Abstract
As malignant breast cancers progress, they acquire the ability to spread to other regions of the body, including bone and lung, but the molecular mechanism underlying the increase in metastatic potential is not fully understood. Here we studied murine 4T1E/M3 highly bone marrow metastatic breast cancer cells, which we established previously. These cells show upregulated expression of bone morphogenetic protein (BMP) 7 and BMP receptors, as well as augmented phosphorylation of Smad1/5/8. Both anchorage-independent cell growth measured in colony forming assays and cell migration measured in wound healing assays were suppressed in 4T1E/M3 cells following treatment with a neutralizing anti-BMP7 antibody or knockdown of BMP7 gene expression. In addition, metastasis of 4T1E/M3 cells to the spine and lung and intracellular levels of phosphorylated Smad1/5/8 were suppressed by knocking down BMP7. Conversely, overexpression of BMP7 in the weakly metastatic parental 4T1E cells augmented their anchorage-independent growth, migration and metastasis to spine and lung. Taken together, our results strongly suggest that augmented autocrine BMP7 signaling leads to increases in the anchorage-independent cell growth, migration and metastatic potential in our bone marrow metastatic breast cancer model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bone is a frequent target of breast cancer metastasis. Therefore, to better understand the mechanism by which breast cancer metastasizes to bone marrow, we previously established the 4T1E/M3 highly bone marrow metastatic murine breast cancer cell line [1]. Then using these cells, we showed that the chemokine CCL2/MCP-1 negatively regulates their growth, migration and metastasis [2].
Bone morphogenetic proteins (BMPs) are secreted signaling molecules belonging to the transforming growth factor β (TGF-β) superfamily and play a variety of important biological roles during embryonic development and organ morphogenesis, and are also involved in the regulation of cell proliferation, differentiation, apoptosis and chemotaxis [3, 4]. In addition, BMP7 (also called osteogenic protein-1) can induce osteogenic differentiation of newborn rat calvarial and osteosarcoma cells [5], and overexpression of BMP4, BMP6 and BMP7 mRNA has been detected in bone-metastasized prostate cancers [6]; in fact, levels of BMP7 mRNA are higher in bone-metastasized prostate cancer than in bone itself [7].
BMPs evoke their cellular effects via specific type I and II serine/threonine receptors [8]. The activated BMP type I receptor phosphorylates specific receptor-regulated Smad1/5/8 proteins, which then form heteromeric complexes with a common partner, Smad4 [9]. These heteromeric Smad complexes translocate into the nucleus where they regulate the transcription of target genes [10]. The purpose of this study was to assess the involvement of BMP 7 signaling in the metastatic activity of our highly metastatic 4T1E/M3 cells.
Materials and methods
Animals
Seven- to eight-week-old female BALB/c mice were purchased from Japan Clea (Tokyo, Japan).
4T1E/M3 cell culture
The highly bone metastatic mouse 4T1E/M3 breast cancer cell line was established as described previously [1]. Briefly, 4T1 murine breast cancer cells were initially transfected with the neomycin resistance gene for in vitro selection. After selecting the transfectants (4T1E cells) in medium containing 120 μg/ml G418 (Invitrogen, Carlsbad, CA, USA), they were injected into the tail vein of BALB/c mice (1 × 106 cells/mouse). Twelve days later, bone marrow cells were collected from the femurs and tibias and cultured in the medium containing 120 μg/ml G418. The growing cells were then intravenously injected into a new set of mice. This cycle was repeated three times to establish the 4T1E/M3 cells. 4T1E/M3 and other cells were cultured in RPMI 1640 medium supplemented with 2 mM l-glutamine, 1.5 g/l sodium bicarbonate, 4.5 g/l glucose, 10 mM HEPES, 1.0 mM sodium pyruvate and 10% fetal calf serum (Cancera International Inc., Ontario, Canada) at 37°C under a humidified atmosphere containing 5% CO2.
Cell proliferation assay
Control/EM3, BMP7(-)/EM3, control/4T1E or BMP7(+)/4T1E cells (1 × 104/ml, 200 μl/well) were seeded into 96-well plates and cultured as described above. On the same day, or 1–4 days later, 10 μl of Cell Counting Kit-8 reagent (a modified MTT assay system; Dojin Laboratories, Tokyo, Japan) were added to each well, and the plates were incubated for an additional 4 h, after which the absorbance at 450 nm was measured using a microplate reader (Model 680, Bio Rad Laboratories, Inc., Tokyo, Japan).
DNA microarray analysis
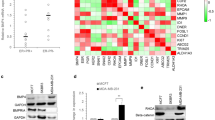
Total RNA was isolated from 4T1E and 4T1E/M3 cells using an RNeasy Midi Kit (QIAGEN, MD, USA) according to the manufacture’s protocol and then treated with an RNase-free DNase set (QIAGEN) to remove any contaminating chromosomal DNA. Thereafter, 2-μg samples were used for microarray analysis performed by Takara Biotechnology Ltd. (Osaka, Japan) using a GeneChip mouse genome 430 2.0 array (Affymetrix Co., 34,000 genes were analyzed). Fluorescence intensities were measured for each probe, and expression profiles were analyzed using Affymetrix Microarray Suite ver. 5 software. Values were calculated by comparing each probe pair between the experimental (4T1E/M3) and baseline (4T1E) arrays, and changes were expressed as the log 2 ratio.
