysis of DNA sequence data sets. Mol Ecol Notes 2:621–624
Flagstad Ø, Syvertsen PO, Stenseth NC, Stacy JD, Olsaker I, Røed KH, Jakobsen KS (2000) Genetic variability in Swayne’s Hartebeest, an endangered antelope of Ethiopia. Conserv Biol 14:254–264
Frankham R (1995) Effective population size/adult population size ratios in wildlife: a review. Genet Res Camb 66:95–107
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Fu YX, Chakraborty R (1998) Simultaneous estimation of all the parameters of a stepwise mutation model. Genetics 150:487–497
Fynn RWS, Bonyongo MC (2010) Functional conservation areas and the future of Africa’s wildlife. Afr J Ecol 49:175–188
Fyumagwa RD, Runyoro V, Horak IG, Hoare R (2007) Ecology and control of ticks as disease vectors in wildlife in Ngorongoro Crater, Tanzania. S Afr J Wildl Res 37:79–90
Gaggiotti OE, Lange O, Rassmann K, Gliddon C (1999) A comparison of two indirect methods for estimating average levels of gene flow using microsatellite data. Mol Ecol 8:1513–1520
Galvin KA, Thornton PK, Boone RB, Sunderland (2004) Climate variability and impacts on east African livestock herders: the Maasai of Ngorongoro conservation area, Tanzania. Afr J Range Forage Sci 21:183–189
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
George AR (1975) Buffalo increase and seasonal use of Ngorongoro Crater. E Afr Wildl J 13:385–387
Goldstein DB, Ruiz-Linares A, Cavalli-Sforza LL, Feldman MW (1995) An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices. Release 2.9.3.2. Available via http://www.unil.ch/izea/softwares/fstat.html. Accessed on 2009
Grant PR, Grant BR (1992) Demography and the genetically effective sizes of two populations of Darwin finches. Ecology 73:766–784
Halley DJ, Vandewalle MEJ, Mari M, Taolo C (2002) Herd switching and long-distance dispersal in female African buffalo (Syncerus caffer). Afr J Ecol 40:97–99
Harley EH, Baumgarten I, Cunningham J, O’Ryan C (2005) Genetic variation and population structure in remnant populations of black rhinoceros, Diceros bicornis in Africa. Mol Ecol 14:2981–2990
Heller R, Lorenzen ED, Okello JBA, Masembe C, Siegismund HR (2008) Mid-Holocene decline in African buffalos inferred from Bayesian coalescent-based analyses of microsatellites and mitochondrial DNA. Mol Ecol 17:4845–4858
Heller R, Okello JBA, Siegismund H (2010) Can small wildlife conservancies maintain genetically stable populations of large mammals? Evidence for increased genetic drift in geographically restricted populations of Cape buffalo in East Africa. Mol Ecol 19:1324–1330
Hilborn R, Arcese P, Borner M, Hando J, Hopcraft G, Loibooki M, Mduma S, Sinclair ARE (2006) Effective enforcement in a conservation area. Science 314:1266
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Iain JG, Alison JH, Marco F (2004) The management of wild large herbivores to meet economic, conservation and environmental objectives. J Appl Ecol 41:1021–1031
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kappes SM, Keele JW, Stone RT, McGraw RA, Sonstegard TS, Smith TPL, Lopez-Corrales NL, Beattie CW (1997) A second generation linkage map of the bovine genome. Genome Res 7:235–249
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Kemp SJ, Brezinsky L, Teale AJ (1993) A panel of bovine, ovine and caprine polymorphic microsatellites. Anim Genet 25:363–365
Kijazi A, Mkumbo S, Thompson DM (1997) Human and livestock population trends. In: Thompson DM (ed) Multiple land-use: the experience of the Ngorongoro conservation area, Tanzania. IUCN, Gland, pp 167–180
Kimura M, Crow JF (1964) The number of allele that can be maintained in a finite population. Genetics 49:725–738
Kimura M, Ohta T (1978) Stepwise mutation model and distribution of allelic frequencies in a finite population. Proc Natl Acad Sci USA 75:2868–3170
Korte L (2009) Herd-switching in adult female African forest buffalo (Syncerus caffer nanus). Afr J Ecol 47:125–127
Kurji F (1981) Human population trends within and around the Ngorongoro conservation area: the demographic settings, BRARUP Research Report, No. 44
Kurji F (1985) Population and conservation in the Serengeti-Part 1, Institute of Resource Assessment Research Paper, No. 9
Loibooki M, Hofer H, Campbell K, East LE (2002) Bush meat hunting by communities adjacent to the Serengeti National Park, Tanzania: the importance of livestock ownership and alternative sources of protein and income. Environ Conserv 29:391–398
Lorenzen ED, Simonsen BOT, Kat PW, Arctander P, Siegismund HR (2006a) Hybridization between subspecies of waterbuck (Kobus ellipsiprymnus) in zones of overlap with limited introgression. Mol Ecol 15:3787–3799
Lorenzen ED, Arctander P, Siegismund HR (2006b) Regional genetic structuring and evolutionary history of the impala Aepyceros melampus. J Hered 97:119–132
Lynn S (2000) The effects of conservation policy and ecology on pastoral land use patterns: a case study of Maasai land use in Northern Tanzania. MSc Thesis, Colorado State University, Fort Collins
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
McCabe JT, Mollel N, Tumaini A (1997) Food security and the role of cultivation. In: Thompson DM (ed) Multiple land-use: the experience of the Ngorongoro conservation area, Tanzania. IUCN, Gland, pp 397–416
McNaughton SJ, Campbell K (1991) Long-term ecological research in African ecosystems. In: Risser PG (ed) Long-term research. Wiley, Chichester, pp 173–189
Meertens HCC, Ndege LJ, Enserink HJ (1995) Dynamics in farming systems: changes in time and space in Sukumaland. Royal Tropical Institute, Arusha
