Abstract
In 2009, malignant melanoma was responsible for approximately 9,000 deaths in the US. These deaths are often associated with aggressive metastasis to secondary sites such as the lungs. Epidemiological and animal studies suggest that obesity is a risk factor for melanoma. Others have shown that B16BL6 melanoma cells metastasize more aggressively in obese ob/ob than in lean mice. However, the mechanism by which obesity promotes B16BL6 melanoma metastasis in ob/ob mice has not been identified. In the present study, we used serum obtained from control and ob/ob leptin-deficient obese mice to determine if obese serum increases the aggressive phenotype of melanoma cells. Results showed that ob/ob serum has higher levels of resistin, insulin, tPAI1, IL-6, TNF-α, and MCP-1 compared to control serum. We showed that ob/ob serum increases the invasive ability of B16BL6 melanomas. To further determine the mechanism by which ob/ob serum increases the invasive ability of melanomas, we determined the effect of ob/ob and control serum on genes associated with the epithelial-to-mesenchymal transition (EMT). Cancer cells with a mesenchymal phenotype have a higher metastatic ability. Snai1 and Twist are genes that are strongly associated with EMT and metastasis of melanomas. Our results showed that ob/ob serum increases the expression of Snai1 and Twist. Moreover, ob/ob serum increased matrix metalloproteast 9 (MMP9) activity and decreased the expression of E-cadherin and the metastasis suppressor gene Kiss1. In summary, results suggest that obesity may increase the metastatic ability of melanoma by promoting a mesenchymal cell phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Malignant melanoma is a type of skin cancer arising from melanocytes; it represents 4% of all skin cancers, however, it accounts for 75% of skin cancer-related deaths according to the American Cancer Society [1]. Similarly, in 2009, about 121,840 new cases of melanoma were reported, and approximately 9,000 people died from this disease in the United States [1]. Currently, treatments available for metastatic melanoma are very inefficient and almost nonexistent. Metastatic melanoma is one of the most deadly and evasive types of cancer. In fact, melanoma patients with metastatic melanoma typically survive for only about 6–9 months after diagnosis [2]. Therefore, there is a need to understand the mechanism by which melanomas metastasize.
Epidemiological and animal studies suggest that obesity is a risk factor for melanoma [3–5]. Studies by Dennis et al. showed that obesity increases the risk of developing subcutaneous melanoma [3]. Moreover, Samanic et al. showed that obesity increases the risk of developing malignant melanoma [4]. In animal studies, Mori et al. demonstrated that obesity promotes pulmonary metastasis of B16BL6 melanoma in obese ob/ob mice [5]. However, the mechanism by which obesity promotes melanoma metastasis is not thoroughly known.
Obesity may increase the metastatic ability of melanoma by promoting a mesenchymal cell phenotype. Studies show that cancer cells with a mesenchymal phenotype have a higher ability to metastasize [6]. The transcription factors Snai1 and Twist are genes that promote the epithelial-to-mesenchymal transition (EMT) [7, 8]. Moreover, Snai1 and Twist have been shown to increase the ability of cancer cells to migrate, invade and metastasize [9]. Furthermore, overexpression of Snai1 and Twist increases cell motility and leads to a significant loss of cell-to-cell adhesion [10]. The mesenchymal phenotype is also associated with higher expression of matrix metalloproteases (MMPs) such as matrix metalloproteast 9 (MMP9) [11]. MMPs play a key role in the metastatic cascade, because they allow cancer cells to degrade the extracellular matrix (ECM), and thus, allow the cancer cells to invade the basement membrane from where cancer cells can eventually metastasize to secondary sites [12].
Alternatively, obesity may promote melanoma metastasis by not only promoting a mesenchymal phenotype but also by simultaneously leading to a decrease in the expression of genes known to inhibit metastasis. Metastasis suppressor genes encode proteins that have the ability to hinder the establishment of metastases [13]. Metastasis suppressor genes, such as Kiss1, have the ability to inhibit the metastatic ability of melanomas [14]. Thus, it is feasible that obesity increases the metastatic ability of melanomas by increasing the expression of pro-metastatic genes such as Snai1 and Twist and also by decreasing the expression of genes that inhibit melanoma metastasis, such as Kiss1.
In the current study, we determined the effects of control and obese serum on the invasiveness of B16BL6 melanoma cells. Results show that ob/ob serum increased the ability of B16BL6 melanoma cells to invade. The higher invasive ability of melanoma was associated with increased expression of Snai1, Twist, and MMP9. Furthermore, ob/ob serum increased the expression of Snai1 in the nucleus and down-regulated E-cadherin expression at the cell membrane of B16BL6 cells. Ob/ob serum also increased MMP9 activity. Additionally, ob/ob serum decreased the expression of the metastasis suppressor gene Kiss1. Overall, our studies suggest that obesity may increase the aggressiveness of melanomas by promoting a mesenchymal cell phenotype and by down-regulating the expression of the metastasis suppressor gene Kiss1.
