Abstract
The loggerhead sea turtle (Caretta caretta) is a cosmopolitan sea turtle species and is listed by IUCN as Vulnerable globally. The Southwest Atlantic is an important regional management unit of C. caretta worldwide due to the distinctive mitochondrial DNA (mtDNA) lineage promoted by recent radiation within the Atlantic-Mediterranean region. However, due to the low resolution of mtDNA, the population structure of C. caretta SWA has not been well understood in the previous studies using only mtDNA. Our study encloses data from literature and a long-term genetic survey (1999 to 2021) distributed through four great nesting areas for the Southwest Atlantic to assess the genetic diversity and the population structure of the C. caretta, using both mtDNA and 15 microsatellite loci. The results demonstrate that the genetic diversity indexes of the Southwest Atlantic C. caretta reflect distinct compositions at a population level due to variation at an individual level. The SSRs results identified well-established and significant spatial population structure between nesting areas. Unique genetic patterns were identified for those females from studied areas of the Southwest Atlantic, and it may be related to their philopatric behavior and high relatedness. Thus, this study deeply evaluated the molecular ecology of Southwest Atlantic C. caretta and provides, for the first time, a fine-scale and long-term resolution of the genetic diversity and population structure due to the use of microsatellite data that must be considered for further studies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The loggerhead sea turtle Caretta caretta (Linnaeus, 1758) is listed as Vulnerable globally by International Union for Conservation of Nature (IUCN) (Casale and Tucker 2017) and Brazil (Brasil 2022). The C. caretta is a cosmopolitan species (Marcovaldi and Marcovaldi 1999) that is widely studied (Uller and Olsson 2008, Clusa et al. 2016; Lockley et al. 2020), and presents around the world several conservation programs that also benefit other species in their natural habitats, defining them as an umbrella taxon (Rees et al. 2016). For example, since the 1980s, the Projeto Tamar—a Sea Turtle Conservation Program—monitors and protects Brazilian nesting and foraging areas of sea turtles against predation, coastal development, degradation of sandbank areas, and bycatch (Marcovaldi and Marcovaldi 1999; Marcovaldi et al. 2006; Marcovaldi and Chaloupka 2007; Santos et al. 2011; Pike 2013; López-Mendilaharsu et al. 2020). However, some aspects of its life history (e.g., migratory behavior, male genetic contribution, the origin of juveniles from neritic feeding areas) are unknown and/or poorly understood due to the difficulty of tracking sea turtles in marine habitats (Bjorndal and Bolten 2008; Luschi and Casale 2014; Stewart et al. 2019) or estimating the connectivity between populations (e.g., using a mark–recapture method and/or satellite telemetry) (McClellan and Read 2007; Soares et al. 2021). Understanding those mechanisms for C. caretta populations is crucial, especially because it exhibits inter-population dynamics and variations (like natal homing, philopatric behavior, and use of distinct feeding and developmental areas) that warrant population-specific management and conservation measures (Carreras et al. 2007; Monzón-Argüello et al. 2010; Wallace et al. 2010; Baltazar-Soares et al. 2020).
Molecular studies using the control region (D-loop) of the mitochondrial genome (mtDNA; matrilineal inheritance) around the world have been performed to improve the knowledge of the loggerhead life history, identifying lineages for females, evaluating their phylogeography and population structure, and the origin of feeding mixed-stocks (e.g., Bjorndal and Bolten 2008; Reis et al. 2010; Shamblin et al. 2014; Reid et al. 2019; Baltazar-Soares et al. 2020; Vilaça et al. 2021). Two great mtDNA lineages of loggerhead have been recognized (Bowen et al. 1994) which diverged 2–4 million years ago (Duchene et al. 2012), and are distributed in the Atlantic-Mediterranean region and the Indo-Pacific region (Matsuzawa et al. 2016). Additionally, Wallace et al. (2010) established, based on mtDNA, that the Atlantic-Mediterranean lineage is composed of four Regional Management Units (RMU) being two on the North Atlantic (Northeast of the USA coast and one on the Northwest of the north coast of Africa), one on the Mediterranean-Greece coast, and another on the Southwest Atlantic coast (SWA). On the other hand, only a few studies are using nuclear DNA loci (nDNA; biparental inheritance) like microsatellite loci (SSRs: Simple Sequences Repeats) which have provided greater sensitivity and fine-scale at the population level than D-loop (e.g., Moore and Ball 2002; Shamblin et al. 2007, 2011; Monzón-Argüello et al. 2010; Carreras et al. 2018).
The SWA encloses important nesting areas for sea turtles, including the coast of Sergipe (SE), Bahia (BA), Espírito Santo (ES), and Rio de Janeiro (RJ) states (Marcovaldi and Chaloupka 2007; Lima et al. 2012; Marcovaldi et al. 2017; Colman et al. 2020). In the previous genetic studies using only D-loop, there were divergent results concerning the number of management units (MUs) for SWA C. caretta. When using short haplotypes (~ 380 bp), it was suggested the existence of two MUs, being the Southern stock (RJ + ES) and Northern stock (BA + SE) (Reis et al. 2010), but when using long haplotypes (~ 800 bp) was suggested three distinct MUs, being the Northeastern coast (SE + BA), the ES, and the RJ (Southeastern coast) (Shamblin et al. 2014). Knowing the number of loggerheads MUs is important, because understanding population boundaries and connectivity by direct observations to properly manage isolated rookeries is very difficult (Komoroske et al. 2017). Therefore, these findings reinforce the great importance of improving our understanding of population genetics of the SWA C. caretta by incorporating biparental inheritance molecular markers to advance conservation and management programs worldwide (e.g., Pike 2013; Lopez et al. 2015; Rees et al. 2016; Monteiro et al. 2019).
Therefore, in this study, we investigated the population structure and possible genetic composition changes along space in the SWA loggerheads from four nesting areas/rookeries (RJ, ES, BA, and SE) using the D-loop of the mtDNA and, for the first time, from 15 microsatellite loci of the nDNA. Thus, we elucidate the uncertainties that remained using increased sampling and nuclear DNA. To reach that, we compared our population structure results (mtDNA and SSRs) with those from previous studies (Reis et al. 2010; Shamblin et al. 2014), which indicated that each nesting area may be considered genetically independent MU). The genetic diversity results revealed complex dynamics and significant spatial population structure (based on nDNA) for SWA loggerheads that may be related to its philopatric behavior, which improves our understanding of the spatial distribution of the SWA loggerheads populations. This demonstrates the importance of using molecular markers with biparental inheritance (such as SSRs) to detect more accurate and refined data that may underpin management units (MUs) boundaries and conservation measures.
Materials and methods
Ethical and research permits
This study was conducted under the authorization, strict control, and permission of the Instituto Chico Mendes de Conservação da Biodiversidade and conducted under SISBIO license numbers #65,543–3 and #42,760. Sampling of C. caretta individuals was performed minimizing animal suffering when obtaining a tissue sample for genetic analyses. For this study, we also obtained permission from the Sistema Nacional de Gestão do Patrimônio Genético e do Conhecimento Tradicional Associado (SisGen) on the number #A32C980.
