Abstract
The Cape Verde Islands harbour the second largest nesting aggregation of the globally endangered loggerhead sea turtle in the Atlantic. To characterize the unknown genetic structure, connectivity, and demographic history of this population, we sequenced a segment of the mitochondrial (mt) DNA control region (380 bp, n = 186) and genotyped 12 microsatellite loci (n = 128) in females nesting at three islands of Cape Verde. No genetic differentiation in either haplotype or allele frequencies was found among the islands (mtDNA F ST = 0.001, P > 0.02; nDNA F ST = 0.001, P > 0.126). However, population pairwise comparisons of the mtDNA data revealed significant differences between Cape Verde and all previously sequenced Atlantic and Mediterranean rookeries (F ST = 0.745; P < 0.000). Results of a mixed stock analysis of mtDNA data from 10 published oceanic feeding grounds showed that feeding grounds of the Madeira, Azores, and the Canary Islands, in the Atlantic Ocean, and Gimnesies, Pitiüses, and Andalusia, in the Mediterranean sea, are feeding grounds used by turtles born in Cape Verde, but that about 43% (±19%) of Cape Verde juveniles disperse to unknown areas. In a subset of samples (n = 145) we evaluated the utility of a longer segment (~760 bp) amplified by recently designed mtDNA control region primers for assessing the genetic structure of Atlantic loggerhead turtles. The analysis of the longer fragment revealed more variants overall than in the shorter segments. The genetic data presented here are likely to improve assignment and population genetic analyses, with significant conservation and research applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The highly migratory loggerhead (Caretta caretta) sea turtle is globally endangered (IUCN 2007), and occurs in temperate and tropical waters worldwide. Following hatching from nesting beaches, sea turtles enter the ocean. Relatively little is known about the location of post-hatchling and small pelagic juveniles during the subsequent “lost years”, where they may circulate in oceanic gyres and drift with currents (Bolten 2003). However, loggerhead turtles born at Florida (USA) rookeries are known to inhabit the pelagic zone of the North Atlantic gyre system for several years (Carr 1986). Next, an unknown proportion of juvenile turtles recruits to neritic habitats closer to their natal beaches, a behaviour known as juvenile natal homing (Bolten 2003; Bowen et al. 2004), while others continue to feed in the high seas (Musick and Limpus 1997). Both pelagic and neritic feeding grounds are usually “mixed stocks” drawn from various different rookeries (Bolten et al. 1998; Laurent et al. 1998; Bowen et al. 2004; Bass et al. 2004; Carreras et al. 2006). At sexual maturity, some adults switch from subadult to adult foraging habitats (Bolten 2003). Thereafter, individuals undergo breeding migrations between feeding and nesting habitats, with females generally exhibiting philopatry and a high degree of nesting site fidelity (Bolten 2003; Schroeder et al. 2003).
While North Atlantic and Mediterranean loggerhead turtles are a relatively well-studied systems in population genetics and connectivity research (Bolten et al. 1998; Encalada et al. 1998; Laurent et al. 1998; Bowen et al. 2004, 2005; Carreras et al. 2006, 2007; Lee 2008), key gaps in sampling still remain. The second largest nesting aggregation of the globally endangered loggerhead sea turtle (IUCN 2007) in the Atlantic Ocean is found at the Cape Verde Islands, which is also the only major rookery in Western Africa for this species. The archipelago is situated about 600 km west of Senegal in the eastern Atlantic, and is comprised of 10 volcanic islands and 5 islets (Fig. 1). The vast majority of nesting activity occurs on Boavista Island (90%) but females also nest on Sal, San Vicente, Santa Luzia, San Nicolau, and Maio Islands (Marco et al. 2008), and sporadically throughout the rest of the archipelago (López-Jurado, unpublished data).
The Cape Verde loggerhead turtle nesting aggregation faces serious conservation threats (Brongersma 1995; Ross 1995; López-Jurado and Andreu 1998; López-Jurado et al. 2000; Fretey 2001; López-Jurado and Liria 2007). These include substantial illegal harvest of eggs and adult females on the beaches, along with the loss of both males and females in the surrounding ocean waters through harvest or incidental capture in fisheries (López-Jurado et al. 2000; Marco et al. 2008). During 2007, it was estimated that about 1,150 females were killed or captured around the whole archipelago (Marco et al. 2008). Furthermore, males are captured due to a belief in the aphrodisiacal powers of their genitals. Other areas, linked to Cape Verde through migration and dispersal, are likely to be affected by these activities as well, underscoring the need to understand the connectivity of this rookery. These may include coastal areas from Mauritania to Sierra Leone, as well as the oceanic waters of Gambia, Guinea Konakry and Guinea Bissau, which satellite tracking studies have shown to constitute important feeding grounds for adult female loggerheads of Cape Verde (Hawkes et al. 2006).
In sea turtles, the contrast between mitochondrial (mtDNA) and nuclear (nDNA) results and among different life stages highlights the complex population structure of these organisms and the need to examine multiple genetic markers and life stages (Bowen et al. 2005). Samples from most major loggerhead turtle rookeries, albeit with significant exceptions such as the Cape Verde Islands, have been sequenced at the mitochondrial DNA control region (Encalada et al. 1998; Bowen et al. 2005; Carreras et al. 2007; Reis et al. in press), revealing significant genetic differentiation among most rookeries. More recent control region research identified five rookery groups in the Western Atlantic (Bowen et al. 2005). In the eastern Mediterranean, Carreras et al. (2007) found four independent units, most of them characterized by one exclusive haplotype. Nuclear DNA microsatellite analyses of both nesting and feeding areas have been carried out to a lesser extent, revealing lower levels of population differentiation. It is suggested that this may indicate male-mediated gene flow and/or mating during overlapping migrations (Bowen et al. 2005; Carreras et al. 2007).