RNA isolation and quantitative RT-PCR
Total RNA was isolated from 4T1E, 4T1E/M3, control/EM3, BMP7(−)/EM3, control/4T1E or BMP7(+)/4T1E cells using an RNeasy Mini Kit (QIAGEN) according to the manufacture’s protocol. cDNA was then synthesized using 1-μg samples of the total RNA with a First Strand cDNA Synthesis Kit (Roche Diagnostics) in a PTC-200 Peltier thermal cycler (Bio-Rad MJ Research, Waltham, MA, USA). The RT protocol entailed incubation at 25°C for 10 min, 55°C for 30 min, 85°C for 5 min and 0°C for 5 min. Specific forward and reverse primers were designed using Primer 3 software (Genetyx Co., Tokyo, Japan) and synthesized by Fasmac Co. Ltd. (Kanagawa, Japan). The primer sequences (5′–3′) and the estimated sizes of the PCR products are shown in Table 1. The quantitative RT-PCR (qRT-PCR) reactions were set up according to the Light Cycler manual (Roche Diagnostics) using a Light Cycler Fast Start DNA Master SYBR Green I Kit (Roche Diagnostics). The PCR protocol entailed denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 10 s, annealing at 59°C for 10 s, extension at 72°C for 11 s; and a final cooling step at 40°C for 30 s. For each reaction, the crossing point (defined as the cycle number at which the noise band intersects the fluorescence curves) was determined using the “Second Derivative Maximum Method” in the Light Cycler software (ver. 3.52, Roche Diagnostics). Relative mRNA levels were calculated using the software with a standard curve constructed using various concentrations of a 1:1 mixture of the RT product and then normalized to GAPDH mRNA.
Western blotting
4T1E, 4T1E/M3, control/EM3, BMP7(−)/EM3, control/4T1E or BMP7(+)/4T1E cells were seeded onto 6-cm dishes and treated with Complete Lysis buffer (Roche Diagnostics) following the manufacture’s protocol, after which the protein obtained was measured using a BCA Protein Assay Kit (Pierce, Rockford, IL, USA). The proteins were then resolved by SDS-PAGE and transferred to i-Blot Gel Transfer Stacks PVDF membranes (Invitrogen) using an i-Blot Dry Blotting System (Invitrogen). After blocking the membranes with blocking solution (GE Healthcare) or skim milk, they were incubated for 1 h or overnight at room temperature with anti-mBMP7 (1/1000, rabbit polyclonal IgG, ab56023; Abcam, San Francisco, CA), anti-BMPRIB (1/200, rabbit polyclonal IgG, sc-25455), anti-BMPR2 (1/200, goat polyclonal IgG, sc-5682; Santa Cruz Biotech Inc., Santa Cruz, CA, USA), anti-p-Smad1/5/8 (1/1000, rabbit polyclonal IgG, #9511; Cell signaling Tech. Inc., Danvers, MA, USA), anti-Smad1/5/8 (1/200, rabbit polyclonal IgG, sc-6031R, Santa Cruz), anti-Smad 4 (1/200, rabbit polyclonal IgG, sc-7154, Santa Cruz) or anti-β-actin (1/2000, rabbit polyclonal IgG, sc-7210, Santa Crus) diluted in Can Get signal immunoreaction Enhancer Solution 1 (Toyobo, Tokyo, Japan) or TBS-T. The membranes were then washed 3 times with TBS-T and incubated for 1 h at room temperature with either HRP-anti-rabbit IgG (1/5000, GE Healthcare) or HRP-anti-goat IgG (1/5000, GE Healthcare) diluted in Can Get signal immunoreaction Enhancer Solution 2 (Toyobo) or TBS-T. Finally, the membranes were treated with ECL Plus or Prime detection reagent (GE Healthcare) according to manufacture’s protocol, and the chemiluminescence was detected using Chemi Doc XRS (Bio-Rad Laboratories. Inc.). Quantitative analysis of the Western blot bands was carried out using Quantity One software Ver.3.0 (Bio-Rad Laboratories. Inc.).
RNA interference
For vector-based RNAi analysis, we synthesized miRNA oligo DNA (AGCTGTACAAGTAAGCTAAGCACTTCGTGGCCGTCGATCGTTTAAAGGGAGGTAGTGAGTCGACCAGTGGATCCTGGAGGCTTGCTGAAGGCTGTATGCTGAGTAGTAGGCAGCATAGCCTTGTTTTGGCCACTGACTGACAAGGCTATTGCCTACTACTCAGGACACAAGGCCTGTTACTAGCACTCACATGGAACAAATGGCCCAGATCTGGCCGCACTCGAGATATCTAGACCCAGCTTTCTTGTACAAAGTGGTTG) using Bmp7 BLOCK-iT™ miR RNAi Select (Invitrogen), Mmi 505271_top_Bmp7 (TGCTG AGTAGTAGGC AGCATAGCCT TGTTTTGGCC ACTGACTGAC AAGGCTATTG CCTACTACT) and Mmi505271_bot_Bmp7 (CCTG AGTAGTAGGC AATAGCCTTG TCAGTACAGT GGCCAA ACAA GGCTATGCTG CTGCCTACTA CTC) (Invitrogen). The sequence was inserted into an expression vector to obtain pcDNA6.2-GW/EmGFP-miR-mBMP7, the sequence of which was verified by Life Technologies, Inc. (Tokyo, Japan). The plasmid pcDNA6.2-GW/EmGFP-miR-neg, which carries a scrambled sequence and cannot target any known vertebrate gene, served as a control non-targeting vector. To knock down the targeted genes, 4T1E/M3 cells were transfected with the vector using Lipofectamine LTX (Invitrogen) following the manufacturer’s protocol to yield stable control/EM3 and BMP7(−)/EM3 clones.