Methods and procedures
Cancer cells and cell culture reagents
The highly metastatic B16BL6 melanoma cells were kindly provided by Dr. Isiah J. Fidler, University of Texas at MD Anderson, Houston TX. The non-metastatic B16F1 melanoma cells were purchased from American Type Culture Collection (ATCC, Chicago, IL). They were maintained in Dulbecco’s modified minimum essential medium (Invitrogen, Carlsbad, CA), pH 7.4, containing 10% heat-inactivated fetal bovine serum (Invitrogen) and 1% antibiotic–antimycotic solution (CellGro, Manassas, VA). The cells were grown at 37°C in a humidified atmosphere of 5% CO2. For cell culture studies, B16BL6 cells were treated with either 5% ob/ob mice serum or 5% ob/+ heterozygous control mice serum. Serums from mice for each group (control or ob/ob) were pooled together after the serum analysis was completed.
Obese ob/ob mice
All animal procedures and methods employed in our studies were approved by the Animal Care and Use Committee at the University of Texas at Austin. Pathogen free male ob/ob and ob/+ heterozygous control C57BL/6J mice were purchased from The Jackson Laboratory (JAX, Bar Harbor, Maine) at 6–8 weeks old. They were housed according to NIH guidelines (National Research Council, 1996). They were singly housed and maintained on water and a chow diet (Research Diets, New Brunswick, NJ) for 15 weeks (n = 12). We measured body weight, food consumption, and liquid consumption on a weekly basis. After 15 weeks on the diet, mice were euthanized using CO2 and blood was drawn by cardiac puncture before cervical dislocation was performed on each mice. The blood was allowed to coagulate at room temperature for 30 min. Serum was collected and frozen in liquid nitrogen. Liver and visceral adipose tissues were similarly collected and flashfrozen in liquid nitrogen for RNA analysis at the time of killing. Both tissue and serum were stored at −80°C until analyzed.
Body composition analyses in mice
Body composition was determined in non-anesthetized mice using an EchoMRI composition analyzer system (Echo Medical Systems, Houston, TX) as instructed by the manufacturer. Percent body fat was determined at 10 weeks into the study (n = 12).
Blood glucose levels
To determine if ob/ob mice were hyperglycemic compared to control mice, we measured blood glucose levels after fasting. At the 8th week of the study, mice were fasted for 7 h and their blood glucose levels were determined using a glucometer elite (Bayer, Elkhart, IN) (n = 12).
Glucose tolerance test (GTT)
To determine the effect of obesity on glucose clearance, we performed the GTT assay. Briefly, mice were fasted for 14 h and then injected with 20% glucose in PBS intraperitoneally. Then, a razor blade was used to make a small cut at the tail vein of each mouse and glucose was measured in the blood using a glucometer elite (Bayer, Elkhart, IN) at 0, 15, 30, 60, and 120 min post-injection. GTT was examined at the 9th week of the study (n = 12).
Insulin tolerance test (ITT)
The ITT assay tests how efficiently endogenous glucose is cleared from the blood as a response to insulin administration. ITT was performed after mice were fasted for 7 h. They were then given an intraperitoneal injection of 0.75 U kg−1 insulin in PBS (Insulin: Sigma Aldrich, St. Louis, MO). A razor blade was used to make a small cut at the tail vein of each mouse, and blood glucose was measured using a glucometer elite (Bayer, Elkhart, IN) at 0, 15, 30, 60, and 120 min post-injection. Area under the curve (AUC) was calculated to determine insulin sensitivity. ITT were examined at the 8th week of the study (n = 12).
Serum analysis
To determine obesity’s effect on systemic factors in the blood, we measured leptin, insulin, interleukin 6 (IL-6), Tumor necrosis factor-alpha (TNF-α), resistin, tissue plasminogen activator inhibitor-1 (tPAI-1), and monocyte chemotactic protein-1 (MCP-1) in the serum of each mouse at week 10 of the study. We measured these factors with the Millipore’s multiplex MAP mouse serum adipokine panel kit (Millipore, Billerica, MA) as instructed by the manufacturer (n = 12).
Invasion assay
We used the Boyden chamber assay to determine the effects of obese mice sera on the ability of B16BL6 melanoma cells to invade [15]. Briefly, the top chamber membrane was coated with a 1:10 dilution of BD Matrigel™ (BD Biosciences, Becton Drive Franklin Lakes, NJ), then, 5 × 104 B16BL6 melanoma cells were placed in the top chamber in serum-free DMEM with 0.1% bovine serum albumin (BSA). The lower chamber was filled with DMEM containing 5% ob/ob serum or 5% control serum. Cells were allowed to migrate from the top side towards the bottom side of each camber for 28 h. After 28 h, cells that remained on the top side were removed using a Q-tip. Cells that invaded to the bottom side were fixed and stained using Siemens Diff-Quick stain set (Siemens, Malvern, PA). Stained cells were visualized and quantified by microscopy. To determine the average number of cells that migrated for each well, we counted three random fields in each well at 200×. Each treatment had three wells per experiment, with each experiment being independently performed three times (n = 3).