Using molecular data, Bolker et al. (2007) introduced the “many-to-many” mixed stock analysis (MSA) method, which simultaneously estimates the origins and destinations of individuals in a metapopulation using Bayesian analysis. By considering the entire structure of the population, results can for the first time be given in a “rookery-centric” way, as the proportions of individuals from each rookery going to each foraging ground. The accuracy of this analysis is known to be limited by factors including incomplete sampling of potential sources, insufficient resolution of the markers used to infer the genetic structure, and insufficient levels of differentiation (Chapman 1996; Pella and Masuda 2001, 2005; Bolker et al. 2003, 2007). The mere size of Cape Verde rookery and its geographic location suggest that at least some of the pelagic Atlantic juvenile loggerhead turtles originally assigned to other sources in previous MSAs, could in fact be from this Macaronesian rookery (Bolten et al. 1998; Laurent et al. 1998; Carreras et al. 2006). Another potentially confounding factor in MSA is lack of resolution in the genetic data. For example, haplotypes that appear to be the same when only a small segment is sequenced may in fact turn out to be different when the analysis is expanded to a longer region. Abreu-Grobois et al. (2006) developed control region primers that expand the target area of formerly used primers (Norman et al. 1994) by an additional ~380 bp, and found that some haplotypes previously considered the same were in fact different, thus emphasizing the need for further investigation.
In this study, we analysed mtDNA control region sequences and nDNA microsatellite genotypes in adult females nesting at three islands of the Cape Verde archipelago. We used the “many-to-many” mixed stock analysis approach (Bolker et al. 2007) to reanalyze published data from oceanic feeding grounds for this species (Bolten et al. 1998; Carreras et al. 2006; Revelles et al. 2007), including the previously uncharacterized Cape Verde rookery. Finally, employing the newly designed mtDNA control region primers (Abreu-Grobois et al. 2006), we sequenced individuals from Cape Verde along with samples from Georgia, USA, two populations whose composition appeared to be dominated by the same CC-A1 mitochondrial haplotype. The objectives of this study were to: (1) characterize the population structure and demographic history of the loggerheads nesting in Cape Verde with mitochondrial and nuclear loci; (2) place the Cape Verde islands into the genetic context of Atlantic-Mediterranean nesting populations; (3) examine 10 oceanic feeding grounds in order to define important areas for recruitment to the Cape Verde nesting population; (4) evaluate whether mtDNA control region haplotypes of the Atlantic are informatively split into different subhaplotypes when examining the longer control region sequences; and (5) consider conservation applications.
Materials and methods
Sample collection
Tissue samples were collected during two nesting seasons between June and September (2004–2005) from Boavista in the Cape Verde Islands. In addition, samples were obtained from two other islands in Cape Verde, Sal and Santa Luzia, during the 2005 nesting season. Blood samples were collected from females nesting at Blackbeard Island, Georgia (USA) from June 04 to 11, 2001, and from June 24 to 28, 2002. All samples were stored in 96% ethanol or lysis buffer (Table 1).
Laboratory procedures
Genomic DNA was isolated from these samples using the DNeasy Tissue Kit (QIAGEN®) following manufacturer’s protocols or using phenol/chloroform extractions (Hillis et al. 1996).
Mitochondrial DNA
A 380 base pair (bp) fragment of the mtDNA control region was amplified by the polymerase chain reaction (PCR) in the totality of the samples (n = 203; Table 1) using the primers TCR5 (5′-TTGTACATCTACTTATTTACCAC-3′) and TCR6 (5′-GTACGTACAAGTAAAACTACCGTATGCC-3′) designed by Norman et al. (1994). We selected a subgroup of samples (total n = 145: Cape Verde, n = 128; and Georgia, n = 17; Table 1) with which to test the utility and applications of more recently designed primers that amplify a ~760 bp of the mtDNA control region (Abreu-Grobois et al. 2006). The LCM15382 (5′-GCTTAACCCTAAAGCATTGG-3′) and H950 (5′-GTCTCGGATTTAGGGGTTTG-3′) primers amplify a segment that completely encompasses the shorter region amplified by the TCR5/TCR6 primer pair (Norman et al. 1994), but leave out the highly repetitive area at the 3′ end of the d-loop (Abreu-Grobois et al. 2006).
For both primer pairs, PCR reactions were typically performed in 20–25 μl volumes under the following conditions: an initial denaturation step at 94°C for 2 min; followed by 40 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min; with a final extension at 72°C for 5 min. PCR products were purified using ExoSap (Amersham) or Agencourt AMPure (Beckman Coulter). Cycle sequencing reactions were conducted with Big Dye fluorescent dye-terminator (Applied Biosystems) and the fragments were analysed on an automated sequencer (Applied Biosystems Inc. model 3100 or 3730). Chromatograms were aligned using the Bioedit Sequence Alignment Editor v.7.0.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), or the program Sequencher v.3.1.2 (Gene Codes Corporation).
Nuclear DNA
Samples collected during the 2005 nesting season in Cape Verde (n = 128, Table 1) were analysed using twelve previously described microsatellite loci designed for loggerhead turtles (Cc2–Cc32; Monzón-Argüello et al. 2008). Each locus was PCR amplified using the protocol described in Monzón-Argüello et al. (2008). Fragment sizes were scored using an ABI 3100 automated sequencer with LIZ 500 (Applied Biosystems) used as an internal fluorescent size standard. The results were analysed using GENEMAPPER v.3.5 (Applied Biosystems).
Data analysis
Mitochondrial DNA: shorter segment (~380 bp)
Genetic diversity, differentiation and demographic history of Cape Verde
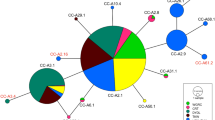
We classified mitochondrial sequence segments amplified by the TCR5/TCR6 primer pair (~380 bp) according to the standardized nomenclature of the Archie Carr Center for Sea Turtle Research (ACCSTR; http://accstr.ufl.edu/ccmtdna.html). Previously undescribed haplotypes were submitted to the ACCSTR to be assigned a name under the standardized nomenclature, and deposited in GenBank. Since the longer sequence contains the shorter segment amplified by primers TCR5/TCR6, we compared results from these shorter segments with those amplified by the new primers designed by Abreu-Grobois et al. (2006). We constructed statistical parsimony haplotype networks (Templeton et al. 1992; Posada and Crandall 2001) to depict patterns of genetic variation between the haplotypes using the software TCS v.1.21 (Clement et al. 2000).