Study area and sampling
Our study encloses a long-term genetic survey (1999 to 2021) distributed through four main nesting areas (RJ, ES, BA, and SE) of the SWA, spanning approximately 1400 km of coastline, which ES and SE are separated by 960 km, ES and BA by 800 km, BA and SE by 160 km, BA and RJ by 1120 km, RJ and SE by 1280 km, and ES and RJ by 320 km (Fig. 1A). The sampling of loggerhead individuals usually occurred during the nesting season (from September to March) with opportunistic encounters during night surveys along nesting beaches. For all individuals, we collected a piece of 6 mm of epithelial tissue with a biopsy punch from the base of the neck and the beginning of the shoulder. Each sample was preserved in ethanol 100% in microtubes of 1.5 mL. In ES, sampling occurred along the Povoação, Guriri, and Comboios beaches between the 2017/18, 2018/19, 2019/20, and 2020/21 nesting seasons. This ES area is located around the mouth of the Doce River and the adjacent continental shelf which is one of the main nesting areas of adult female loggerhead turtles in the SWA (Marcovaldi et al. 2016). In BA, sampling occurred in Camaçari and Mata de São João during 2005/06, 2006/07, and 2008/09 nesting seasons, and in Arembepe beach during the 2019/20 nesting season. In SE, sampling occurred on Estância and Pirambu beaches during the 2008/09, 2009/10, and 2010/11 nesting seasons. Additionally, tissue samples from ES, BA, and SE collected between 2004 and 2009 nesting seasons were also provided by the Projeto Tamar. At last, for comparison levels, we compiled genetic information from the long D-loop haplotypes of previous studies (Shamblin et al. 2014). The samples were deposited in a scientific tissue collection at the Federal University of Espírito Santo in Brazil.
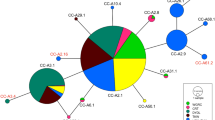
Southwest Atlantic nesting areas [RJ, ES, BA, and SE] of Caretta caretta evaluated in this study demonstrating the (A) D-loop haplotype frequencies distribution, and their (B) network relationships built using NETWORK, based on compiled SWA dataset, which colors correspond to the nesting areas and the pie charts size to the frequency as indicated in the legend (see Table 1 for details)
Laboratory procedures
To access the genetic diversity of the loggerheads sampled, we amplified through Polymerase Chain Reactions (PCRs) the control region D-loop of the mtDNA genome in both strands using the LCM15382 and H950 primers (Abreu-Grobois et al. 2006), and from nDNA through genotyping of 15 microsatellite loci (Shamblin et al. 2007, 2009). In the Núcleo de Genética aplicada à Conservação da Biodiversidade of the Federal University of Espírito Santo in Brazil (NGACB-UFES: https://blog.ufes.br/ngacb/), the genomic DNA (gDNA) of each sample was isolated using a saline protocol of Bruford et al. (1992) and CTAB 2% of Doyle and Doyle (1987). The gDNA was resuspended in ultrapure water and, then, quantified through spectrophotometer NanoDrop ND100 (Thermo Scientific) to verify the DNA concentration (ideal 50 ng/µL) and purity of each sample. Subsequently, the quality of the gDNA was analyzed using electrophoresis in an agarose gel of 1% in UV-transilluminator L-PIX Touch 20 × 20 cm (Loccus©).
The amplification of the D-loop region was carried out following the instructions of Shamblin et al. (2012), which PCR cycling profile conditions were as follows: initial denaturation at 94 °C for 3 min, followed by 35 cycles of denaturation step of 94 °C for 30 s, annealing at 51 °C for 30 s, and extension at 72 °C for 40 s, followed by a final extension at 72 °C for 10 min. To check if there was any contamination, we added negative (containing only PCR’s mix) and positive controls (samples with a positive result that has been amplified before) for each reaction. The genotyping of the 15 microsatellite loci: Cc7G11, Cc1F01, Cc1G02, Cc1G03, CcP7D04, CcP2F11, CcP7C06, CcP8D06, CcP1F09, CcP5C11, CcP1F01, CcP1G03, CcP1B03, CcP5C08, and CcP5H07 (Shamblin et al. 2007, 2009) following the PCR cycling profile conditions of Shamblin et al. (2009), which were directly labeled with fluorescent FAM, PET, NED, and VIC dyes (Shamblin et al. 2011).
Subsequently, all the PCRs products for both target regions (mtDNA and SSRs) were checked through electrophoresis in agarose gel 1%, stained with Gel-Red (Biotium), 100 bp ladder (Ludwig Biotech©), and detected by UV-transilluminator L-PIX Touch 20 × 20 cm (Loccus©). Then, PCR products were purified ExoSAP-IT (Applied Biosystems) to remove surplus reagents, following the manufacturer’s protocol. PCR products were also sequenced in both directions using Big Dye Terminator components (Applied Biosystems) according to the manufacturer’s protocols. The genotyping was performed using a reagent mix with 7.0 µL of formamide, 0.5 µL of fluorescent standard molecular size GeneScan™ 600 LIZ™ Size Standard v2.0 (Applied Biosystems©), and 0.5 µL of the purified PCR’s product. For both sequencing and genotyping, we used the sequencer ABI Pris 3700 Automatic Sequencer (Thermo Fisher Scientific) (Applied Biosystems).
Both strands of the D-loop sequences were edited and a consensus for each individual was generated using Geneious R11.1.5 (Biomatters Ltd; Kearse et al. 2012). Next, an alignment was also carried out under default conditions using ClustalW (Thompson et al. 1996) implemented in Geneious. Further, SSRs alleles were scored and tabulated using Geneious. The identification of null allele, large allele dropout, PCR slippage, and genotyping errors was evaluated by MICRO-CHECKER v2.2.3 (van Oosterhout et al. 2004) with a 95% confidence interval by Monte Carlo simulation. Additionally, deviations from Hardy–Weinberg equilibrium (HWE) and the linkage disequilibrium (LD) between each pair of loci were evaluated through the significance test using the Markov Chain method (10,000 dememorizations, 20 batches, and 5000 iterations per batch) using the software ML-Relate (Kalinowski et al. 2006) and FreeNA (Kawashima et al. 2009), respectively.
mtDNA analyses
To evaluate if there are possible genetic composition changes along space the SWA loggerheads and their genetic diversity, we first identified the D-loop haplotypes from our dataset compared with existing haplotypes from nesting and foraging locations worldwide and, which new long haplotypes were identified through the Archie Carr Center for Sea Turtle Research database (ACCSTR) (available at: http://accstr.ufl.edu/ccmtdna.html). Second, we estimated the genetic diversity by the number of haplotypes (H), the haplotype (h; Nei 1973), and nucleotide (π; Nei and Li 1979) diversities through DnaSP v5 (Librado and Rozas 2009) (Table 1), and we compared among them. Third, we compiled our dataset with those available in Shamblin et al. (2014) naming them as “compiled SWA” (Table 1) to maximize the space scale of sampling and make more feasible comparisons for the optimal rookery clustering for the SWA RMU. Fourth, we inferred the haplotype genetic relationships and their geographic frequency distribution by building a haplotype network using Median-Joining (Saitou and Nei 1987) that was displayed by nesting areas using the NETWORK method presented in Bandelt et al. (1999).