Mouse BMP7 expression vector
Full-length cDNA encoding mouse BMP7 was amplified by PCR using the primers mBMP7 5′w/BamHI (5′-CGGGATCCATGCACGTGCGCTCGCTGCG-3′) and mBMP7 3′w/XhoI: (5′-CCGCTCGAGTGTGGCAGCCACAGGCCCGGA-3′). Plasmid pcDNA3.1/(FLAG)3-(His)6 (Invitrogen) was then digested using BamHI and XhoI, and the mBMP7 cDNA was inserted. This was followed by digestion of plasmid pcDNA3.1w/puromycin [11] using PmeI and insertion of mBMP7 cDNAw/(FLAG)3-(His)6, yielding pcDNA3.1w/puromycin/mBMP7-(FLAG)3-(His)6. The final plasmid constructs were confirmed by sequencing using an ABI310 DNA sequencer (Applied Bioscience). Lipofectamine LTX (Invitrogen) or Fu GENE HD (Roche Diagnostics, Indianapolis, IN, USA) was then used to establish transient control/4T1E or BMP7(+)/4T1E transfectants by transfecting pcDNA3.1w/puromycin or pcDNA3.1w/puromycin/mBMP7-(FLAG)3-(His)6, into 4T1E cells.
Anchorage-independent proliferation assay
Anchorage-independent colony formation in soft agar was assayed as described previously [2]. Briefly, 4T1E, 4T1E/M3, control/EM3, BMP7(−)/EM3, control/4T1E or BMP7(+)/4T1E cells (1 × 104/ml, 2 ml/dish) were seeded into culture medium containing 0.3% agarose (Difco noble agar; BD Diagnostic Systems, Sparks, MD, USA). The agarose-containing culture medium was then layered onto 1 or 0.5% agarose medium (5 ml/dish) in 6-cm dishes. When seeding the cells, neutralizing anti-BMP7 antibody (final concentration, 8, 16 or 32 μg/ml, mouse IgG2B, MAB3541; R&D Systems, Minneapolis, MN, USA) or mouse IgG isotype control antibody (final concentration, 8 μg/ml, MAB002, R&D systems) was added to the medium. The cells were then cultured for 12 days at 37°C under 5% CO2, after which the total numbers of colonies per dish were counted under a microscope. During the 12 days incubation, anti-BMP7 antibody existed in the medium but it was not added again.
Wound healing assay
4T1E, 4T1E/M3, control/EM3, BMP7(-)/EM3, control/4T1E or BMP7(+)/4T1E cells (1 × 105/ml, 1 ml/well) were seeded into 24-well plates and incubated for 2 days. After confluent monolayers had formed, they were wounded by scratching in lines using a plastic pipette tip. The wells were then washed twice to remove any debris and observed and photographed under a microscope. Neutralizing anti-BMP7 antibody or an isotype control (8 μg/ml, R&D system) was also added to the wells at that time. The plates were then incubated for 8 h at 37°C under 5% CO2, after which the cells were photographed, and the distance they had migrated was measured on the photomicrographs. The relative percent wounded area filled through cell migration was then calculated as follows: [(mean wound width − mean remaining width)/mean wound width] × 100 (%).
In vivo experiment
4T1E, 4T1E/M3, control/EM3, BMP7(−)/EM3, control/4T1E or BMP7(+)/4T1E cells were intravenously injected into BALB/c mice (1 × 106/mouse), after which tissue metastasis was monitored and evaluated. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Institute of Advanced Industrial Science and Technology (AIST).
Histology
Sections of lung, liver, heart, kidney, spleen, brain and bone (femur, tibia and spine) harvested from mice were fixed in 10% neutral buffered formalin (Wako, Osaka, Japan), processed, embedded in paraffin, cut into 5–6-μm sections, stained with H&E, and examined and photographed under a microscope. Before embedding the bone, it was decalcified in decalcifying solution A (Wako) following the manufacture’s protocol. The percent metastatic area per lung in the histological sections was calculated using the ROI Manager analysis tool in ImageJ 1.42 (NIH, Bethesda, MD, USA).
Statistical Analysis
Differences in mRNA expression, anchorage-independent growth and wound healing were analyzed using one-way ANOVA followed by Bonferroni’s multiple comparisons test. GraphPad Prism (Version 4; GraphPad Software, San Diego, CA, USA) was used for all statistical analyses.