Genetic differentiation between different seasons from the same location (Boavista 2004 and 2005), and between loggerheads nesting at different islands of Cape Verde, was quantified using the exact test of population differentiation (Raymond and Rousset 1995) and Phi statistics (φST) (Excoffier et al. 1992), with statistical significance obtained over 10,000 permutations. All computations were carried out by the program Arlequin v.3.0 (Excoffier et al. 2005).
Mismatch distributions based on pairwise nucleotide differences among DNA sequences were used to evaluate the demographic history of the Cape Verde rookery using Arlequin v.3.0 (Excoffier et al. 2005). The validity of the estimated mismatch parameters was tested using Arlequin’s sum of square deviations (SSD) test of goodness of fit, comparing observed and expected mismatch distributions (Schneider and Excoffier 1999). We calculated 95% confidence intervals for all parameters with 100 replicates using Arlequin’s parametric bootstrap approach.
Phylogeography of Cape Verde with respect to other Atlantic and Mediterranean loggerhead populations
Haplotype frequencies, haplotype diversity (h) and nucleotide diversity (π) of mtDNA sequences were estimated using Arlequin v.3.0 (Excoffier et al. 2005). Analyses used the Tamura–Nei model designed for control region sequences (Tamura and Nei 1993) to estimate sequence divergence. Genetic differentiation between this population and other Atlantic and Mediterranean rookeries was quantified in comparison to published sequences (Encalada et al. 1998; Bowen et al. 2005; Carreras et al. 2007; Reis et al. in press) using the exact test of population differentiation (Raymond and Rousset 1995) and φST (Excoffier et al. 1992), with statistical significance obtained over 10,000 permutations.
Mixed stock analysis of Atlantic and western Mediterranean loggerhead turtle feeding grounds
With the new data generated by this study, we were able to include all major sources of nesting colonies in the MSA by adding Cape Verde data to the source baseline of published rookery sequences (Encalada et al. 1998; Bowen et al. 2005; Carreras et al. 2007; Reis et al. in press). As noted previously, the absence of this major rookery in previous analyses could have affected their accuracy. Results of population differentiation tests were used to define the rookeries or source populations for the MSA. Fourteen nesting-area samples showed sufficient genetic differentiation to proceed as independent sources in the baseline (Table 2). We then reanalysed previously published data from oceanic feeding grounds in the Atlantic Ocean and Mediterranean Sea (Bolten et al. 1998; Carreras et al. 2006; Monzón-Argüello et al. 2009; Table 1).
Before carrying out the MSA, we used the exact test of population differentiation (Raymond and Rousset 1995) in the program Arlequin v.3.0 (Excoffier et al. 2005) to determine if these groups could be mixed stocks by comparing them to single rookeries. Then, a Bayesian MSA using the “many-to-many” approach (Bolker et al. 2007) was employed to calculate the proportions of individuals going from the Cape Verde rookery to each foraging area. After using the Markov chain Monte Carlo (MCMC) method to obtain the posterior distribution of the parameters of interest, the Gelman–Rubin diagnostic test was used to confirm convergence of the chains to the posterior distribution, with values less than 1.2 (Gelman and Rubin 1992). Prior information about relative rookery size was used in our Bayesian analysis, assuming that the overall contribution of a rookery is proportional to its size. Rookery sizes were taken from Ehrhart et al. (2003) and Margaritoulis et al. (2003) (Table 1). The effect of geographic distance on source contributions to each feeding ground was examined by regressing “many-to-many” MSA estimates on the shortest water distances (km) between Cape Verde and juvenile feeding areas (Table 1).
Nuclear DNA
Genetic diversity, differentiation and demographic history of Cape Verde
First, we used the software Micro-Checker (Van Oosterhout et al. 2004) to check for null alleles or scoring errors. We then calculated the number of alleles per locus (k), observed (H o) and expected (H e) heterozygosities, and polymorphic information content (PIC) with Cervus v.3.0 (Marshall et al. 1998). We tested for deviations from Hardy–Weinberg (HW) equilibrium via Markov chain permutation (Guo and Thompson 1992) using Arlequin v.3.0 (Excoffier et al. 2005). Sequential Bonferroni corrections were conducted using a global P value of 0.05 (Rice 1989). We tested for differentiation between islands using F ST and R ST values, as well as the exact test of population differentiation, with Markov chain permutations (Guo and Thompson 1992) as implemented by the Arlequin v.3.0 program (Excoffier et al. 2005).
Furthermore, population structure was assessed using the program STRUCTURE v.2.2 (Pritchard et al. 2000), which employs a Bayesian clustering method to identify the most likely number of populations (K) assuming no particular population structure a priori. We followed the search strategy described in Evanno et al. (2005), and 20 runs were carried out for each value of K (from 1 to 5). We set the length of the burn-in period to 10,000 and Markov Chain Monte Carlo (MCMC) to 100,000, as preliminary tests showed that the results did not change substantially with longer values. We employed the ad hoc statistic ∆K (Evanno et al. 2005) to detect the number of clusters in our sample.
We used the program Bottleneck v.1.2 (Cornuet and Luikart 1996) to test for recent bottlenecks using the two-phased model (TPM) with 90% stepwise and 10% infinite allele mutations, recommended for microsatellite markers (Luikart et al. 1998). Results were also evaluated under the stepwise mutation model (SMM).
Results
Mitochondrial DNA: shorter segment (~380 bp)
Genetic diversity, differentiation and demographic history of Cape Verde
Five haplotypes were revealed, including the previously undiscovered haplotype CC-A47 (GenBank accession number EU091309). The CC-A1 haplotype was found in the vast majority of the individuals (68% relative frequency), while other haplotypes and their frequencies were as follows: CC-A17 (28%), CC-A47 (2%), CC-A2 (1%) and CC-A11 (1%).