Fifth, using our D-loop dataset plus RJ data (from Shamblin et al. 2014), we evaluated the population structure by pairwise FST (Nei 1977) comparing the nesting areas of the SWA. Complementary, we then performed additional comparisons by Analysis of Molecular Variance Analysis (AMOVA; Excoffier et al. 1992), and pairwise and global FST using the compiled SWA dataset (Table 1). We tested the following hypotheses to identify the optimal rookery clustering for SWA: (1) the Northern stock (SE + BA) is genetically distinct from the Southern stock (ES + RJ) as suggested by Reis et al. (2010); (2) the Northeastern coast (SE + BA) is genetically distinct from the ES and also from RJ coast as suggested by Shamblin et al. (2014); (3) the area SE + BA + ES is genetically distinct from the RJ nesting area; and (4) each nesting area being a genetic resource population. We performed all population structure analyses using a D-loop in Arlequin 3.0 (Excoffier et al. 2005), and a significance of P value was computed with 1,000 permutations.
Microsatellite analyses
Further, using our SSRs dataset, we estimated the genetic diversity indexes by the number of alleles (A), the allelic richness (AR), the observed (Ho) and expected heterozygosity (He), private alleles (PA), and inbreeding coefficient (FIS; Brown 1970) in diveRsity and PopGenReport packages (Keenan et al. 2013; Adamack and Gruber 2014) for R (Team 2021). Then, we estimated the relatedness among individuals (r) using the algorithm of Lynch and Ritland (1999), with reference allele frequencies calculated and mean values within and between each population and its 95% confidence interval, in GenAlex 6.5 (Peakall and Smouse 2012).
We used four distinct methods to properly evaluate the possible genetic composition changes along space in the SWA loggerheads. First, we used AMOVA to comprehend the partitioning of genetic variation structure at distinct population levels (FST, FIT, and FIS). Second, a pairwise FST (multilocus; Slatkin 1995) comparing the nesting areas of the SWA, and a significance of P value was computed with 1,000 permutations for all combinations in GenAlex. Third, the population structure was also evaluated using the Bayesian clustering approach by a Discriminant Analysis of Principal Components (DAPC; Jombart et al. 2010). The DAPC was chosen, because equalizes the genetic variation among the populations using Principal Components analysis (PCA) and validates results by hierarchical clustering using discriminant analysis (DA) through a Bayesian Information Criterion (BIC; Jombart et al. 2010), and it also allows the use of loci that deviate from HWE, non-panmictic populations, use of samples related by descent, and low structuring indices (FST < 0.3), which fits our study system. DAPC was performed using the adegenet R package (Jombart 2008) in R (Team 2021) and the steps are provided in Supplementary Material. Structure was performed in parallel, but divergent results was obtained (Fig. S2). Finally, we performed the Principal Coordinates Analysis (PCoA) to find the major population genetic patterns of each analyzed area that plots the major axes of population variation, revealing the most principal components that separate among them (Orlóci 2013) using as prior the pairwise FST in GenAlex, which significance of P value was computed with 1000 permutations. Unfortunately, it was not possible to perform these SSRs analyses using samples from ES: 2017/18 nesting season due to high missing genotypes, and neither for RJ samples, because we obtained only the D-loop data from Shamblin et al. (2014).
Results
Genetic diversity
For this study, we sampled 251 individuals including one dead male, and 250 adult females, which were successfully sequenced. From Shamblin et al. (2014), we compiled 131 D-loop 867 bp haplotypes. Then, we built the “compiled SWA” totalizing 382 sequences, with h was 0.487 and π ranging being 0.00064 (Table 1).
We identified eight haplotypes for the SWA overall, being A4.2 the most frequent (65.7%), followed by A4.1 (28.5%) (Table 1; Fig. 1A). The A4.3 (N = 5) and A25.1 (N = 7) were identified only in the ES nesting area (Table 1; Fig. 1A). Additionally, we identified three new haplotypes the A4.4, A4.5, and A4.6 (see * in Table 1; Fig. 1A, B). We sampled only one male in the 2018/19 nesting year in Guriri beach (ES) and it bears the haplotype A4.2. The haplotype network resulted in a star pattern where the A4.2 is the central and the most frequent haplotype and the others originated from it by only one mutation step each (Fig. 1B).
From the 15 SSRs, only four deviated from the HWE after Bonferroni correction, and only one locus (CcP2F11) presented > 10% null allele frequency (Table S1). Overall, we retrieved high genetic diversities (A = 132, AR = 8.80, mean r = 27.426, HO = 0.816, HE = 0.841, mean PA = 31.3, and FIS = 0.031), where the HO was higher than HE for almost all populations, and the FIS was in general low and sometimes negative indicating outbreeding, which opposed to the relatedness (r) that were moderate-high for all (Table 1). The pairwise r between the nesting areas was higher between the ES and BA (r = 32.103) followed by BA and SE (r = 31.792), and ES and SE (r = 29.940) (Fig. S1).
Population structure
The D-loop and SSRs’ population structure analyses yielded different results. Using only our D-loop dataset (SWA overall) (N = 251), neither pairwise FST nor AMOVA yielded significant differences (Tables 2, 3). However, for compiled SWA (N = 382), two pairwise FST comparisons were significant (P value ≤ 0.05) involving RJ versus ES and RJ versus BA (Table 2). The additional AMOVA comparisons using the compiled SWA dataset (N = 381) did not confirm the hypothetical stock structure (all FCT values were non-significant) but were significant among the four discrete populations from SWA (global FST = 0.019, P = 0.046) (Table S2).
On the other hand, using the SSRs dataset, all pairwise FST comparisons and AMOVA were highly significant (P value ≤ 0.001) (Table 2), being the population structure detected in all three population levels being the highest variability (72%) observed within individuals (FIS = 0.278) (Table 3).
DAPC detected four genetic clusters based on the lowest BIC explaining 97% of the genetic differentiation across the individuals (FIT = 0.260, P value ≤ 0.001) (without population assigned) (Fig. 2A). Genetic clusters k2 and k3 were found in all nesting areas, while k1 was found in the ES and BA, and k4 was exclusive for the ES nesting area (Fig. 2B). While, when individuals were clustered in their respective nesting areas, the DAPC demonstrates that there is a more homogeneous distribution within the areas (attributed to distinct colors: yellow to SE, brown to ES, and black to BA in Fig. 2C) and low admixture (sharing genetic pattern ≥ 50%) between them except for 22 individuals (Fig. 1C), one individual of SE shared patterns with ES, three individuals of ES shared patterns with SE, seven individuals of ES shared patterns with BA, 11 individuals of BA shared patterns with ES, and there were no sharing patterns among SE and BA (Fig. 2C). PCoA identified 99.83% of genetic variation between areas demonstrating great genetic distance among them (Fig. 1D). The sharing of genetic clusters by DAPC between the nesting areas (Fig. 2C) and PCoA (Fig. 2D) results explain the low genetic variation obtained by pairwise FST and AMOVA in all levels, but they were highly significant (P value ≤ 0.001) (Tables 2, 3).