Metzger KL, Sinclair ARE, Hilborn R, Hopcraft JGC (2010) Evaluating the protection of wildlife in parks: the case of African buffalo in Serengeti. Biodivers Conserv 19:3431–3444
NCAA (Ngorongoro Conservation Area Authority) (1999) 1998 Aerial boma count, 1999 people and livestock census, and human population trend between 1954 and 1999 in the NCA. Research and Planning Unit. Ngorongoro Conservation Area Authority, Ngorongoro Crater
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Neumann RP (1998) Imposing wilderness: struggles over livelihood and nature preservation in Africa. University of California, Berkeley
Newmark WD (2008) Isolation of African protected areas. Front Ecol Environ 6:321–328
O’Ryan C, Harley EH, Bruford MW, Beaumont M, Wayne RK, Cherry MI (1998) Microsatellite analysis of genetic diversity in fragmented South African buffalo populations. Anim Conserv 1:85–94
Ogutu ZA (2002) The impact of ecotourism on livelihood and natural resource management in Eselenkei, Amboseli ecosystem, Kenya. Land Degrad Dev 13:251–256
Okello JBA, Wittemyer G, Rasmussen HB, Arctander P, Nyakaana S, Douglas-Hamilton I, Siegismund HR (2008) Effective population size dynamics reveal impacts of historic climatic events and recent anthropogenic pressure in African elephants. Mol Ecol 17:3788–3799
Prins HHT (1996) Ecology and behaviour of the African buffalo. Social inequality and decision making. Champman and Hall, London
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ralph K, Karin EH, Otto H, Josef S, Bernhard S, Christoph S (2006) Genetic effect of transportation infrastructure on Roe Deer populations (Capreolus capreolus). J Hered 98:13–22
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rousset F (2008) GenePop’007: a complete re-implementation of the GenePop software for Windows and Linux. Mol Ecol Resour 8:103–106
Runyoro V, Hofer H, Chausi EB, Moehlman PD (1995) Long-term trends in the herbivore population of the Ngorongoro Crater, Tanzania. In: Sinclair ARE, Arcese P (eds) Serengeti II dynamics management and conservation of an ecosystem. University of Chicago Press, Chicago, pp 146–168
Sidney J (1965) The past and present distribution of some African ungulates. Trans Zool Soc Lond 30:1–397
Simonsen BT, Siegismund HR, Arctander P (1998) Population structure of African buffalo inferred from mtDNA sequences and microsatellite loci: high variation but low differentiation. Mol Ecol 7:225–237
Sinclair ARE (1977) In: George BS (ed) The African buffalo, a study of resource limitation of population. Chicago University Press, Chicago, p 355
Solinas TS, Fries R (1993) Physically mapped, cosmid-derived microsatellite markers as anchor loci on bovine chromosomes. Mamm Genome 4:720–727
Songorwa AN (2004) Human population increase and wildlife conservation in Tanzania: Are the wildlife managers addressing the problem or treating symptoms? AJEAM-RAGEE 9:49–77
Soulé ME, Mills S (1992) Conservation genetics and conservation biology: a troubled marriage. In: Sandlund OT, Hindar K, Brown AHD (eds) Conservation of biodiversity for sustainable development. Scandinavian University Press, Oslo, pp 55–69
Stuwart SN, Adams RJ, Jenkins MD (1990) Biodiversity in sub-Sahara Africa and its islands. IUCN-SSC No. 6 IUCN. University of Chicago Press, Glands
TAWIRI (2008) Aerial census in the Serengeti ecosystem, wet season report, TAWIRI aerial survey report
Thirgood S, Anna M, Sebastian T, Grant H, Ephraim M, Titus M, Morris K, John F, Sinclair ARE, Markus B (2004) Can parks protect migratory ungulates? The case of the Serengeti wildebeest. Anim Conserv 7:113–120
Van Hooft WF, Groen AF, Prins HHT (2000) Microsatellite analysis of genetic diversity in African buffalo (Syncerus caffer) populations throughout Africa. Mol Ecol 9:2017–2025
Van Hooft WF, Groen AF, Prins HHT (2002) Phylogeography of the African buffalo based on mitochondrial and y-chromosomal loci: pleistocene origin and population expansion of the Cape buffalo subspecies. Mol Ecol 11:267–279
Van Hooft WF, Groen AF, Prins HHT (2003) Genetic structure of African buffalo herds based on variation at the mitochondrial D-loop and autonomic microsatellite loci: evidence for male-biased gene flow. Conserv Genet 4:467–477
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wenink PW, Groen AF, Roelke-Parker ME, Prins HHT (1998) African buffalo maintaining high genetic diversity in the major histocompatibility complex in spite of historically known population bottlenecks. Mol Ecol 7:1315–1322
World Resources Institute (Cited as WRI) (1989) An assessment of the resource base that supports the global economy. Basic Books Inc., New York
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Acknowledgments
The SNP, MGR and NCA Managements Authorities are acknowledged for allowing sampling of buffaloes. COSTECH and TAWIRI are acknowledged for offering research permit. Thanks to the Norwegian Programme for Development, Research and Education (NUFU) for funding. Members of the Antelope conservation project are acknowledged for revising the manuscript. Liv Midthjell, Ellen Gunby and Margrethe Jacobsen are acknowledged for their assistance during laboratory work.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ernest, E.M., Haanes, H., Bitanyi, S. et al. Influence of habitat fragmentation on the genetic structure of large mammals: evidence for increased structuring of African buffalo (Syncerus caffer) within the Serengeti ecosystem. Conserv Genet 13, 381–391 (2012). https://doi.org/10.1007/s10592-011-0291-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0291-0