Temporal analysis comparing two consecutive nesting seasons (Boavista, 2004–2005; Table 1) revealed no significant differences (φST = −0.0198, P = 0.99; exact P = 1.00), and consequently the haplotypes from different years were pooled. The comparison of the three locations revealed no genetic structure after sequential Bonferroni correction, using both φST and the exact test (P > 0.02; exact P = 0.105). We therefore pooled all Cape Verde haplotypes for comparison to other rookeries. Furthermore, as discussed below, the mismatch distribution for the Cape Verde islands was incompatible with the model of a sudden demographic expansion (SSD P = 0.030), but exhibited a putative secondary expansion peak probably due to the occurrence of a very rare and distant haplotype (CC-A2).
Phylogeography of Cape Verde with respect to other Atlantic and Mediterranean loggerhead populations
Pairwise comparisons revealed significant differences between Cape Verde and all other Atlantic and Mediterranean nesting populations (Table 2). Haplotype diversity (h) in Cape Verde was among the highest values when compared with all rookeries; however nucleotide diversity (π) was relatively low (Table 1) because all of the Cape Verde haplotypes are closely related.
Mixed stock analysis of Atlantic and western Mediterranean loggerhead turtle feeding grounds
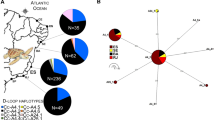
Significant differences were found between each of the Atlantic and Mediterranean feeding grounds and each nesting population. In addition, haplotypes from the foraging grounds were not found together at a single rookery but were instead typical of several widely separated rookeries. Together these findings reject the hypothesis of a single juvenile origin, and corroborate the hypothesis that they constitute mixed stocks. The “many-to-many” rookery-centric approach revealed that turtles born in Cape Verde distribute in Atlantic and Mediterranean waters (including Gimnesies, Madeira, Andalusia, Pitiüses, Azores and the Canary Islands); although point estimates had large standard deviations (SDs). However, a relatively high percentage of turtles nesting at Cape Verde goes to unknown oceanic foraging area/s (43%; Fig. 2). Linear regression revealed no significant relationship between MSA estimates and geographic distance (R 2 = 0.158, F = 1.50, P = 0.255).
Mitochondrial DNA: longer segment (~760 bp)
Ten mtDNA control region haplotypes were found at the Cape Verde rookery through analysis of the longer control region segment (Table 3; Fig. 3). Haplotypes CC-A1.5, CC-A11.2, CC-A17.1, CC-A17.2 and CCA-47.1 were previously undescribed (GenBank accession numbers EU483081, FJ817091, EU403082, EU483083 and EU483084, respectively). The common CC-A1 haplotype split into four different sub-haplotypes (CC-A1.1–CC-A1.5). Interestingly, only one of these sub-haplotypes was found in the samples from Georgia (USA) where it was fixed, and this sequence differed from the three CC-A1 sub-haplotypes found in the Cape Verde population (Fig. 3).
Statistical parsimony network of the ~760 bp sequences. Haplotype colours denote their geographic locations. The size of the haplotypes is approximately proportional to their frequency in the overall sample set, except CC-A1.3, which is drawn to the half of proportional size due to space constraints. Parentheses show the actual number of individuals carrying each haplotype
The level of differentiation between Cape Verde and Georgia was more pronounced in the analysis of the longer than shorter segments, because these two populations no longer shared the common CC-A1 subhaplotype (φST(shorter) = 0.161 vs. φST(longer) = 0.667, P < 0.05). The comparison of the longer sequences with the traditionally used shorter ~380 bp fragments revealed additional informative polymorphic sites, with the highest levels of overall nucleotide diversity found outside of the shorter segments (Fig. 4). The comparison of the three Cape Verde locations based on the longer sequences did not show genetic structure after sequential Bonferroni corrections, using both φST and the global exact test of differentiation among samples (P > 0.072; exact P = 0.024, respectively). Interestingly however, the exact test of population differentiation for the pairwise comparison between Sal and Santa Luzia islands showed significant differentiation (exact P = 0.003).
Nuclear DNA
Genetic diversity, differentiation and demographic history of Cape Verde
All of the loci were highly polymorphic, and overall allele numbers ranged from 3 (Cc32) to 15 (Cc13 and Cc21), with an average of 8.83 alleles per locus (Table 4). The mean number of alleles (k), H o, H e and PIC for each locus is shown in Table 4. Departures from HW equilibrium were not detected for any loci after sequential Bonferroni correction, with the exception of locus Cc30 (χ2 test, P = 0.000). The MICRO-CHECKER software (Van Oosterhout et al. 2004) indicated the possible presence of null alleles and/or stuttering at this locus, which could explain the deviation from HW equilibrium. Cc30 was therefore removed from the analysis. We assumed independence of loci since no linkage disequilibrium was found between marker pairs after sequential Bonferroni corrections (χ2 test, P > 0.01).
Pairwise comparisons between the Boavista, Sal and Santa Luzia islands revealed no significant genetic differentiation after sequential Bonferroni corrections (exact P = 0.292; Table 5). Without using prior information on sampling location, the ∆K ad hoc statistic (Evanno et al. 2005) revealed the most probable number of populations at this rookery (K = 3), although the increment in the mean ln likelihood was very low (K = 1, mean ln likelihood = −4200.83, SD = 0.11; K = 2, mean ln likelihood = −4180.42, SD = 0.95; K = 3, mean ln likelihood = −4160.98, SD = 3.18; Fig. S1 in supplemental information available online). However, the three clusters did not correspond to islands or other evident geographic structures, and the software found each individual could be assigned to each of the three clusters with approximately equal probability, thus failing to conclusively carry out assignment (see Fig. S2 in supplemental information available online). The Wilcoxon test for bottlenecks did not reveal significant heterozygote excess with respect to allele number under either model of evolution. However, the test indicated a heterozygote deficiency consistent with population expansion, as discussed further below (TMP P = 0.008; SMM P = 0.002). Only the stepwise mutation model (SMM) result was significant after sequential Bonferroni corrections.