Population structure of the Caretta caretta of the Southwest Atlantic based on SSRs’ dataset obtained by DAPC (A–C) and PCoA (D). A The identification and distribution of four genetic clusters [k1, k2, k3, and k4]. B the individual assignment probability [vertical bars] to be clustered [0–100%] in the genetic clusters [k1–k4] without population assigned. The colors indicate the shared proportion of the genetic clusters as a legend. C the individual assignment probability [vertical bars] to be clustered [0–100%] in its respective nesting areas [SE, ES, and BA]. The arrow points to the unique sampled male of the study. The colors indicate the shared proportion of the nesting areas as a legend. D PCoA results demonstrate the genetic variation between the SE, ES, and BA nesting areas for which the FST is also given
Discussion
This study evaluated the population structure and the genetic composition changes in the SWA loggerheads along space (RJ, ES, BA, and SE) and time (from 1999 to 2021) using for the first time SSRs dataset and comparing our D-loop dataset with the previous studies. Overall, the SSRs results demonstrate that distinct compositions at populational and individual levels may have generated distinct genetic clusters between the SWA females, distinguishing them by nesting areas. Therefore, our results also attest that each nesting area is genetically independent as MU, contrasting with the previous studies that suggested two (Reis et al. 2010) or three (Shamblin et al. 2014) potential genetic stocks for SWA. Such differences could be associated with the origin of molecular markers used in those studies (matrilineal inheritance, mtDNA) and from ours (SSRs, nDNA), which demonstrate the improvement in using molecular markers with biparental inheritance to refine population genetics analysis and has been well accepted for the sea turtles’ studies worldwide (e.g., Naro-Maciel et al. 2007; Monzón-Argüello et al. 2010; Dutton et al. 2013; Gallego-Garcia et al. 2018; Loisier et al. 2021; Vargas et al. 2022).
Spatial population structure, philopatry, and admixture
A spatial population structure was detected for the SWA loggerheads, using both D-loop and SSRs. Although the “compiled SWA” results for mtDNA alone presented a low resolution to detect population structure signals among the nesting areas, the SSRs’ results detected a highly significant signal of population structure with great genetic variation. The pairwise comparisons corroborated that once the highest values were found between BA and SE, which are about 160 km geographically closer than ES and SE which are about 960 km geographically distant, and ES and BA are about 800 km apart (Table 2). Interestingly, according to the SSRs results, there is a spatial population structure that was evidenced by distinct genetic clusters for each nesting area, and it was not previously detected for other C. caretta populations worldwide (e.g., Moore and Ball 2002; Monzón-Argüello et al. 2010; Carreras et al. 2018; Loisier et al. 2021). We advocate that the philopatric behavior by related SWA females may be generating such spatial structure and has directly reflected in their kinship within each nesting area and diverging among them, which is corroborated by the r results (Table 1; Fig. S1). The philopatry behavior has been genetically explored for the Mediterranean RMU loggerheads populations which have been linked to restricted gene flow among the nesting grounds, and only detected when using SSRs dataset (Clusa et al. 2018; this study). In accordance, our findings attest to the high fidelity of the SWA females to return to their birth areas to nest (Marcovaldi et al. 2016; Baltazar-Soares et al. 2021), which were previously registered by mark–recapture and telemetry methods (Marcovaldi et al. 2010; Barreto et al. 2019), postulated by geomagnetism studies (e.g., Cameron et al. 2019), but for the first time was detected a genetic signal defining their spatial population structure.
Despite the philopatric behavior, the detection of individuals with admixture patterns signalizes to us that there was sharing ancestry by nesting areas. This suggests that may have resulted from the reproduction of parents from different origins and/or the mothers mating with the same males near the nesting beach as already postulated by Marcovaldi et al. (2016). This assumption is corroborated by the detection of 18 admixture individuals among the ES and BA nesting areas with low FST (FST = 0.047). Besides, admixture individuals were identified between areas, which may be attached to their kinship and complex life history with low male-mediated gene flow. Such admixture patterns have also been found for the sea turtle Natator depressus Garman, 1880 (FitzSimmons et al. 2020), and for the C. caretta of the Mediterranean RMU (Clusa et al. 2018) but for the first time for the SWA.
On the other hand, it is uncertain whether the spatial genetic population structure of the C. caretta can be due to the mere lack of gene flow, as the exchange of only one migrant could be sufficient to prevent the accumulation of large genetic differences between populations (Wright 1931; Mills and Allendorf 1996). Because, although we only sampled one male, we suggest that they may also be displaying a degree of fidelity to natal breeding areas (e.g., Clusa et al. 2018; Medeiros et al. 2019) due to the high genetic similarity among those females within the SWA nesting areas and could be more related to some levels of male-mediated gene flow within the same nesting seasons than among consecutive seasons within the same nesting area. However, neither migration and/nor gene flow was previously studied in the SWA due to probably low genetic variation of the D-loop at the individual level (e.g., Reis et al. 2010; Shamblin et al. 2014; Reid et al. 2019). Thus, further studies should deeply investigate the demographic patterns between the populations of these nesting areas using SSRs to solve our proposed hypothesis.
Conservation genetic concerns
The SWA loggerheads’ populations present unique D-loop haplotypes composition that underlies its lineage, which has been the subject of numerous discussions about its origin and expansion compared with other RMU of the Indo-Pacific and Mediterranean-Atlantic (e.g., Wallace et al. 2010; Reid et al. 2019; Baltazar-Soares et al. 2020). Therefore, this study offers a new perspective on the spatial genetic pattern distribution of those SWA lineages, improving our understanding of philopatric behavior using SSRs against mtDNA. Herein, we provided valuable information on the unique genetic clusters at a highly localized geographic scale (within the SWA nesting areas), and attest that the SWA populations are isolated from the other RMU. Also, we demonstrate that each SWA nesting area may be treated as an independent MU for C. caretta conservation as a species, which can contribute to further studies underlying the origin of individuals from feeding grounds or washed ashore.
Besides, at the SWA scale, our population structure results warn on the conservation status of these loggerhead subpopulations, especially from ES nesting area, because it was subjected to several environmental impacts (IBAMA 2015; Fernandes et al. 2016; Segura et al. 2016; Hatje et al. 2017; Almeida et al. 2018; Burritt and Christ 2018; Magris et al. 2019; Coimbra et al. 2020; Lacerda et al. 2020), and has been identified as an important male nursery for the SWA (Marcovaldi et al. 2016). On the ES coast, recently, it has been detected and monitored the presence of heavy metals at distinct trophic levels, being in algae, microcrustaceans (Vergilio et al. 2021), fishes assemblies (Bonecker et al. 2019; Lacerda et al. 2020), and eggs and hatchlings of other sea turtles as the leatherback Dermochelys coriacea (Linnaeus, 1766) (Freire et al. 2021), from immature green turtle Chelonia mydas (Linnaeus, 1758) (Frossard et al. 2020; Miguel et al. 2022), and late juveniles of loggerheads (Cantor et al. 2020). The presence of heavy metals has been also reported for other C. caretta subpopulations worldwide (Jerez et al. 2010; Yipel et al. 2017; Canzanella et al. 2021). The assumed metals bioaccumulation can compromise birth rate, decrease health status in adult individuals, and culminate in declines in census population sizes and effective population size in the next generations (e.g., Bowem et al. 2005; Kobayashi et al. 2017; Erb and Wyneken 2019).