Wounding assay
The wounding assay was used to determine the effects of high and low glucose on the ability of B16BL6 melanoma cells to migrate. This was done to determine if glucose levels can affect the ability of these cells to migrate. Ob/ob mice are hyperglycemic, and thus, it is possible that the high glucose levels found in ob/ob mice may affect the aggressiveness of melanoma cells to metastasize [16]. In the wounding assay, cells are plated to 100% confluency and a scratch is drawn in the middle of the plate. The decrease in gap distance is measured over time and quantified; the larger the gap distance that remains, the less the cells have migrated. Briefly, B16BL6 cells were grown on a 24-well plate until they were confluent. Then, cells were grown in FBS-free DMEM overnight. The next day, wells were washed twice with PBS and a scratch was drawn on each well using a p200 pipette tip. After two more washes with PBS to get rid of cell debris, the following cell culture media was added: 1,000 mg/l glucose DMEM (low glucose) or 4,500 mg/l glucose DMEM (high glucose). Both media was supplemented with 5% FBS. Each well was photographed at the time of treatment (0 h) and after 9 h of incubation (9 h) at 40×. The difference in gap distance was measured (9 h time point–0 h time point) to quantify cell motility. The wounding assay was done in the absence of mitomycin C; however, the assay was terminated after 9 h, which is less time than the 16–18 h needed for B16BL6 cells to double [17]. Thus, cell migration is most likely not due to changes in cell proliferation. Each experiment was repeated three times with each group having three wells per experiment (n = 3).
Quantitative real time PCR (qRT-PCR)
Total RNA was collected from three replicate of cells that were serum starved overnight and then treated with 5% ob/ob serum, 5% control serum, or other obesity factors for 24 h or as indicated. The RNA was extracted using an RNeasy mini kit according to the manufacturer’s instructions (Qiagen). Using 1 μg of RNA for each sample, reverse transcription was performed with the high capacity cDNA reverse transcription kit (Applied Biosystems). Primers for 18S, IL-6, TNF-α, Snai1, Twist, MMP9, and Kiss1 were purchased from Integrated DNA Technologies (IDT, Coralville, IA) and were as follows: 18S F: GCATGGCCGTTCTTAGTTGGTGGA, B: TCTCGGGTGGCTGAACG-CCA; IL-6 F: GCTGGTGACAACCACGGCCT, B: AGCCTCCGACTTGTGAAGTGGT; TNF-α F: CTCTTCAAGGGACAAGGCTG, B: CGGACTCCGCAAAGTCTAAG; Snai1 F: CACCTCCAGA-CCCACTCAGAT, B: CCTGAGTGGGGTGGGAGCTTCC; Twist F: CCACGCTGCCCTCGGA-CAAG, B: CCAGGCCCCCTCCATCCTCC; MMP9 F: GCCCACCGTCCTTTCTTGTTGGA, B: GGGAGAGGTGGTTTAGCCGGTG; Kiss1 F: GCAAGCCTGGGTCTGCAGGG, B: CGACTGC-GGGAGGCACACAG.
Quantitative RT-PCR was performed with a SYBR GreenER qPCR kit (Invitrogen) in a Mastercycler ep realplex real-time PCR thermocycler (Eppendorf North America, Hauppauge, NY). The relative expression levels of target genes were normalized to the housekeeping 18S rRNA. Amplification specificity was confirmed by melting curve analysis. Each gene was measured in quadruplicate and the average ΔCt was taken from the three replicate wells before fold change was calculated using the ΔΔCt method. At least three independent sets of samples were performed for each gene (n = 3).
Immunofluorescence Microscopy
To determine protein localization of Snai1 and E-cadherin, immunoflurescence microscopy was used. Wax pencils were used to mark a closed circle on microscope coverslips. B16BL6 cells were plated into the circles and allowed to attach for 24 h before being FBS-starved overnight. They were subsequently treated with 5% ob/ob serum or 5% control serum for 24 h. Cells were washed twice with PBS and fixed in 4% formalin/PBS for 10 min. Then, cells were subsequently washed twice with PBS before being stored in PBS at 4°C overnight. The next day, cells were permeabilized with 0.1% Triton-×100/PBS and neutralized with 100 μM glycine/PBS, before being treated with antibodies against Snai1 or E-cadherin (Cell Signaling, Danvars, MA and Santa Cruz Biotechnology Inc., Santa Cruz, CA) and appropriate fluorescently-conjugated secondary antibodies (Cell Signaling and Abcam, Cambrigde, UK) as recommended by the manufacturer. After three washes with 0.2% Tween20 in PBS, cells were counterstained with two drops of DAPI/antifade (Millipore) according to the manufacturer’s instructions for detection of cellular nuclei. After 15 min of incubation, coverslips were placed onto microscope slides and sealed with nail polish, and images were taken with the Zeiss Axiovert 200 M fluorescent microscope at UT Austin’s ICMB Core Facility. Images of control and experimental cells were acquired under identical exposure conditions for comparative analysis (n = 3).