Discussion
Population structure of loggerheads nesting in Cape Verde
The shorter mtDNA fragment revealed that females nesting on different islands of Cape Verde constitute a panmictic population. In our study, the two most distant islands are separated by more than two hundred kilometres. Similarly, the nesting population from northeast Florida to North Carolina was found to be genetically homogeneous over about 1,000 km (Bowen and Karl 2007). In contrast, loggerhead nesting populations in south and northeast Florida separated on a scale of 50–100 km are distinct in their mtDNA haplotype frequencies, showing that females are capable of homing on a relatively fine scale (Bowen and Karl 2007). Similar results were obtained in the Japanese islands, where loggerhead nesting colonies separated on a scale of about 400 km were found to be genetically distinct (Hatase et al. 2002). Similarly, analysis of the new ~760 bp sequence within the Cape Verde archipelago did not detect any statistically significant genetic differentiation in any of the global tests performed.
Comparisons using nDNA markers corroborated the global mtDNA results by revealing no significant population structure among islands in Cape Verde. Further, no consistent differentiation was found using Bayesian clustering analysis. Bowen et al. (2005) obtained similar results when comparing rookeries from the eastern coast of USA using microsatellite loci. Since we found no significant population structure within the Cape Verde archipelago when using mtDNA and nDNA markers, we can conclude that Cape Verde is one panmictic population composed of several rookeries.
However, the exact test of population differentiation for two of these islands (Sal and Santa Luzia) yielded significant levels of differentiation when only the longer fragments were analysed. There are two rare haplotypes unique to Sal (CC-A2.1 and CC-A11.2), and the former is indeed very distant from all of the other haplotypes. Furthermore, another rare haplotype (CC-A1.5) present in Boavista and Santa Luzia is absent in Sal. Finally, the second most common haplotype, formerly described as CC-A17, and now split into two subhaplotypes with the analysis of the longer sequence, presents a very different frequency distribution. The first of these haplotypes (CC-A17.1) occurs in Boavista and Santa Luzia with a frequency up to three times higher than in Sal, while the second (CC-A17.2) occurs in Sal with a frequency three times higher than in the other two islands (Table 3).
There are two possible explanations for these results. The first is related to the sample sizes available in this study. Given the fact that the apparent differences between Sal and Santa Luzia are influenced by the presence or absence of rare haplotypes, one could argue that with a larger sample these differences would disappear. The second explanation centers on the possibility of a stricter nest site fidelity of turtles from Sal. Such fine scale homing behaviour of sea turtles has been identified in several populations of the species elsewhere in the world (Bowen and Karl 2007). Stricter homing behaviour on Sal than on the other islands could be due to unknown natural or anthropogenic environmental conditions.
Demographic history of loggerheads nesting in Cape Verde
The demographic history analysis yielded somewhat conflicting results, but the differences are not necessarily contradictory since the two types of markers are likely to be influenced by demographic events acting on different time scales. Mismatch distribution analysis revealed no population expansion following a bottleneck or founding event. The minor peak in the mismatch distribution is likely due to the presence of the very divergent CC-A2 haplotype that appears in several distant rookeries. Consequently, it indicates occasional movements between those rookeries and the archipelago. Analysis of nuclear DNA data did not detect any recent bottleneck in this population. However, it did reveal a heterozygote excess suggestive of population expansion. Both results could be consistent if the population expansion occurred from larger ancestral populations or colonisation events. Microsatellite loci evolve even more rapidly than the mtDNA control region (Parsons et al. 1997; Schug et al. 1998), and this could explain the discrepancy between the stasis and expansion from mtDNA and nDNA results, respectively. Also, the finding of possible population expansion contrasts markedly with the high mortality rate due to ongoing harvest of this population, suggesting that nDNA could be reflecting episodes that occurred in the past, before the relatively recent major declines.
Phylogeography of Cape Verde with respect to other Atlantic and Mediterranean loggerhead populations
The significant differences between Cape Verde and other Atlantic and Mediterranean rookeries indicate that Cape Verde can be considered an independent management unit. A management unit (MU: sensu Moritz 1994) is typically characterized by significant divergence of allele frequencies at nuclear or mitochondrial loci, as well as differences in key demographic features (Moritz 1994; Bowen et al. 2005). As noted above, substantial illegal harvest of eggs and turtles occurs in Cape Verde, especially outside of the areas protected by conservation projects in the Boavista, Sal and Maio Islands (López-Jurado et al. 2000). Thus, the protection and conservation of this distinct population is a priority for the species.
Analysis of the longer sequences distinguished haplotypes previously thought to be the same including CCA-1, which differentiated into four sub-haplotypes. In addition, levels of population differentiation between the two rookeries (Cape Verde and Georgia) analysed with the longer mtDNA sequences were consistently higher than for the shorter segments, although expanding the sample size of the Georgia population would be recommended in future studies. In contrast to haplotype diversity, which increased due to the fact that more polymorphic sites were found, nucleotide diversity using the longer fragment did not increase likely because almost twice the number of nucleotides was screened. Similar results were obtained in hawksbill (Eretmochelys imbricata) and leatherback (Dermochelys coriacea) studies, where additional variable sites and the splitting of common previously lumped haplotypes were revealed using the longer segments (Abreu-Grobois et al. 2006; Velez-Zuazo et al. 2008; Vargas et al. 2008).