However, to date, the relation between the shifts in genetic diversity and unique genetic clusters as a response to environmental stress was not reported for other loggerheads RMUs worldwide (e.g., Bjorndal and Bolten 2008; Shamblin et al. 2014; Carreras et al. 2018; Reid et al. 2019; Loisier et al. 2021). Thus, the loss of genetic diversity detected herein can be detrimental for the SWA population, which we presume may present less resilience, ability to circumvent anthropic impacts, and to remain genetically healthy in the long term due to threats like bycatch, climate changes but also to the environmental accidents that could impair not only maintenance of the next generations but also its survivorship (e.g., Hawkes et al. 2007; Colman et al. 2020; Martín-del-Campo et al. 2021; McCallum 2021; Soares et al. 2021).
Data availability
Sequences associated with the D-loop haplotypes (Cc-A4.5, Cc-A4.4, and Cc-A4.6) are deposited at GenBank by accession numbers MZ466566–MZ466568.
References
Abreu-Grobois FA, Horrocks JA, Formia A, Dutton P, LeRoux R, Vélez-Zuazo X, Soares L, Meylán P (2006) New mtDNA Dloop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis. In: Frick MA et al. (ed) Proceedings of the 26th Annual Symposium on Sea Turtle Biology Book of Abstracts, Island of Crete: Greece, 179 pp
Adamack AT, Gruber B (2014) PopGenReport: simplifying basic population genetic analyses in R. Methods Ecol Evol 5:384–387. https://doi.org/10.1111/2041-210X.12158
Almeida CA, de Oliveira AF, Pacheco AA, Lopes RP, Neves AA, de Queiroz MELR (2018) Characterization and evaluation of sorption potential of the iron mine waste after Samarco dam disaster in Doce River basin–Brazil. Chemosphere 209:411–420. https://doi.org/10.1016/j.chemosphere.2018.06.071
Baltazar-Soares M, Klein JD, Correia SM, Reischig T, Taxonera A, Roque SM, Dos Passos L, Durão J, Lomba JP, Dinis H, Cameron SJK, Stiebens VA, Eizaguirre C (2020) Distribution of genetic diversity reveals colonization patterns and philopatry of the loggerhead sea turtles across geographic scales. Sci Rep 10:18001. https://doi.org/10.1038/s41598-020-74141-6
Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Barreto J, Thomé JC, Baptistotte C, Rieth D, Marcovaldi MÂ, Marcovaldi GGD et al (2019) Reproductive longevity of the loggerhead sea turtle, Caretta caretta, in Espírito Santo, Brazil. Mar Turt Newsl 157: 10–12. http://www.seaturtle.org/mtn/archives/mtn157/mtn157-3.shtml
Bjorndal KA, Bolten AB (2008) Annual variation in source contributions to a mixed stock: implications for quantifying connectivity. Mol Ecol 17:2185–2193
Bonecker ACT, de Castro MS, Costa PG, Bianchini A, Bonecker SLC (2019) Larval fish assemblages of the coastal area affected by the tailings of the collapsed dam in southeast Brazil. Reg Stud Mar Sci 32:100848. https://doi.org/10.1016/j.rsma.2019.100848
Bowem BW, Bass AL, Soares L, Toonen RJ (2005) Conservation implications of complex population structure: lessons from the loggerhead turtle (Caretta caretta). Mol Ecol 14:2389–2402
Bowen BW, Kamezaki N, Limpus CJ, Hughes GR, Meylan AB, Avise JC (1994) Global phylogeography of the loggerhead turtle (Caretta caretta) as indicated by mitochondrial DNA haplotypes. Evol 48:1820–1828
Brasil (2022) Portaria MMA nº 148 de 07 de junho de 2022. Altera os Anexos da Portaria nº 443, de 17 de dezembro de 2014, da Portaria nº 444, de 17 de dezembro de 2014, e da Portaria nº 445, de 17 de dezembro de 2014, referentes à atualização da Lista Nacional de Espécies Ameaçadas de Extinção. Diário Oficial da União de 08.06.2022, Edição 108, Seção 1, p 74. available at: https://www.icmbio.gov.br/cepsul/images/stories/legislacao/Portaria/2020/P_mma_148_2022_altera_anexos_P_mma_443_444_445_2014_atualiza_especies_ameacadas_extincao.pdf
Brown AHD (1970) The estimation of Wright’s fixation index from genotypic frequencies. Genetica 41:399–406
Burritt RL, Christ KL (2018) Water risk in mining: Analysis of the Samarco dam failure. J Clean Prod 178:196–205
Cameron SJ, Baltazar-Soares M, and Eizaguirre C (2019) Region-specific magnetic fields structure sea turtle populations. bioRxiv, 630152. https://doi.org/10.1101/630152
Cantor M, Barreto AS, Taufer RM, Giffoni B, Castilho PV, Maranho A et al (2020) High incidence of sea turtle stranding in the southwestern Atlantic Ocean. ICES J Mar Sci 77:1864–1878. https://doi.org/10.1093/icesjms/fsaa073
Canzanella S, Danese A, Mandato M, Lucifora G, Riverso C, Federico G, Gallo P, Esposito M (2021) Concentrations of trace elements in tissues of loggerhead turtles (Caretta caretta) from the tyrrhenian and the Ionian coastlines (Calabria, Italy). Environ Sci Pollut Res 28:26545–26557. https://doi.org/10.1007/s11356-021-12499-4
Carreras C, Pascual M, Cardona L, Aguilar A, Margaritoulis D, Rees A et al (2007) The genetic structure of the loggerhead sea turtle (Caretta caretta) in the mediterranean as revealed by nuclear and mitochondrial DNA and its conservation implications. Conserv Genet 8:761–775. https://doi.org/10.1007/s10592-006-9224-8
Carreras C, Pascual M, Tomás J, Marco A, Hochscheid S, Castillo JJ et al (2018) Sporadic nesting reveals long distance colonisation in the philopatric loggerhead sea turtle (Caretta caretta). Sci Rep 8:1435. https://doi.org/10.1038/s41598-018-19887-w
Casale P and Tucker AD (2017) Caretta caretta (amended version of 2015 assessment). The IUCN Red List of Threatened Species 2017: .T3897A119333622. https://dx.doi.org/https://doi.org/10.2305/IUCN.UK.2017-2.RLTS.T3897A119333622.en. Downloaded on 03 April 2021.