Results
BMP7 expression is significantly increased in 4T1E/M3 metastatic breast cancer cells
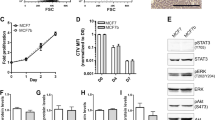
It has been reported that BMP7 is overexpressed in breast cancer, and that the increased expression is associated with the occurrence of bone metastasis [12]. In the same vein, bone-metastasized prostate cancers show upregulated expression of BMP4, BMP6 and BMP7 mRNA [6]. We therefore used a DNA microarray to compare the expression of BMP family genes in our highly metastatic 4T1E/M3 cells with their expression in the weakly metastatic parental 4T1E cells. We found that, as compared to 4T1E cells, levels of BMP7 gene expression were upregulated in 4T1E/M3 cells (log fold = 4.9; Table 2). Subsequent qRT-PCR analysis confirmed that expression of BMP7 mRNA is significantly (4.8 times) increased in 4T1E/M3 cells (Fig. 1A), and western blot analysis showed that BMP7 protein is also increased (2.4 times) in 4T1E/M3 cells (Fig. 1B, C).
Establishment of BMP7 knockdown 4T1E/M3 cells and expression of BMP7 A Quantitative real-time RT-PCR (qRT-PCR) analysis of BMP7 expression. Levels of the indicated mRNAs in 4T1E, 4T1E/M3, control/EM3 and BMP7(−)/EM3 cells are shown relative to the level of GAPDH mRNA. Values are means ± SD; **P < 0.001. B Western blot analysis of BMP7 in lysates of 4T1E, 4T1E/M3, control/EM3 and BMP7(−)/EM3 cells. C Relative density of the bands in B. *P < 0.01
4T1E/M3 cells express higher levels of BMPR1B and BMPR2
We then examined the mRNA and protein expression of BMP receptors using PCR and western blotting, respectively. As shown in Fig. 2A, expression of BMPR1B and BMPR2 mRNAs was significantly upregulated in 4T1E/M3 cells (shadowed columns), as compared to the parental 4T1E cells (solid columns): 1.8 times (BMPR1B) and 2.2 times (BMPR2) increase, respectively. Likewise, levels of BMPR1B (1.9 times) and BMPR2 (1.6 times) proteins were also increased in 4T1E/M3 cells (Fig. 2B, C). As for other receptors, Acvr1, Acvr2a, and Acvr2b, there was no significant difference of the mRNA expression between that in 4T1E and 4T1E/M3 (data not shown).
Expression of BMP receptors A qRT-PCR analysis of BMPR mRNA expression in 4T1E and 4T1E/M3 cells. Levels of BMPRIA, -IB and -2 mRNA are shown relative to that of GAPDH mRNA. Black bars represent 4T1E; hatched bars represent 4T1E/M3. Values are means ± SD; *P < 0.01, ns Not significant. B Western blot analysis of BMPR expression. Expression of BMPRIB and -2 in lysates of 4T1E and 4T1E/M3 cells was measured by western blotting. C Relative density of the bands in B. *P < 0.01
Smad1/5/8 is phosphorylated in 4T1E/M3 cells
Because we found that 4T1E/M3 cells express both BMP7 and its receptor components (i.e., BMPR1B/BMPR2) at higher levels than 4T1E cells, we also compared their intracellular signaling. When we analyzed the cell lysates, we detected higher levels of phosphorylated (p)-Smad1/5/8 in 4T1E/M3 cells than in 4T1E cells (Fig. 3). On the other hand, similar levels of unphosphorylated Smad1/5/8 and Smad 4 were detected in 4T1E/M3, and 4T1E cells (Fig. 3).
Knocking down BMP7 gene expression suppresses both regulatory Smad activation and cell proliferation
To assess the effects of the augmented BMP7 expression in 4T1E/M3 cells, we established a stable transfectant clone, BMP7(−)/EM3, in which BMP7 was knocked by miR RNAi. Then using qRT-PCR, we confirmed that BMP7 mRNA was expressed at a lower level in BMP7(−)/EM3 cell than in the mock transfectants (control/EM3 cells) (Fig. 1A). Consistent with that finding, western blotting showed that BMP7 protein was also expressed at a lower level in BMP7(−)/E/M3 cell than in control/EM3 cells or in untransfected 4T1E/M3 cells (Fig. 1B). Relative density of the band from BMP7(−)/EM3 was less than half of that from control/EM3 (Fig. 1C). Furthermore, levels of p-Smad1/5/8 were clearly lower in BMP7(−)/EM3 cells than in control/M3 cells, while levels of unphosphorylated Smad1/5/8 were not changed (Fig. 3). Proliferation of BMP7(−)/EM3 cells was slightly slower than that of control/EM3 cells (Fig. 4A).