Dispersion of juveniles from the Cape Verde population
Caution is recommended when interpreting the results of this MSA, since point estimates had large SDs, and not all rookeries have been adequately sampled. Therefore, the value of this analysis in the present context is more to show the presence or absence of Cape Verde juveniles in the different Atlantic and Mediterranean foraging grounds, rather than the actual proportion of them in each area. The rookery-centric “many-to-many” MSA revealed that turtles born in Cape Verde distribute in both Atlantic (Madeira, Azores, and the Canary Islands) and Mediterranean (Gimnesies, Pitiüses, and Andalusia) feeding areas, although their abundance at each feeding ground is not determined by geographic distance (straight line distance) in this archipelago. However, straight line distance may not be an adequate measure of sea turtles movements (Naro-Maciel et al. 2007). For example, Carreras et al. (2006) found that genetic structuring in the Mediterranean could be explained by the pattern of sea surface currents and water masses, where the foraging grounds off the North-African coast and the Gimnesies Islands are shown to be inhabited mainly by turtles from Western Atlantic nesting stocks. Our results corroborate this hypothesis, as juveniles from Cape Verde distribute in the Mediterranean exclusively in the feeding grounds surrounded by Atlantic currents (Gimnesies, Pitiüses, and Andalusia).
The high and unambiguous percentage of juveniles going to unknown areas reveals the need to investigate additional oceanic areas, for example in the western and south Atlantic (e.g. Caribbean region, Venezuela, southwestern Africa, central south Atlantic, and east coast of South America). Further, due to recent findings of temporal variation in the genetic composition of feeding grounds (Bjorndal and Bolten 2008), additional long term studies of these areas is needed. Finally, it is important to consider that juveniles going to these unknown areas could be the ones that do not survive to join the foraging groups (juveniles that are incidentally killed or harvested), and are lost to the metapopulation.
We cannot use the longer segment at this time to determine the relationships to other rookeries, or the origins of turtles in the pelagic in-water groups, due to insufficient sequencing of the longer segments at the source rookeries. The analysis of other loggerhead nesting populations as well as feeding areas with the longer fragment will allow for assessment of the advantages of splitting common haplotypes, potentially contributing to more accurate MSAs and estimates of population genetic variation and differentiation. However, the increased number of orphan or rare haplotypes in future analysis may also indicate the need for larger sample sizes for accurate analysis.
Conservation applications
There are several conservation implications of our study. First, as noted above, Cape Verde can be considered as one management unit. In 2008, the Cape Verde government developed a conservation management plan to protect and conserve sea turtles there, the Plano Nacional para a Conservação das Tartarugas Marinhas em Cabo Verde. The results of this study indicates that management activities included in the plan, such as the translocation of eggs from Boavista to other islands of this archipelago, where nesting activity has decreased or even ceased due to human predation, may help to ensure the long-term viability of this population without altering its genetic structure. However, ultimately a long-term solution to the conservation threats at Cape Verde that includes intensive sea turtle protection and community work is needed.
Second, the mitochondrial evidence shows the isolation of Cape Verde from all other Atlantic and Mediterranean nesting populations and indicates that conservation of Cape Verde, where 90% of nesting activity occurs on the east coast of Boavista Island, is a priority for the species. Currently, the Cape Verde loggerhead nesting aggregation is the only major rookery in the eastern Atlantic (Fretey 2001). In light of serious threats to this rookery, we recommend increased focus on its conservation.
To enhance conservation, we also recommend additional research, particularly expanding the genetic analysis throughout Macaronesia and the west coast of Africa to include unsampled areas. For example, although occasional nesting activity has been reported at other African locations such as Mauritania or Senegal (Fretey 2001), the connectivity between these nesting populations and the Cape Verde rookery remains unknown and should be investigated. In addition, for comprehensive threat assessment, further studies of oceanic areas are needed in order to identify high seas feeding grounds or possible threats from areas connected to Cape Verde through migration. Finally, we recommend that genetic monitoring be continuously conducted in order to assess the effectiveness of the conservation program. At the very least, routine annual sampling should be implemented as a standard procedure so as to build up a comprehensive time-series of samples. The long-term gathering of ecological data and DNA samples will safeguard future scientific studies of the rookery by ensuring a solid basis on which to implement new methods of analysis in the future. This will provide a means of evaluating the genetic consequences of current management activities and other human impacts.
References
Abreu-Grobois FA, Horrocks J, Formia A, LeRoux R, Velez-Zuazo X, Dutton P, Soares L, Meylan P, Browne D (2006) New mtDNA Dloop primers which work for a variety of marine turtle species may increase the resolution capacity of mixed stock analysis. In: Frick M, Panagopoulou A, Rees AF, Williams K (eds) Book of abstracts of the 26th annual symposium on sea turtle biology and conservation, Crete, Greece, 3–8 April 2006, p 179
Bass AL, Epperly SP, Braun-McNeill J (2004) Multi-year analysis of stock composition of a loggerhead turtle (Caretta caretta) foraging habitat using maximum likelihood and Bayesian methods. Conserv Genet 5:783–796