Clusa M, Carreras C, Pascual M, Gaughran SJ, Piovano S, Avolio D et al (2016) Potential bycatch impact on distinct sea turtle populations is dependent on fishing ground rather than gear type in the Mediterranean Sea Mar Biol 163:1–10
Clusa M, Carreras C, Cardona L, Demetropoulos A, Margaritoulis D, Rees AF, Hamza AA, Khalil M, Levy Y, Turkozan O, Aguilar A, Pascual M (2018) Philopatry in loggerhead turtles Caretta caretta: beyond the gender paradigm. Mar Ecol Prog Ser 588:201–213. https://doi.org/10.3354/meps12448
Coimbra KTO, Alcântara E, de Souza Filho CR (2020) Possible contamination of the Abrolhos reefs by Fundao dam tailings, Brazil-New constraints based on satellite data. Sci Total Environ 733:138101. https://doi.org/10.1016/j.scitotenv.2020.138101
Colman LP, Lara PH, Bennie J, Broderick AC, de Freitas JR, Marcondes A et al (2020) Assessing coastal artificial light and potential exposure of wildlife at a national scale: the case of marine turtles in Brazil. Biodivers Conserv 29:1135–1152. https://doi.org/10.1007/s10531-019-01928-z
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Duchene S, Frey A, Alfaro-Núñez A, Dutton PH, Gilbert MTP, Morin PA (2012) Marine turtle mitogenome phylogenetics and evolution. Mol Phylogenet Evol 65:241–250. https://doi.org/10.1016/j.ympev.2012.06.010
Dutton PH, Roden SE, Stewart KR, LaCasella E, Tiwari M, Formia A, Thomé JC, Livingstone SR, Eckert E, Chacon-Chaverri D, Rivalan P, Allman P (2013) Population stock structure of leatherback turtles (Dermochelys coriacea) in the Atlantic revealed using mtDNA and microsatellite markers. Conserv Genet 14:625–636. https://doi.org/10.1007/s10592-013-0456-0
Erb V, Wyneken J (2019) Nest-to-Surf Mortality of Loggerhead Sea Turtle (Caretta caretta) Hatchlings on Florida’s East Coast. Front Mar Sci 6:271. https://doi.org/10.3389/fmars.2019.00271
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491. https://doi.org/10.1093/genetics/131.2.479
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Fernandes GW, Goulart FF, Ranieri BD, Coelho MS, Dales K, Boesche N et al (2016) Deep into the mud: ecological and socio-economic impacts of the dam breach in Mariana, Brazil. Nat Conservacao 14:35–45. https://doi.org/10.1016/j.ncon.2016.10.003
FitzSimmons NN, Pittard SD, McIntyre N, Jensen MP, Guinea M, Hamann M et al (2020) Phylogeography, genetic stocks, and conservation implications for an Australian endemic marine turtle. Aquat Conserv 30:440–460. https://doi.org/10.1002/aqc.3270
Freire JB, Sousa RM, Taufner K, Carneiro MT, Filgueiras HR, Lenz D, Ferreira PD, Endringer CD (2021) Metals in eggs and hatchlings of Dermochelys coriacea (Testudines) in Espírito Santo, Brazil and their relation to hatching and emergence success. Mar Ecol 42:e12633. https://doi.org/10.1111/maec.12633
Frossard A, Vieira LV, Carneiro MTWD, Gomes LC, Chippari-Gomes AR (2020) Accumulation of trace metals in eggs and hatchlings of Chelonia mydas. J Trace Elem Med Bio 62:126654. https://doi.org/10.1016/j.jtemb.2020.126654
Gallego-García N, Forero-Medina G, Vargas-Ramírez M, Caballero S, Shaffer HB (2018) Landscape genomic signatures indicate reduced gene flow and forest-associated adaptive divergence in an endangered neotropical turtle. Mol Ecol 28:2757–2771. https://doi.org/10.1111/mec.15112
Hatje V, Pedreira RM, de Rezende CE, Schettini CAF, de Souza GC, Marin DC, Hackspacher PC (2017) The environmental impacts of one of the largest tailing dam failures worldwide. Sci Rep 7:10706. https://doi.org/10.1038/s41598-017-11143-x
Hawkes LA, Broderick AC, Godfrey MH, Godley BJ (2007) Investigating the potential impacts of climate change on a marine turtle population. Glob Change Biol 13:923–932. https://doi.org/10.1111/j.1365-2486.2007.01320.x
IBAMA (2015) Laudo técnico Preliminar: Impactos Ambientais Decorrentes do Desastre Envolvendo O Rompimento da Barragem de Fundão, Em Mariana, Minas Gerais [Preliminary Technical Report: Environmental Impacts of the Disaster Involving the Disruption of Fundão Dam in Mariana, Minas Gerais].
Jerez S, Motas M, Cánovas RA, Talavera J, Almela RM, Del Río AB (2010) Accumulation and tissue distribution of heavy metals and essential elements in loggerhead turtles (Caretta caretta) from Spanish mediterranean coastline of Murcia. Chemosphere 78:256–264. https://doi.org/10.1016/j.chemosphere.2009.10.062
Jombart T (2008) Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Kalinowski ST, Wagner AP, Taper ML (2006) ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6:576–579. https://doi.org/10.1111/j.1471-8286.2006.01256.x
Kawashima R, Ji Y, Maruyama K (2009) FreeNA: a multi-platform framework for inserting upper-layer network services. IEICE T Inf Syst 92:1923–1933. https://doi.org/10.1587/transinf.E92.D.1923
Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S et al (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: an R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788. https://doi.org/10.1111/2041-210X.12067
Kobayashi S, Wada M, Fujimoto R, Kumazawa Y, Arai K, Watanabe G, Saito T (2017) The effects of nest incubation temperature in embryos and hatchlings of the loggerhead sea turtle: implications of sex difference for survival rates during early life stages. J Exp Mar Biol Ecol 486:274–281. https://doi.org/10.1016/j.jembe.2016.10.020
Komoroske LM, Jensen MP, Stewart KR, Shamblin BM, Dutton PH (2017) Advances in the application of genetics in marine turtle biology and conservation. Front Mar Sci 4:156. https://doi.org/10.3389/fmars.2017.00156
Lacerda D, dos Santos Vergilio C, da Silva Souza T, Costa LHV, Rangel TP, de Oliveira BCV et al (2020) Comparative metal accumulation and toxicogenetic damage induction in three neotropical fish species with distinct foraging habits and feeding preferences. Ecotox Environ Saf 195:110449. https://doi.org/10.1016/j.ecoenv.2020.110449
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Lima EPE, Wanderlinde J, de Almeida DT, Lopez G, Goldberg DW (2012) Nesting ecology and conservation of the loggerhead sea turtle (Caretta caretta) in Rio de Janeiro, Brazil. Chelonian Conserv Biol 11:249–254. https://doi.org/10.2744/CCB-0996.1
Lockley EC, Lockley EC, Fouda L, Correia SM, Taxonera A, Nash LN, Fairweather K, Reischig T, Durão J, Dinis H, Roque SM, Lomba JP, Passos L, Cameron SJK, Stiebens VA, Eizaguirre C (2020) Long-term survey of sea turtles (Caretta caretta) reveals correlations between parasite infection, feeding ecology, reproductive success and population dynamics. Sci Rep 10:18569. https://doi.org/10.1038/s41598-020-75498-4