Proliferation, anchorage-independent growth and migration of BMP7 knockdown 4T1E/M3 cells. A Growth curves for control/EM3 and BMP7(-)/EM3 cells measured in MTT assays. Values are means ± SD; *P < 0.001. B, C, D Anchorage-independent growth of BMP7(−)/EM3 cells measured in colony formation assays. 4T1E/M3, control/EM3 and BMP7(−)/EM3 cells (1 × 104 cells/dish, 3 dishes/cell type) were cultured for 12 days in medium containing 0.3% agarose layered on 0.5% agarose. B Representative photomicrograph of colonies. C Values are the average size of the colonies in each dish ± SD; **P < 0.001; D Values are the average numbers of colonies per dish ± SD; **P < 0.001. E, F Migration of BMP7(−)/EM3 cells measured in wound-healing assays. Confluent monolayers of control/EM3 and BMP7(−)/EM3 cells in 24-well plates were wounded by scratching and then cultured for an additional 8 h. E Representative photomicrograph of the monolayers and wounds. F Migration was assessed on the basis of percent wound area recovered. Data are means ± SD; ***P < 0.0001
BMP7(−)/EM3 cells show reduced potential for anchorage-independent growth and cell migration
High potentials for anchorage-independent cell growth and cell migration are well-known in vitro properties related to cancer metastasis and malignancy. Moreover, we previously demonstrated that 4T1E/M3 cells exhibit greater anchorage-independent cell growth and migration than the parental 4T1E cells [1]. We therefore next used colony formation and wound-healing assays to evaluate the effect of BMP7 suppression on anchorage-independent cell growth and cell migration, respectively. We found that BMP7(−)/EM3 cells showed significantly less anchorage-independent growth (Fig. 4B–D) and migration than control/EM3 cells (Fig. 4E, F). The size of the colonies formed by BMP7(−)/EM3 cells was smaller than those formed by control/EM3 cells (Fig. 4B), mean colony size of BMP7(−)/EM3 and control/EM3 was 63.7 and 134.7 μm, respectively (Fig. 4C). BMP7(−)/EM3 cells formed significantly fewer colonies than control/EM3 cells, mean colony number of BMP7(−)/EM3 and control/EM3 was 26.3 and 75.3, respectively (Fig. 4D). In addition, wound healing, reflecting cell migration, was significantly downregulated in BMP7(−)/EM3 cells, as compared to control/EM3 cells (Fig. 4E, F), mean percent wounded area filled of control/EM3 and BMP7(−)/EM3 was 50.1 and 36.1, respectively (Fig. 4F).
Neutralizing anti-BMP7 antibody suppresses anchorage-independent growth and migration of 4T1E/M3 cells
When a neutralizing anti-BMP7 antibody was added to colony formation and wound-healing assays, both the migration of 4T1E/M3 cells (Fig. 5A) and their anchorage-independent growth (Fig. 5B, C) were significantly suppressed. Mean percent wounded area filled after the addition of control and anti-BMP7 antibodies was 77.0 and 51.0, respectively (Fig. 5A). Because this result suggested that 8 μg/ml antibody was sufficient to suppress cellular migration (Fig. 5A), colony formation was also evaluated under this condition. We found that the anti-BMP7 antibody significantly suppressed anchorage-independent growth of 4T1E/M3 cells: both the size (Fig. 5B) and number (Fig. 5C) of the colonies formed were clearly diminished by treatment with anti-BMP7. Mean size of the colonies after the addition of control and anti-BMP7 antibodies was 123.8 and 63.5 μm, respectively (Fig. 5B) and mean number of those was 48.3 and 21.6, respectively (Fig. 5C). Taken together, these results suggest that BMP7 secreted by 4T1E/M3 cells exerts autocrine effects that enhance the growth and migration of 4T1E/M3 cells.
Effects of neutralizing anti-BMP7 antibody on the migration and anchorage-independent growth of 4T1E/M3 cells. A Effect of anti-BMP7 antibody on 4T1E/M3 cell migration. Neutralizing anti-BMP7 antibody (black bars) or control IgG (hatched bars) was added at the indicated concentrations to migration assays. Values are means ± SD; **P < 0.001. B, C Effects of neutralizing anti-BMP7 antibody (Ab) on anchorage-independent growth of 4T1E/M3 cells in soft agar. 4T1E or 4T1E/M3 cells (1 × 104 cells/dish, 3 dishes/cell type) were left untreated or treated with neutralizing anti-BMP7 antibody or control Ab (8 μg/ml as IgG) for 12 days in medium containing 0.3% agarose layered on 0.5% agarose. B Values are the average size of the colonies in each dish ± SD; **P < 0.001. C Values are the average numbers of colonies per dish ± SD; *P < 0.01
BMP7(−)/EM3 cells show reduced metastasis to bone marrow and lung
To investigate the effects of BMP7 knockdown on bone metastasis, we intravenously injected BMP7(−)/EM3 or control/EM3 cells into syngenic female BALB/c mice via the tail vein. We then evaluated metastasis to various organs histologically. As shown in Table 3, injection of BMP7(−)/EM3 cells resulted in a rather lower incidence of spinal metastasis than injection of control/EM3 cells (60 vs. 80%, respectively, P < 0.01). No metastasis was found in liver, heart, kidney, spleen or brain although small foci could have been missed in soft tissue sites. There was no significant difference of the size of spinal metastasis lesions. BMP7(−)/EM3 cells also metastasized to lung, but the number of metastatic tumors and the size of the area affected by those tumors were significantly smaller than in mice administered control/EM3 cells: the mean percent metastatic area per lung was 9.8 ± 2.35% in BMP7(−)/EM3-injected mice, but was 70.4 ± 3.26% in control/EM3-injected mice (Table 4, P < 0.01). Representative microscopic images of well developed metastatic tumors in a lung from a control/EM3-injected mouse and of sparsely distributed tumors in a lung from a BMP7(−)/EM3-injected mouse are shown in Fig. 6.