Bjorndal KA, Bolten AB (2008) Annual variation in source contributions to a mixed stock: implications for quantifying connectivity. Mol Ecol 17:2185–2193
Bolker BM, Okuyama T, Bjorndal KA, Bolten AB (2003) Sea turtle stock estimation using genetic markers: accounting for sampling error of rare genotypes. Ecol Appl 13:763–775
Bolker BM, Okuyama T, Bjorndal KA, Bolten AB (2007) Incorporating multiple mixed stocks in mixed stock analysis: “many -to-many” analyses. Mol Ecol 16:685–695
Bolten AB (2003) Active swimmers-passive drifters: the oceanic juvenile stage of loggerheads in the Atlantic system. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution Press, Washington, DC, pp 63–78
Bolten AB, Bjorndal KA, Martins HR, Dellinger T, Biscoito MJ, Encalada SE, Bowen BW (1998) Transatlantic developmental migrations of loggerhead sea turtles demonstrated by mtDNA sequence analysis. Ecol Appl 8:1–7
Bowen BW, Karl SA (2007) Population genetics and phylogeography of sea turtles. Mol Ecol 16(23):4886–4907
Bowen BW, Bass AL, Chow S-M, Bostrom M, Bjorndal KA, Bolten AB, Okuyama T, Bolker BM, Epperly S, Lacasella E, Shaver D, Dodd M, Hopkins-Murphy SR, Musick JA, Swingle M, Rankin-Baransky K, Teas W, Witzell WN, Dutton PH (2004) Natal homing in juvenile loggerhead turtles (Caretta caretta). Mol Ecol 13:3797–3808
Bowen BW, Bass AL, Soares L, Toonen J (2005) Conservation implications of complex population structure: lessons from the loggerhead turtle (Caretta caretta). Mol Ecol 14:2389–2402
Brongersma LD (1995) Marine turtles of the eastern Atlantic Ocean. In: Bjorndal KA (ed) Biology and conservation of sea turtles. Smithsonian Institution Press, Washington, DC, pp 407–416
Carr A (1986) New perspectives on the pelagic stage of sea turtle development. NOAA Technical Memorandum NMFS-SEFC-190. U. S. Department of Commerce, p 36
Carreras C, Pon S, Maffucci F, Pascual M, Barceló A, Bentivegna F, Cardona L, Alegre F, SanFélix M, Fernández G, Aguilar A (2006) Genetic structuring of immature loggerhead sea turtles (Caretta caretta) in the Mediterranean Sea reflects water circulation patterns. Mar Biol 149(5):1269–1279
Carreras C, Pascual M, Cardona L, Aguilar A, Margaritoulis D, Rees A, Turkozan O, Levy Y, Gasith A, Aureggi M, Khalil M (2007) The genetic structure of the loggerhead sea turtle (Caretta caretta) in the Mediterranean as revealed by nuclear and mitochondrial DNA and its conservation implications. Conserv Genet 8:761–775
Chapman RW (1996) A mixed stock analysis of the green turtle: the need for null hypotheses. In: Bowen BW, Witzell WN (eds) Proceedings of the international symposium on sea turtle conservation genetics. NOAA Technical Memorandum NMFS-SEFSC-396, pp 137–146
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Ehrhart LM, Bagley DA, Redfoot WE (2003) Loggerhead turtles in the Atlantic Ocean: geographic distribution, abundance, and population status. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution, Washington, DC, pp 157–174
Encalada SE, Bjorndal KA, Bolten AB, Zurita JC, Schroeder B, Possardt E, Sears CJ, Bowen BW (1998) Population structure of loggerhead turtle (Caretta caretta) nesting colonies in the Atlantic and Mediterranean as inferred from mitochondrial DNA control region sequences. Mar Biol 130:567–575
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Smouse P, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Laval LG, Schneider S (2005) Arlequin ver. 3: an integrated software package for population genetics data analysis. Evol Bioinf Online 1:47–50
Fretey J (2001) Biogéographie et conservation des tortues marines de la côte Atlantique de l’Afrique [Biogeography and conservation of marine turtles of the Atlantic Coast of Africa]. CMS Technical Series Publication No. 6. UNEP/CMS Secretariat, Bonn
Gelman A, Rubin DB (1992) Inference from iterative simulation using multiple sequences (with discussion). Stat Sci 7:457–511
Guo SW, Thompson EA (1992) A Monte Carlo method for combined segregation and linkage analysis. Am J Hum Genet 51:1111–1126
Hatase H, Kinoshita M, Bando T, Kamezaki N, Sato K, Matsuzawa Y, Goto K, Omuta K, Nakashima Y, Takeshita H, Sakamoto W (2002) Population structure of loggerhead turtles, Caretta caretta, nesting in Japan: bottlenecks on the Pacific population. Mar Biol 141:299–305
Hawkes LA, Broderick AC, Coyne MS, Godfrey MH, López-Jurado LF, López-Suarez P, Merino SE, Varo-Cruz N, Godley BJ (2006) Phenotypically linked dichotomy in sea turtle foraging requires multiple conservation approaches. Curr Biol 16:990–995
Hillis DM, Mable BK, Larson A, Davis SK, Zimmer EA (1996) Nucleic acids IV: sequencing and cloning. In: Hillis DM, Moritz C, Mable BK (eds) Molecular systematics. Sinauer Associates, Sunderland, pp 321–381
IUCN (2007) Red list of threatened species. www.iucnredlist.org
Laurent L, Casale P, Bradai MN, Godley BJ, Gerosa G, Broderick AC, Schroth W, Scierwater B, Levy AM, Freggi D, Abd El-Mawla EM, Hadoud DA, Gomati HE, Domingo M, Hadjichristophorou M, Kornaraky L, Demirayak F, Gautier CH (1998) Molecular resolution of marine turtle stock composition in fishery by-catch: a case study in the Mediterranean. Mol Ecol 7:1529–1542
Lee PLM (2008) Molecular ecology of marine turtles: new approaches and future directions. J Exp Mar Biol Ecol 356:25–42
López-Jurado LF, Andreu AC (1998) Caretta caretta (Linnaeus, 1758). In: Salvador A (ed) Reptiles. Fauna Ibérica, vol 10. Museo Nacional de Ciencias Naturales, CSIC, Madrid, pp 44–56 (in Spanish)
López-Jurado LF, Liria A (2007) Programa de reintroducción de la tortuga boba en Canarias. Quercus 255:20–25 (in Spanish)
López-Jurado LF, Évora C, Cabrera I, Cejudo D, Alfama P (2000) Proposals for the conservation of marine turtles on the Island of Boavista (Republic of Cabo Verde, western Africa). In: Kalb HJ, Wibbels T (comps) Proceedings of the 19th annual symposium on sea turtle conservation and biology. NOAA Technical Memorandum NMFS-SEFSC-443. South Padre Island, TX, 1999, pp 204–205