Loisier A, Savelli MP, Arnal V, Claro F, Gambaiani D, Sénégas JB et al (2021) Genetic composition, origin and conservation of loggerhead sea turtles (Caretta caretta) frequenting the French mediterranean coasts. Mar Biol 168:52. https://doi.org/10.1007/s00227-021-03855-6
Lopez GG, Saliés EDC, Lara PH, Tognin F, Marcovaldi MA, Serafini TZ (2015) Coastal development at sea turtles nesting ground: efforts to establish a tool for supporting conservation and coastal management in northeastern Brazil. Ocean Coast Manage 116:270–276. https://doi.org/10.1016/j.ocecoaman.2015.07.027
López-Mendilaharsu M, Giffoni B, Monteiro D, Prosdocimi L, Vélez-Rubio GM, Fallabrino A et al (2020) Multiple-threats analysis for loggerhead sea turtles in the southwest Atlantic Ocean. Endanger Species Res 41:183–196. https://doi.org/10.3354/esr01025
Luschi P, Casale P (2014) Movement patterns of marine turtles in the mediterranean sea: a review. Ital J Zool 81:478–495. https://doi.org/10.1080/11250003.2014.963714
Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152:1753–1766. https://doi.org/10.1093/genetics/152.4.1753
Magris RA, Marta-Almeida M, Monteiro JAF, Ban NC (2019) A modeling approach to assess the impact of land mining on marine biodiversity: assessment in coastal catchments experiencing catastrophic events (SW Brazil). Sci Total Environ 659:828–840. https://doi.org/10.1016/j.scitotenv.2018.12.238
Marcovaldi MA, Chaloupka M (2007) Conservation status of the loggerhead sea turtle in Brazil: an encouraging outlook. Endanger Species Res 3:133–143. https://doi.org/10.3354/esr003133
Marcovaldi MÂ, Lopez GG, Soares LS, Lima EH, Thomé JC, Almeida AP (2010) Satellite-tracking of female loggerhead turtles highlights fidelity behavior in northeastern Brazil. Endanger Species Res 12:263–272. https://doi.org/10.3354/esr00308
Marcovaldi MA, López-Mendilaharsu M, Santos AS, Lopez GG, Godfrey MH, Tognin F et al (2016) Identification of loggerhead male producing beaches in the south Atlantic: implications for conservation. J Exp Mar Biol Ecol 477:14–22. https://doi.org/10.1016/j.jembe.2016.01.001
Marcovaldi MÂ, Santos AS, Lara PH, López-Mendilaharsu M (2017) Novel Research Techniques Provide New Insights to the Sea Turtle Life Cycle. Advances in Marine Vertebrate Research in Latin America. Springer, Cham, pp 169–195
Marcovaldi MA, Sales G, Thomé JC, da Silva ACCD, Gallo BM, Lima EHSM et al (2006) Sea turtles and fishery interactions in Brazil: identifying and mitigating potential conflicts. Mar Turt Newsl 112: 4–8. http://www.seaturtle.org/mtn/archives/mtn112/mtn112p4.shtml
Martín-del-Campo R, Calderón-Campuzano MF, Rojas-Lleonart I, Briseño-Dueñas R, García-Gasca A (2021) Congenital malformations in sea turtles: puzzling interplay between genes and environment. Animals 11:444. https://doi.org/10.3390/ani11020444
Matsuzawa Y, Kamezaki N, Ishihara T, Omuta K, Takeshita H, Goto K et al (2016) Fine-scale genetic population structure of loggerhead turtles in the Northwest Pacific. Endanger Species Res 30:83–93. https://doi.org/10.3354/esr00724
McCallum ML (2021) Turtle biodiversity losses suggest coming sixth mass extinction. Biodivers Conserv 30:1257–1275. https://doi.org/10.1007/s10531-021-02140-8
McClellan CM, Read AJ (2007) Complexity and variation in loggerhead sea turtle life history. Biol Lett 3:592–594. https://doi.org/10.1098/rsbl.2007.0355
Medeiros L, Monteiro DS, Botta S, Proietti MC, Secchi ER (2019) Origin and foraging ecology of male loggerhead sea turtles from southern Brazil revealed by genetic and stable isotope analysis. Mar Biol 166:76. https://doi.org/10.1007/s00227-019-3524-2
Miguel C, Costa PG, Bianchini A, Luzardo OLP, Vianna MRM, de Deus Santos MR (2022) Health condition of chelonia mydas from a foraging area affected by the tailings of the collapsed dam in southeast Brazil. Sci Total Environ 821:153353. https://doi.org/10.1016/j.scitotenv.2022.153353
Mills LS, Allendorf EW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 6:1509–1518. https://doi.org/10.1046/j.1523-1739.1996.10061509.x
Monteiro CC, Carmo HM, Santos AJ, Corso G, Sousa-Lima RS (2019) First record of bioacoustic emission in embryos and hatchlings of hawksbill sea turtles (Eretmochelys imbricata). Chelonian Conserv Biol 18:273–278. https://doi.org/10.2744/CCB-1382.1
Monzón-Argüello C, Rico C, Naro-Maciel E, Varo-Cruz N, López P, Marco A, López-Jurado LF (2010) Population structure and conservation implications for the loggerhead sea turtle of the Cape Verde Islands. Conserv Genet 11:1871–1884. https://doi.org/10.1007/s10592-010-0079-7
Moore MK, Ball RM (2002) Multiple paternity in loggerhead turtle (Caretta Caretta) nests on melbourne beach, florida: a microsatellite analysis. Mol Ecol 11:281–288. https://doi.org/10.1046/j.1365-294X.2002.01426.x
Naro-Maciel E, Becker JH, Lima EH, Marcovaldi MA, DeSalle R (2007) Testing dispersal hypotheses in foraging green sea turtles (Chelonia mydas) of Brazil. J Hered 98:29–39. https://doi.org/10.1093/jhered/esl050
Nei M (1973) Analysis of gene diversity in subdivided populations. P Natl A Sci 70:3321–3323. https://doi.org/10.1073/pnas.70.12.3321
Nei M (1977) F-statistics and analysis of gene diversity in subdivided populations. Ann Hum Genet 41:225–233. https://doi.org/10.1111/j.1469-1809.1977.tb01918.x
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. PNAS 76:5269–5273. https://doi.org/10.1073/pnas.76.10.5269
Orlóci L (2013) Multivariate analysis in vegetation research. Springer
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in excel. Population genetic software for teaching and research—an update. Bioinformatics 28:2537–2539. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pike DA (2013) Forecasting range expansion into ecological traps: climate-mediated shifts in sea turtle nesting beaches and human development. Glob Change Biol 19:3082–3092. https://doi.org/10.1111/gcb.12282
Rees AF, Alfaro-Shigueto J, Barata PCR, Bjorndal KA, Bolten AB, Bourjea J, Godley BJ (2016) Are we working towards global research priorities for management and conservation of sea turtles? Endanger Species Res 31:337–382. https://doi.org/10.3354/esr00801
Reid BN, Naro-Maciel E, Hahn AT, FitzSimmons NN, Gehara M (2019) Geography best explains global patterns of genetic diversity and postglacial co-expansion in marine turtles. Mol Ecol 28:3358–3370. https://doi.org/10.1111/mec.15165