Western blot analysis
To determine the total protein expression of E-cadherin in B16BL6 cells in response to DMEM, control serum or ob/ob serum treatments, western blot analysis was carried out on whole cell lysates. For this purpose, B16BL6 cells were grown to about 70% confluency in 10 cm3 plates. Cells were rinsed with PBS and then FBS-starved overnight. Then, cells were treated with control DMEM, 5% control serum, or 5% ob/ob serum in DMEM for 24 h. After the various treatments, cells were rinsed twice with PBS, trypsinized, and centrifuged at 10,000 rpm for 5 min at 4°C. The supernatant was discarded, and the pellets were stored in −80°C overnight. The next day, cells were washed twice with PBS before being lysed in 300 μl RIPA buffer (Thermo Scientific, Waltham, MA) supplemented with a protease inhibitor cocktail (Roche, South San Francisco, CA). After 45 min incubation on ice, cells were centrifuged at 10,000 rpm for 10 min at 4°C. The supernatant was collected and protein concentration was measured using the Bradford assay. 50 μg of protein was loaded and run on an SDS–PAGE gel at 100–150 V for 1 h. The gel was subsequently transferred to a PVDF membrane, blocked in 5% milk, washed numerous times in TBST, and probed for E-cadherin (Santa Cruz Biotechnology Inc.) and β-actin (Cell Signaling) and appropriate HRP-conjugated secondary antibodies (Santa Cruz Biotechnology Inc.) before incubation with ECL and film exposure.
Zymogen gel assay
To determine MMP9 activity of B16BL6 cells after different treatments, we performed the zymogen gel assay. Briefly, B16BL6 cells were treated with 5% ob/ob serum or 5% control serum for 24 h. 1.5 mL of this media was subsequently collected and concentrated using a centrifugal protein concentrator (Millipore). 20 μl of protein concentrate was mixed with a 2× zymogen loading dye to a final volume of 40 μl. This sample was loaded onto a zymogen gel (Bio-Rad Laboratories, Hercules, CA) to determine MMP9 activity as described previously [18]. Three separate assays were performed (n = 3).
Statistical analysis
All experiments were analyzed for significance using the independent Student’s t-test in SPSS (PAWS version 18). P-values ≤0.05 were considered significant, and all data is represented as the mean ± SEM.
Results
Ob/ob mice
Others have shown that ob/ob mice are hyperglycemic and insulin resistant [19, 20]. We verified these previous findings. Final body weights for control mice was 33.73 g while ob/ob mice had a final body weight of 56.65 g (P < 0.05). Results showed that ob/ob mice had significantly higher body fat levels than control mice (Fig. 1a). We also showed that ob/ob mice are hyperglycemic and hyperinsulinemic compared to control mice (Figs. 1b, 2a). Our results similarly showed that ob/ob mice were insulin resistant as determine by the GTT and ITT assay (Fig. 1c, d). The GTT assay measures how quickly injected glucose is cleared from the blood. The ITT assay tests how quickly endogenous glucose is cleared from the blood in response to insulin administration. Results showed that ob/ob mice cleared blood glucose at a significant lower rate than control mice, thus, suggesting that they are indeed insulin resistant (P < 0.05).
Systemic factors in ob/ob mice. Mice were given water and food ad lib for 15 weeks. At week 10, serum was collected and analyzed for circulating factors. Ob/ob mice had a undetectable leptin, higher levels of insulin, tPAI-1, resistin, b monocyte chemoattractant protein-1 (MCP-1), c IL-6, and TNF-α compared to control mice (mean ± SEM, n = 12 mice/group, *P < 0.05). At the end of the study, adipose tissue and liver were collected and analyzed using qRT-PCR for inflammatory factors. d Ob/ob mice had higher levels of IL-6 and TNF-α in adipose tissue. Ob/ob mice had higher levels of TNF-α in the liver; however, IL-6 levels in the liver were similar to those of control mice (mean ± SEM, n = 3 mice/group, *P < 0.05)
Adipokine and pro-inflammatory factors in ob/ob mice
As expected, systemic levels of leptin in ob/ob were undetectable (Fig. 2a). However, other adipokines such as resistin and tPAI-1 were significantly higher in ob/ob mice (Fig. 2a). In agreement with previous reports, we show that ob/ob mice have higher levels of pro-inflammatory factors; systemic MCP-1, IL-6, and TNF-α were significantly higher in ob/ob mice (Fig. 2b, c). Additionally, IL-6, and TNF-α were elevated in the adipose tissue of ob/ob mice, however, only TNFα was elevated in the livers of these mice (Fig. 2d).