Histological examination of the lungs of mice injected by BMP7(−)/EM3 cells. Shown are representative histological sections of lung. Cancer metastasis in lung samples collected 10 days after injection of control/EM3 or BMP7(−)/EM3 cells (1 × 106 cells/mouse) was detected using H&E staining. Images are shown at magnifications of ×4 and ×40; the bars indicate 500 and 25 μm, respectively
Overexpression of BMP7 in parental 4T1E cells enhances anchorage-independent cell growth and migration
To further investigate the effects of BMP7, we transiently transfected 4T1E cells with a mouse BMP7 expression vector (BMP7(+)/4T1E cells), and confirmed that the transfectants expressed higher levels (141 times) of BMP7 than mock transfectants (control/4T1E; Fig. 7A). When assessed in MTT assays, the rate of BMP7(+)/4T1E cell proliferation was slightly higher than control (Fig. 7B). Moreover, colony formation and wound-healing assays showed anchorage-independent growth (Fig. 8A–C) and migration (Fig. 8D, E) to be significantly enhanced in BMP7(+)/4T1E cells. Both size (Fig. 8A, B) and the number (Fig. 8C) of the colonies formed by BMP7(+)/4T1E cells were larger than those formed by control/4T1E cells. Mean size of the control/4T1E and BMP(+)/4T1E colonies was 74.2 and 167.9 μm, respectively (Fig. 8B) and mean number of those was 24.3 and 44.0, respectively (Fig. 8C). Mean percent wounded area filled in control/4T1E and BMP(+)/4T1E cells was 45.2 and 65.7, respectively (Fig. 8E).
Establishment of BMP7-overexpressing 4T1E cells and their growth. A qRT-PCR analysis of BMP7 mRNA expression in mock transfected 4TE1 cells (control/4T1E) and in BMP7(+)/4T1E cells. Levels of BMP7 mRNA are shown relative to that of GAPDH mRNA. Values are means ± SD; ***P < 0.0001. B Growth curves for the control/4T1E and BMP7(+)/4T1E cells measured in MTT assays. *P < 0.05, ***P < 0.001
Anchorage-independent growth and migration of 4T1E cells overexpressing BMP7. A, B, C Anchorage-independent growth of BMP7(−)/EM3 cells measured in colony formation assays. Mock transfected control/4T1E cells and BMP7(+)/4T1E cells (1 × 104 cells/dish, 3 dishes/cell type) were cultured for 12 days in medium containing 0.3% agarose layered on 0.5% agarose. A Representative images of colonies. Bars 100 μm. B Values are the average size of the colonies in each dish ± SD; **P < 0.001; C Values shown are the average number of colonies per dish ± SD; *P < 0.01. D, E Migration of BMP7(+)/4T1E cells measured in wound-healing assays. Confluent monolayers of control/EM3 and BMP7(−)/EM3 cells in 24-well plates were wounded by scratching and then cultured for an additional 8 h. D Representative photomicrographs of the monolayers and wounds. E Migration was assessed on the basis of percent wound area recovered. Data are means ± SD; **P < 0.001
BMP7(+)/4T1E cells show enhanced metastatic activity
We then assessed the metastatic activity of BMP7(+)/4T1E cells 10 days after injecting them into mice via the tail vein. Histological analysis showed that the incidences of spinal metastasis were greater in mice administered BMP7(+)/4T1E (67%) than in mice administered control/4T1E cells (20%) (Table 3). No metastasis was found in liver, heart, kidney, spleen or brain. However, small foci could have been missed in soft tissue sites. There was no significant difference of the size of spinal metastasis lesions. Metastasis to lung was observed in all the mice injected (Table 3), but the lung tumors formed by BMP7(+)/4T1E cells were much larger and more invasive: in mice administered by BMP7(+)/4T1E cells, the mean percent metastatic area per lung was 73.8 ± 2.76%, but it was only 12.5 ± 1.87% (P < 0.01) in mice administered control/4T1E cells (Table 4). Representative photographs are shown in Fig. 9.
Histological examination of lungs from mice injected with BMP7(+)/4T1E cells. Cancer metastasis in lung samples collected 10 days after injection of control/4T1E or BMP7(+)/4T1E cells (1 × 106 cells/mouse) was detected using H&E staining. Representative images are shown at magnifications of ×4 and ×40; the bars indicate 500 and 25 μm, respectively
Discussion
Bone metastasis in breast cancer patients poses a serious clinical problem, as it causes significant disease-specific morbidity and mortality. We recently established the mouse 4T1E/M3 breast cancer cells, which exhibit greater bone metastasis and malignancy than the parental 4T1E cells, as judged by their greater lethality, anchorage-independent cell growth and cell migration [1]. In the present study, we have shown that there is a strong correlation between the BMP7 signaling and the metastatic potential of these cells.