Reis EC, Soares LS, Vargas SM, Santos FR, Young RJ, Bjorndal KA, Bolten AB, Lôbo-Hajdu G (2010) Genetic composition, population structure and phylogeography of the loggerhead sea turtle: colonization hypothesis for the Brazilian rookeries. Conserv Genet 11:1467–1477. https://doi.org/10.1007/s10592-009-9975-0
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Santos RG, Martins AS, da Nobrega FJ, Horta PA, Pinheiro HT, Torezani E et al (2011) Coastal habitat degradation and green sea turtle diets in Southeastern Brazil. Mar Pollut Bull 62:1297–1302. https://doi.org/10.1016/j.marpolbul.2011.03.004
Segura FR, Nunes EA, Paniz FP, Paulelli ACC, Rodrigues GB, Braga GUL, dos Pedreira Filho W, R., Barbosa Jr, F., Cerchiaro, G., Silva, F.F., Batista, B.L., (2016) Potential risks of the residue from Samarco’s mine dam burst (Bento Rodrigues, Brazil). Environ Pollut 218:813–825. https://doi.org/10.1016/j.envpol.2016.08.005
Shamblin BM, Faircloth BC, Dodd M, Wood-Jones ALICIA, Castleberry SB, Carroll JP, Nairn CJ (2007) Tetranucleotide microsatellites from the loggerhead sea turtle (Caretta Caretta). Mol Ecol Resour 7:784–787. https://doi.org/10.1111/j.1471-8286.2007.01701.x
Shamblin BM, Faircloth BC, Dodd MG, Bagley DA, Ehrhart LM, Dutton PH, Frey A, Nairn CJ (2009) Tetranucleotide markers from the loggerhead sea turtle (Caretta Caretta) and their cross-amplification in other marine turtle species. Conserv Genet 10:577–580. https://doi.org/10.1007/s10592-008-9573-6
Shamblin BM, Dodd MG, Williams KL, Frick MG, Bell R, Nairn CJ (2011) Loggerhead turtle eggshells as a source of maternal nuclear genomic DNA for population genetic studies. Mol Ecol Resour 11:110–115. https://doi.org/10.1111/j.1755-0998.2010.02910.x
Shamblin BM, Bolten AB, Bjorndal KA, Dutton PH, Nielsen JT, Abreu-Grobois FA et al (2012) Expanded mitochondrial control region sequences increase resolution of stock structure among North Atlantic loggerhead turtle rookeries. Mar Ecol Progress Ser 469:145–160. https://doi.org/10.3354/meps09980
Shamblin BM, Bolten AB, Abreu-Grobois FA, Bjorndal KA, Cardona L, Carreras C, Nel R (2014) Geographic patterns of genetic variation in A broadly distributed marine vertebrate: new insights into loggerhead turtle stock structure from explanded mitochondrial Dna sequences. PLoS ONE 9:E85956. https://doi.org/10.1371/journal.pone.0085956
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139(1):457–462
Soares LS, Bjorndal KA, Bolten AB, Wayne ML, Castilhos JC, Weber MI et al (2021) Reproductive output, foraging destinations, and isotopic niche of olive ridley and loggerhead sea turtles, and their hybrids, in Brazil. Endanger Species Res 44:237–251. https://doi.org/10.3354/esr01095
Stewart KR, LaCasella EL, Jensen MP, Epperly SP, Haas HL, Stokes LW, Dutton PH (2019) Using mixed stock analysis to assess source populations for at-sea bycaught juvenile and adult loggerhead turtles (Caretta caretta) in the north-west Atlantic. Fish Fish 20:239–254. https://doi.org/10.1111/faf.12336
Team RDC (2021) R Programming. R Development Core Team. https://www.r-project.org/
Thompson J, Gibson T, and Higgens D (1996) Clustal W version 1.6. EMBL: 1.
Uller T, Olsson M (2008) Multiple paternity in reptiles: patterns and processes. Mol Ecol 17:2566–2580. https://doi.org/10.1111/j.1365-294X.2008.03772.x
van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Vargas SM, Barcelos AC, Rocha RG, Guimarães P, Amorim L, Martinelli A, Santos FR, Erickson J, Marcondes ACJ, Ludwig S (2022) Genetic monitoring of the critically endangered leatherback turtle (Dermochelys coriacea) in the South West Atlantic. Reg Stud Mar Sci 25:102530. https://doi.org/10.1016/j.rsma.2022.102530
Vergilio C, Lacerda DS, da Silva ST, de Oliveira BCV, Fioresi VS, de Souza VV et al (2021) Immediate and long-term impacts of one of the worst mining tailing dam failure worldwide (Bento Rodrigues, Minas Gerais, Brazil). Sci Total Environ 756:143697. https://doi.org/10.1016/j.scitotenv.2020.143697
Vilaça ST, Piccinno R, Rota-Stabelli O, Gabrielli M, Benazzo A, Matschiner M et al (2021) Divergence and hybridization in sea turtles: inferences from genome data show evidence of ancient gene flow between species. Mol Ecol 30:6178–6192. https://doi.org/10.1111/mec.16113
Wallace BP, DiMatteo AD, Hurley BJ, Finkbeiner EM, Bolten AB, Chaloupka MY et al (2010) Regional management units for marine turtles: a novel framework for prioritizing conservation and research across multiple scales. PLoS ONE 5:e15465. https://doi.org/10.1371/journal.pone.0015465
Yipel M, Tekeli İO, İşler CT, Altuğ ME (2017) Heavy metal distribution in blood, liver and kidneys of Loggerhead (Caretta caretta) and green (Chelonia mydas) sea turtles from the Northeast mediterranean sea. Mar Pollut Bull 125:487–491. https://doi.org/10.1016/j.marpolbul.2017.08.011
Acknowledgements
The present study was carried out as part of the Aquatic Biodiversity Monitoring Program, Ambiental Area I, established by the Technical-Scientific Agreement (DOU number 30/2018) between FEST/RRDM and Renova Foundation. This study was supported in part by the IBAMA/ICMBio, the Fundação Projeto TAMAR, CAPES, FAPES, and the Federal University of Espírito Santo. We thank FEST for the scholarships, the Marcos Daniel Institute, Tommy Magalhães, and Fundação Projeto TAMAR for the fieldwork, Luciano Soares, Juliana Justino, Monique Nascimento, and the Núcleo de Genética aplicada à Conservação da Biodiversidade for technical support.
Funding
This work was supported by the IBAMA/ICMBio, Fundação Projeto Tamar, and the work of Sarah Maria Vargas was supported by Fest/RRDM and Renova Foundation, 30/2018.
Author information
Authors and Affiliations
Contributions
SL and SMV designed the study; ACB, JE, LA, and PRLG carried out field and laboratory work; SL analyzed the data and wrote the manuscript; SL, LM, and SMV reviewed the drafts; SMV supervised and coordinated the project; All authors approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no known competing financial interests or personal relationships that could have appeared to influence the publication of this paper.
Additional information
Responsible Editor: C. Eizaguirre .
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ludwig, S., Amorim, L., Barcelos, A.C. et al. Going deeper into the molecular ecology of the Southwest Atlantic Caretta caretta (Testudinata: Cheloniidae), what do microsatellites reveal to us?. Mar Biol 170, 78 (2023). https://doi.org/10.1007/s00227-023-04212-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00227-023-04212-5