Ob/ob serum increased the invasiveness of B16BL6 melanoma
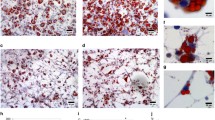
To determine if the factors that increased the metastatic phenotype of B16BL6 melanoma cells in ob/ob mice were found in the blood, we determined the ability of serum from ob/ob and control mice to affect the invasive ability of melanoma cells in cell culture conditions. For this purpose, we tested the effect of ob/ob serum on the ability of B16BL6 cells to invade through a Matrigel-coated chamber compared to control serum treatment. Results showed that B16BL6 cells exposed to ob/ob serum were more invasive than B16BL6 cells exposed to control serum (Fig. 3a).
Ob/ob serum increases B16BL6 cell invasion and high glucose increases cell migration. B16BL6 cells were exposed to 5% ob/ob serum or 5% control serum for the invasion assay. a The images shown are pictures of cells that invaded during the 24 h period. Number of B16BL6 cells that invaded in the ob/ob and the control serum over the 24 h period. Next, B16BL6 cells were treated with DMEM media containing either high (4,500 mg/l) or low (1,000 mg/l) glucose, and cell motility was determined using the wounding assay after 9 h. High glucose exposure increased the ability of B16BL6 cells to migrate. b Representative pictures of cell motility and quantification of the assay results (mean ± SEM, n = 3 separate experiments with three replicate for each condition, *P < 0.05)
High glucose increased B16BL6 melanoma cell migratory ability
Evidence suggests that high systemic glucose levels can increased the risk of cancers [16, 21]. Thus, it is possible that high levels of glucose in ob/ob mice can affect the metastatic ability of melanoma cells. Using the wounding assay in cell culture conditions, we determined the effects of a high glucose media on the ability of B16BL6 cells to migrate compared to low glucose media. Results showed that high glucose levels increased the ability of B16BL6 cells to migrate compared to low glucose (Fig. 3b). Thus, it is possible that once the melanoma cells reach the blood, the high levels of growth factors (e.g., insulin), pro-inflammatory factors (e.g., IL-6) in conjunction with high glucose levels promotes the metastatic ability of melanoma cells.
Ob/ob serum increases the expression of Snai1, Twist, and MMP9
To better understand the mechanism by which ob/ob serum affects the invasive phenotype of melanomas, we determined the effect of ob/ob serum on the expression of genes associated with the EMT. The mesenchymal phenotype in cancer cells is associated with an strong ability of cancer cells to invade and metastasize [6]. The gene expression of both Snai1 and Twist transcription factors has been established to be strongly associated with the mesenchymal phenotype of cancers such as melanomas [22, 23]. To determine if the metastatic ability of B16BL6 melanoma cells were associated with the expression levels of Snai1 and Twist, we measured their levels in highly metastatic B16BL6 and non-metastatic B16F1 melanoma cells. Our results showed that Snai1 and Twist are up-regulated while E-cadherin is down-regulated in highly metastatic B16BL6 cells compared to non-metastatic B16F1 melanoma cells (Fig. 4a). To determine if the expression of Snai1 is modulatable by the presence and absence of serum, we exposed the highly metastatic B16BL6 to either no serum or 10% fetal bovine serum for 24 h. Results showed that Snai1 expression was increased in B16BL6 cells treated with 10% FBS compared to FBS-starved cells (Fig. 4b). Thus, these results showed that expression of Snai1 can be modulated by different serum.