The mechanism by which upregulated BMP7 enhances the growth, migration and metastatic potential of 4T1E/M3 cells remains unclear at present. One possibility is that BMP7 modulates other factors to induce growth and migration. It has been reported, for example, that BMP7 signaling modulates wound healing through regulation of the extracellular matrix protein matrilin 2 [13]. Another possibility is that BMP7 modulates the secretion of matrix metalloproteinases (MMPs), which in turn regulate cell migration and metastasis. BMP4 was previously reported to control the expression of MMPs in melanoma cells [14].
It was also previously reported that BMP7 reverses TGF-β1-induced epithelial to mesenchymal transition (EMT) through re-induction of E-cadherin (epithelial marker) [15, 16]. On the other hand, BMPs reportedly induce EMT in PC-3 prostate cancer cells [17].
The role played by BMP7 in tumor progression and metastasis is complex. Our present results are consistent with those of several earlier studies in which tumor cells autonomously expressing high levels of BMP7 showed enhanced migration and metastasis [12, 18]. In human breast tumor cells, BMP7 expression correlated with their metastatic potential and a poor prognosis [19]. BMP7 is also strongly expressed in colorectal cancer cells, where its levels were found to correlate with the invasiveness of the cells and liver metastasis [20]. The level of BMP7 expression also correlates with the metastatic potential of melanoma cells [21]. All of these results suggest that upregulated expression of BMP7 contributes to the metastatic potential of tumor cells. The 4T1E/M3 breast cancer cells we studied here were originally established as highly metastatic cells through in vivo injection cycling. We have now shown that they endogenously express high levels of BMP7. Furthermore, the results of our present BMP7 knockdown and overexpression experiments clearly show that augmented autocrine BMP7 signaling increases the metastatic potential of these cells, and enhances anchorage-independent cell proliferation and cell migration.
Notably, BMP7 appears to exert the opposite effects in some cells. For example, intravenous administration of BMP7 significantly inhibits intrabone growth of MDA-231 human breast cancer cells in nude mice [22]. In addition, BMP7 reduces the invasiveness and migration of human melanoma cells [23], as well as the motility of lung cancer cells [24]. We speculate that because BMP7 was administered to the cells as an exogenous factor in some studies, the resultant cellular signaling may differ from the autocrine signaling. Consistent with that idea, studies have shown that exogenously added signaling molecules may act differently than endogenously expressed ones [25, 26]. Alternatively, it may be that the effect of BMP7 is dependent on cell type [27]. BMP7 reportedly promotes prostate cancer cell migration and invasion, but the biological responses to BMP7 were cell type-specific [16]. The effects of BMP7 on prostate cancer cells also reportedly depend on cell phenotype and the tumor microenvironment [28]. Similarly, TGF-β is reported to have diverse and conflicting effects on tumor progression [29]. Considering the fact that breast cancer patient themselves represent a heterogenous group of cancers, there is a benefit of our model to unravel mechanisms behind variability.
Little is known about the effect of BMP7 to lung metastasis. However, because overexpression of BMP7 on 4T1E cells promoted migration and anchorage-independent growth activity, and knockdown of BMP7 in 4T1E/M3 cells reduced these activities, it seems reasonable that BMP7 promoted the metastasis not only to bone but also to lung.
Further study of the mechanisms by which BMP7 expression and signaling are modulated in tumor cells is needed and should provide new and deeper insight into the mechanisms underlying breast cancer metastasis as well as strategies for its effective prevention.
References
Takahashi M, Furihata M, Akimitsu N, Watanabe M, Kaul S, Yumoto N, Okada T (2008) A highly bone marrow metastatic murine breast cancer model established through in vivo selection exhibits enhanced anchorage-independent growth and cell migration mediated by ICAM-1. Clin Exp Metastasis 25:517–529
Takahashi M, Miyazaki H, Furihata M, Sakai H, Konakahara T, Watanabe M, Okada T (2009) Chemokine CCL2/MCP-1 negatively regulates metastasis in a highly bone marrow-metastatic mouse breast cancer model. Clin Exp Metastasis 26:817–828
Chen D, Zhao M, Mundy GR (2004) Bone morphogenetic proteins. Growth Factors 4:233–241
Ye L, Lewis-Russell JM, Kyanaston HG, Jiang WG (2007) Bone morphogenetic proteins and their receptor signaling in prostate cancer. Histol Histopathol 10:1129–1147
Asahina I, Sampath TK, Hauschka PV (1996) Human osteogenic protein-1 induces chondroblastic, osteoblastic, and/or adipocytic differentiation of clonal murine target cells. Exp Cell Res 222:38–47