Luikart G, Allendorf FW, Cornuet J-M, Sherwin WB (1998) Distortion of allele frequency distributions provides a test for recent population bottlenecks. J Hered 89:238–247
Marco A, López O, Abella E, Varo N, Martins S, Gaona P, Sanz P, López-Jurado LF (2008) Massive capture of nesting females in severely threatening the CaboVerdian loggerhead population. In: 28th Annual symposium on sea turtle biology and conservation, Loreto, Baja California Sur, Mexico, 19–26 January 2008
Margaritoulis D, Argano R, Baran I, Bentivegna F, Bradai MN, Camiñas JA, Casale P, de Metrio G, Demetropoulos A, Gerosa G, Godley BJ, Haddoud DA, Houghton J, Laurent L, Lazar B (2003) Loggerhead turtles in the Mediterranean Sea: present knowledge and conservation perspectives. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution Press, Washington, DC, pp 175–198
Marshall TC, Slate J, Kruuk LEB, Pemberton JM (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Monzón-Argüello C, Muñoz J, Marco A, López-Jurado LF, Rico C (2008) Twelve new polymorphic microsatellite markers from the loggerhead sea turtle (Caretta caretta) and cross-species amplification on other marine turtle species. Conserv Genet 9:1045–1049
Monzón-Argüello C, Rico C, Carreras C, Calabuig P, Marco A, López-Jurado LF (2009) Variation in spatial distribution of juvenile loggerhead turtles in the eastern Atlantic and western Mediterranean Sea. J Exp Mar Biol Ecol 373:79–86
Moritz C (1994) Defining “evolutionary significant units” for conservation. Trends Ecol 8:1063–1067
Musick JA, Limpus CJ (1997) Habitat utilization and migration in juvenile sea turtles. In: Lutzand PL, Musick JA (eds) The biology of sea turtles. CRC Press, Boca Raton, pp 137–163
Naro-Maciel E, Becker JH, Lima EHSM, Marcovaldi MA, DeSalle R (2007) Testing dispersal hypotheses in foraging green sea turtles (Chelonia mydas) of Brazil. J Hered 98:29–39
Norman JA, Moritz C, Limpus CJ (1994) Mitochondrial DNA control region polymorphisms: genetic markers for ecological studies of marine turtles. Mol Ecol 3:363–373
Parsons TJ, Muniec DS, Sullivan K, Woodyatt N, Alliston-Greiner R, Wilson MR, Berry DL, Holland KA, Weedn VW, Gill P, Holland MM (1997) A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet 15:363–367
Pella J, Masuda M (2001) Bayesian methods for analysis of stock mixtures from genetic characters. Fish Bull 99:151–167
Pella J, Masuda M (2005) Classical discriminant analysis, classification of individuals, and source population composition of mixtures. In: Cadrin SX, Friedland KD, Waldman JR (eds) Stock identification methods: application in fishery science. Elsevier Academic Press, Burlington, pp 517–552
Posada D, Crandall K (2001) Intraspecific genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) An exact test for population differentiation. Evolution 49:1280–1283
Reis EC, Soares LS, Vargas SM, Santos FR, Young RJ, Bjorndal KA, Bolten AB, Lôbo-Hajdu G (in press) Genetic composition, population structure and phylogeography of the loggerhead sea turtle: colonization hypothesis for the Brazilian rookeries. Conserv Genet. doi:10.1007/s10592-009-9975-0
Revelles M, Carreras C, Cardona L, Marco A, Bentivegna F, Castillo JJ, de Martino G, Mons JL, Smith MB, Rico C, Pascual M, Aguilar A (2007) Evidence for an asymmetrical size exchange of loggerhead sea turtles between the Mediterranean and the Atlantic through the Straits of Gibraltar. J Exp Biol Ecol 349:261–271
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Ross JP (1995) Historical decline of loggerhead, ridley and leatherbacks sea turtles. In: Bjorndal KA (ed) Biology and conservation of sea turtles. Smithsonian Institution Press, Washington, DC, pp 189–195
Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152:1079–1089
Schroeder BA, Foley AM, Bagley DA (2003) Nesting patterns, reproductive migrations, and adult foraging areas of loggerhead turtles. In: Bolten AB, Witherington BE (eds) Loggerhead sea turtles. Smithsonian Institution, Washington, DC, pp 114–124
Schug MD, Hutter CM, Wetterstrand KA, Gaudette MS, Mackay TFC, Aquadro CF (1998) The mutation rates of di-, tri- and tetranucleotide repeats in Drosophila melanogaster. Mol Biol Evol 15:1751–1760
Tamura K, Nei M (1993) Estimation of the number of substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Templeton AR, Crandall KA, Sing CF (1992) A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics 132:619–633
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vargas SM, Araújo FCF, Monteiro DS, Almeida AP, Soares LS, Santos FR (2008) Genetic diversity and origin of leatherback turtles (Dermochelys coriacea) from the Brazilian coast. J Hered 99:215–220
Velez-Zuazo X, Ramos WD, Van Dam RP, Diez CE, Abreu-Grobois A, McMillan WO (2008) Dispersal, recruitment and migratory behaviour in a hawksbill sea turtle aggregation. Mol Ecol 17:839–853
Acknowledgements
We thank P. Calabuig, O. López, P. Sanz, S. Merino, A. Herrero, C. Almeida, Ll. Ballell, P. García, the monitors and volunteers of Cabo Verde Natura 2000, and students of ISECMAR for their contributions to sampling and field collection. A. Liria, D. Cejudo, X. Vélez-Zuazo, and B. Bolker helped us with data analysis and interpretations. We also thank P. Lee for helpful comments on earlier versions on this manuscript. Thanks to B. Bowen for his helpful comments on analysis of longer sequences. We are grateful to the Cabo Verde Ministry of the Environment (General Direction for the Environment), INDP (National Fisheries Institution), the Canary Islands government, Instituto Canario de Ciencias Marinas, Estación Biológica de Doñana, Fundación BBVA, and Junta de Andalucía for helping with the field and laboratory equipment. CMA was supported by a PhD grant from the Canary Islands government.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Monzón-Argüello, C., Rico, C., Naro-Maciel, E. et al. Population structure and conservation implications for the loggerhead sea turtle of the Cape Verde Islands. Conserv Genet 11, 1871–1884 (2010). https://doi.org/10.1007/s10592-010-0079-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-010-0079-7