B16BL6 gene expression and MMP9 activity. B16F1 or B16BL6 cells were treated with DMEM, 10% FBS, 5% ob/ob serum or 5% control serum for 24 h as indicated and analyzed by qRT-PCR or immunofluorescence microscopy. a Metastatic B16BL6 cells have higher Snai1 and Twist mRNA levels than non-metastatic B16F1 cells. Conversely, B16BL6 cells express less E-cadherin than B16F1 cells. b B16BL6 cells treated with 10% FBS express higher Snai1 mRNA levels than FBS-starved cells. c Ob/ob serum increased Snai1, Twist, and MMP9 mRNA levels. Conversely, ob/ob serum decreased the expression of the metastasis suppressor Kiss1 (mean ± SEM, n = 4 separate experiments with three replicates for each condition, * is P < 0.05). d Nuclear expression of Snai1 and localization of E-cadherin in B16BL6 cells. White arrow indicates cell–cell junction (n = 3). e Total E-cadherin expression in B16BL6 cells. f Active MMP9 in B16BL6 cells (n = 3)
Next, we exposed the highly metastatic B16BL6 melanoma to ob/ob serum to determine if obese sera would increase the expression of Snai1 and Twist. Results showed that the ob/ob obese serum significantly increased the expression of both Snai1 and Twist (Fig. 4c). Through immunofluorescence microscopy, we also determined that ob/ob serum increased Snai1 expression in the nucleus of B16BL6 cells and correspondingly decreased E-cadherin expression at the cell membrane compared to control serum (Fig. 4d). We confirmed a decrease in total E-cadherin protein expression in B16BL6 cells after ob/ob serum treatment (Fig. 4e). Thus, we showed that ob/ob serum increases Snai1 expression and decreases the expression of E-cadherin, which are hallmarks of EMT [8].
In some cells, the MMP9 expression is regulated by Snai1 [24]. MMP9 allows cancer cells to invade surrounding tissue by degrading the ECM [25]. Since, ob/ob serum increased the ability of melanomas to invade and also upregulated the expression of Snai1, we expected the obese serum to increase the expression of MMP9. Indeed, results showed that ob/ob serum increased the expression of MMP9 in B16BL6 cells (Fig. 4c). Furthermore, ob/ob serum modestly increased the amount of active MMP9 compared to control serum (Fig. 4f).
Effects of ob/ob serum on Kiss1 expression
Alternatively, obesity may increase the ability of melanoma cells to metastasize by promoting a mesenchymal phenotype and also by decreasing the expression of genes known to inhibit metastasis. Metastasis suppressors genes can slow down or prevent metastasis [14]. Kiss1 is a metastasis suppressor known to inhibit melanoma metastasis [26]. Our results showed that ob/ob serum decreased the expression of Kiss1 in B16BL6 cells (Fig. 4c).
Effects of insulin, resistin, and TNF-α on Snai1, Kiss1, and MMP9 expression
To determine which obesity-related factors found in the serum of ob/ob mice affected the expression of Snai1, KiSS1, and MMP9; we treated B16BL6 melanoma cells with purified insulin, resistin, and TNF-α to measure the expression levels of these genes. We showed that resistin treatment increased Snai1 and MMP9 expression, though not significant, while it decreased Kiss1 levels significantly (Fig. 5a). Kiss1 was decreased by insulin treatment while MMP9 was up-regulated (Fig. 5b). Lastly, we showed that Kiss1 expression decreased in a time-dependent manner after TNF-α treatment (Fig. 5c). Thus, these obesity-related factors may additively contribute to the modulation of the aggressiveness of B16BL6 melanoma and the EMT phenotype.
Effects of obesity factors on Snai1, Kiss1, and MMP9 expression. B16BL6 cells were treated with resistin (100 ng/mL), insulin (1 and 6 ng/mL) and TNF-α (1 ng/mL) for 24 h unless indicated otherwise, to determine the expression of Snai1, Kiss1, and MMP9 through qRT-PCR. a Effects of resistin on Snai1, MMP9 and Kiss1 mRNA expression. b Effects of insulin on Snai1, MMP9 and Kiss1 mRNA expression. c Effects of TNF-α on Kiss1 mRNA expression (mean ± SEM, n = 3 separate experiments with three replicate for each condition, *P < 0.05)
Discussion
Consistent with previous reports, we show that leptin-deficient ob/ob mice exhibit numerous obesity-related symptoms such as excess body weight, excess body fat, insulin resistance, hyperinsulinemia, and chronic inflammation [19]. Moreover, ob/ob mice had a local (e.g. adipose and liver tissue) and systemic inflammatory environment as reflected by high levels of IL-6, TNF-α, resistin, tPAI-1, and MCP-1 in the serum compared to control mice. We also showed that ob/ob mice have higher glucose levels than control mice, and that higher glucose levels increased the ability of melanomas to migrate in cell culture conditions. Hence, the higher levels of glucose in ob/ob mice may not only meet the higher nutritional requirements of melanoma cells, but also increase their ability to migrate and invade to secondary sites. Thus, the pro-cancerous environment found in the blood of ob/ob mice may allow melanoma cells to enhance their survival and to establish metastases in secondary tissue. This was clearly demonstrated by Mori et al. who showed that B16BL6 melanoma cells established much more numerous and grossly larger metastases in the lungs of obese ob/ob mice than in control mice [5].