Autzen P, Robson CN, Bjartell A, Malcolm AJ, Johnson MI, Neal DE, Hamdy FC (1998) Bone morphogenetic protein 6 in skeletal metastases from prostate cancer and other common human malignancies. Br J Cancer 78:1219–1223
Masuda H, Fukabori Y, Nakano K, Takezawa Y, CSuzuki T, Yamanaka H (2003) Increased expression of bone morphogenetic protein-7 in bone metastatic prostate cancer. Prostate 54:268–274
Miyazono K, Kamiya Y, Morikawa M (2010) Bone morphogenetic protein receptors and signal transduction. J Biochem 147:35–51
Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K (2002) Two major Smad pathways in TGF-beta superfamily signalling. Genes Cells 12:1191–1204
Massagué J, Blain SW, Lo RS (2000) TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103:295–309
Asada M, Honda E, Imamura T (2006) Construction of pcDNA 3.1-based vectors with blasticidin and puromycin resistant markers. Anal Biochem 352:305–307
Alarmo EL, Pärssinen J, Ketolainen JM, Savinainen K, Karhu R, Kallioniemi A (2009) BMP7 influences proliferation, migration, and invasion of breast cancer cells. Cancer Lett 275:35–43
Ichikawa T, Suenaga Y, Koda T, Ozaki T, Nakagawara A (2008) DeltaNp63/BMP-7-dependent expression of matrilin-2 is involved in keratinocyte migration in response to wounding. Biochem Biophys Res Commun 369:994–1000
Rothhammer T, Braig S, Bosserhoff AK (2008) Bone morphogenetic proteins induce expression of metalloproteinases in melanoma cells and fibroblasts. Eur J Cancer 44:2526–2534
Zeisberg M, Hanai J, Sugimoto H, Mammoto T, Charytan D, Strutz F, Kalluri R (2003) BMP-7 counteracts TGF-beta1-induced epithelial-to-mesenchymal transition and reverses chronic renal injury. Nat Med 9:964–968
Rees JR, Onwuegbusi BA, Save VE, Alderson D, Fitzgerald RC (2006) In vivo and in vitro evidence for transforming growth factor-beta1-mediated epithelial to mesenchymal transition in esophageal adenocarcinoma. Cancer Res 66:9583–9590
Yang S, Zhong C, Frenkel B, Reddi AH, Roy-Burman P (2005) Diverse biological effect and Smad signaling of bone morphogenetic protein 7 in prostate tumor cells. Cancer Res 65:5769–5777
Rothhammer T, Poser I, Soncin F, Bataille F, Moser M, Bosserhoff AK (2005) Bone morphogenic proteins are overexpressed in malignant melanoma and promote cell invasion and migration. Cancer Res 65:448–456
Alarmo EL, Korhonen T, Kuukasjärvi T, Huhtala H, Holli K, Kallioniemi A (2008) Bone morphogenetic protein 7 expression associates with bone metastasis in breast carcinomas. Ann Oncol 19:308–314
Motoyama K, Tanaka F, Kosaka Y, Mimori K, Uetake H, Inoue H, Sugihara K, Mori M (2008) Clinical significance of BMP7 in human colorectal cancer. Ann Surg Oncol 15:1530–1537
Rothhammer T, Wild PJ, Meyer S, Bataille F, Pauer A, Klinkhammer-Schalke M, Hein R, Hofstaedter F, Bosserhoff AK (2007) Bone morphogenetic protein 7 (BMP7) expression is a potential novel prognostic marker for recurrence in patients with primary melanoma. Cancer Biomark 3:111–117
Buijs JT, Henriquez NV, van Overveld PG (2007) Bone morphogenetic protein 7 in the development and treatment of bone metastases from breast cancer. Cancer Res 67:8742–8751
Na YR, Seok SH, Kim DJ et al (2009) Bone morphogenetic protein 7 induces mesenchymal -to-epithelial transition in melanoma cells, leading to inhibition of metastasis. Cancer Sci 100:2218–2225
Chen J, Ye L, Xie F, Yang Y, Zhang L, Jiang WG (2010) Expression of bone morphogenetic protein 7 in lung cancer and its biological impact on lung cancer cells. Anticancer Res 4:1113–1120
Uruno T, Oki J, Ozawa K, Miyakawa K, Ueno H, Imamura T (1999) Distinct regulation of myoblast differentiation by intracellular and extracellular fibroblast growth factor-1. Growth Factors 17:93–113
Schoner W, Scheiner-Bobis G (2007) Endogenous and exogenous cardiac glycosides: their roles in hypertension, salt metabolism, and cell growth. Am J Physiol Cell Physiol 293:C509–C536
Fournier PG, Guise TA (2007) BMP7: a new bone metastases prevention? Am J Pathol 171:739–743
Morrissey C, Brown LG, Pitts TE, Vessella RL, Corey E (2010) Bone morphogenetic protein 7 is expressed in prostate cancer metastases and its effects on prostate tumor cells depend on cell phenotype and the tumor microenvironment. Neoplasia 12:192–205
Padua D, Massagué J (2009) Roles of TGFbeta in metastasis. Cell Res 19:89–102
Acknowledgments
We thank Ms. Yoko Ezaki for her technical assistance. This work was supported in part by the Japan Science and Technology Agency (JST).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sakai, H., Furihata, M., Matsuda, C. et al. Augmented autocrine bone morphogenic protein (BMP) 7 signaling increases the metastatic potential of mouse breast cancer cells. Clin Exp Metastasis 29, 327–338 (2012). https://doi.org/10.1007/s10585-012-9453-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-012-9453-9