Altogether, the serum of obese ob/ob mice may increase the metastatic phenotype of melanoma. The invasive ability of a cancer cell is an important functional phenotype necessary for successful metastasis [27]. Supporting Mori el al.’s results, we showed that ob/ob serum increased the invasive ability of B16BL6 melanoma cells compared to control serum. To understand the molecular mechanisms by which ob/ob serum increased cell invasion, we determined its effect on the expression of genes linked to cell invasion and metastasis. In fact, we showed that ob/ob serum increased the expression of Snai1 and Twist, genes that are associated with the mesenchymal phenotype [28]. We focused our attention on these transcription factors because they have been shown to modulate EMT and the aggressive phenotype of melanoma cells [29, 30]. Melanoma cells with an epithelial phenotype are less aggressive than those with a mesenchymal phenotype [31]. Cancer cells that transition towards a mesenchymal phenotype often lose the expression of cell adhesion proteins that keep the cells anchored together and prevent them from breaking away from the primary tumor [32]. In some cancers, the transcription factors Snai1 and Twist decreases the expression of E-cadherin, which is a protein found in the cell membrane that plays a key role in cell adhesion: it allows cells within tissues to adhere to each other [33, 34]. Our results showed that treatment of melanoma cells with ob/ob serum increased nuclear localization of Snai1 and decreased the amount of E-cadherin at the cell membrane.
Factors found in the serum of ob/ob mice such as IL-6 and TNF-α have been shown to increase the expression of Snai1 [35, 36]. Thus, it is feasible that the pro-inflammatory environment found in ob/ob mice promotes a mesenchymal phenotype by increasing the expression of Snai1 and Twist to affect the metastatic ability of B16BL6. Indeed, we showed that resistin and insulin also affected the expression of Snai1 and MMP9. Others have shown that overexpression of Snai1 and Twist can increase both the mesenchymal phenotype and the metastatic ability of melanoma cells [10, 22, 29]. Moreover, Snai1 also increases the expression of MMP9, a protein that can degrade ECM proteins; the degradation of the ECM proteins may allow cancer cells to escape into the blood or invade secondary tissues in order to establish metastases [12, 24]. In our studies, we showed that ob/ob serum increased the expression of MMP9 and its activity in B16BL6 melanoma cells, which may explain why B16BL6 melanoma cells have a higher invasive ability in the presence of ob/ob serum.
Besides affecting genes associated with EMT, obese serum may also increase the metastatic phenotype of melanoma cells by decreasing the expression of genes known to inhibit metastasis. Metastasis suppressors are genes that have been identified to stop or slow down metastasis [37]. In our study, we measured the expression levels of Kiss1, which was originally discovered for its ability to suppress the metastatic ability of two human melanoma cell lines [38]. Studies suggest that one anti-metastatic function of Kiss1 may be to decrease the expression of MMPs, such as MMP9 [25, 12]. Interestingly, others show that Kiss1 is also down-regulated in cancer cells that acquire a mesenchymal phenotype [22]. We found that treatment of B16BL6 melanoma cells with ob/ob serum decreased the expression of Kiss1. Furthermore, we show that both resistin and insulin decreased Kiss1 expression and increased MMP9 expression in B16BL6 melanoma. Similarly, TNF-α treatment decreased Kiss1 expression in a time-dependent manner. Thus, systemic obesity-related factors collectively may modulate EMT and the aggressive phenotype of melanoma cells. In summary, as Fig. 6 depicts, our results suggest that systemic factors found in the blood of ob/ob mice may contribute towards increasing the aggressiveness of melanoma cells to metastasize by promoting a mesenchymal phenotype and by decreasing the expression of the metastasis suppressor gene Kiss1.
Proposed mechanism by which ob/ob serum increases the metastatic ability of B16BL6 melanoma. Obesity is characterized by high systemic levels of glucose, IL-6, and TNF-α. These factors may increase the mesenchymal phenotype by increasing the expression of Snai1, Twist, and MMP9, and thereby increase the invasive ability of B16BL6 melanoma cells. Moreover, such pro-cancerous environment may simultaneously decrease the expression of metastasis suppressor such as Kiss1, and thus further increasing the metastatic phenotype of melanoma
References
American Cancer Society (2009) Cancer facts and figures 2009. Atlanta, Georgia
Klimek VM, Wolchok JD, Chapman PB et al (2000) Systemic chemotherapy. Clin Plast Surg 27(3):451–461 ix-x
Dennis L, Lowe J, Lynch C et al (2008) Cutaneous melanoma and obesity in the agricultural health study. Ann Epidemiol 18(3):214–221
Samanic C, Chow W, Gridley G et al (2006) Relation of body mass index to cancer risk in 362, 552 Swedish men. Cancer Causes Control 17(7):901–909
Mori A, Sakurai H, Choo M et al (2006) Severe pulmonary metastasis in obese and diabetic mice. Int J Cancer 119(12):2760–2767
Bonnomet A, Brysse A, Tachsidis A et al (2010) Epithelial-to-mesenchymal transitions and circulating tumor cells. J Mammary Gland Biol Neoplasia 15(2):261–273
Peinado H, Olmeda D, Cano A (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer 7(6):415–428
Kang Y, Massagué J (2004) Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118(3):277–279
Foubert E, De Craene B, Berx G (2010) Key signalling nodes in mammary gland development and cancer. The Snail1-Twist1 conspiracy in malignant breast cancer progression. Breast Cancer Res 12(3):206
Yang J, Mani SA, Donaher JL et al (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117(7):927–939
Przybylo JA, Radisky DC (2007) Matrix metalloproteinase-induced epithelial-mesenchymal transition: tumor progression at Snail’s pace. Int J Biochem Cell Biol 39(6):1082–1088
Kessenbrock K, Plaks V, Werb Z (2010) Matrix metalloproteinases: regulators of the tumor microenvironment. Cell 141(1):52–67
Steeg P (2003) Metastasis suppressors alter the signal transduction of cancer cells. Nat Rev Cancer 3(1):55–63
Stafford L, Vaidya K, Welch D (2008) Metastasis suppressors genes in cancer. Int J Biochem Cell Biol 40(5):874–891
Albini A, Iwamoto Y, Kleinman H et al (1987) A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res 47(12):3239–3245
Stattin P, Björ O, Ferrari P et al (2007) Prospective study of hyperglycemia and cancer risk. Diabetes Care 30(3):561–567
Ganapathi R, Grabowski D, Schmidt H et al (1987) Characterization in vitro and in vivo of progressively adriamycin-resistant B16-BL6 mouse melanoma cells. Cancer Res 47(13):3464–3468
Wallert and Provost Lab (2007) MMP zymogen gel assay protocol
Lindström P (2007) The physiology of obese-hyperglycemic mice [ob/ob mice]. Sci World J 7:666–685
Dubuc PU (1976) The development of obesity, hyperinsulinemia, and hyperglycemia in ob/ob mice. Metabolism 25(12):1567–1574
Stocks T, Lukanova A, Johansson M et al (2008) Components of the metabolic syndrome and colorectal cancer risk; a prospective study. Int J Obes (Lond) 32(2):304–314
Dissanayake SK, Wade M, Johnson CE et al (2007) The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J Biol Chem 282(23):17259–17271
Ansieau S, Morel AP, Hinkal G et al (2010) TWISTing an embryonic transcription factor into an oncoprotein. Oncogene 29(22):3173–3184
Jordà M, Olmeda D, Vinyals A et al (2005) Upregulation of MMP-9 in MDCK epithelial cell line in response to expression of the Snail transcription factor. J Cell Sci 118(Pt 15):3371–3385
Yan C, Wang H, Boyd D (2001) KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha -induced block of p65/p50 nuclear translocation. J Biol Chem 276(2):1164–1172
Shirasaki F, Takata M, Hatta N et al (2001) Loss of expression of the metastasis suppressor gene KiSS1 during melanoma progression and its association with LOH of chromosome 6q16.3–q23. Cancer Res 61(20):7422–7425
Liotta LA, Kohn EC (2001) The microenvironment of the tumour-host interface. Nature 411(6835):375–379
Thiery J, Acloque H, Huang R et al (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139(5):871–890
Kuphal S, Palm HG, Poser I et al (2005) Snail-regulated genes in malignant melanoma. Melanoma Res 15(4):305–313
Poser I, Domínguez D, de Herreros AG et al (2001) Loss of E-cadherin expression in melanoma cells involves up-regulation of the transcriptional repressor Snail. J Biol Chem 276(27):24661–24666
Alonso SR, Tracey L, Ortiz P et al (2007) A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res 67(7):3450–3460
Tse JC, Kalluri R (2007) Mechanisms of metastasis: epithelial-to-mesenchymal transition and contribution of tumor microenvironment. J Cell Biochem 101(4):816–829
Pećina-Slaus N (2003) Tumor suppressor gene E-cadherin and its role in normal and malignant cells. Cancer Cell Int 3(1):17
Vesuna F, van Diest P, Chen JH et al (2008) Twist is a transcriptional repressor of E-cadherin gene expression in breast cancer. Biochem Biophys Res Commun 367(2):235–241
Sullivan N, Sasser A, Axel A et al (2009) Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28(33):2940–2947
Wu Y, Deng J, Rychahou P et al (2009) Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell 15(5):416–428
Smith S, Theodorescu D (2009) Learning therapeutic lessons from metastasis suppressor proteins. Nat Rev Cancer 9(4):253–264
Lee J, Miele M, Hicks D et al (1996) KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst 88(23):1731–1737
Acknowledgements
This work was supported by the American Cancer Society grant ACS RSG CNE-113703 and by grants from the National Institutes of Health: National Cancer Society grant NCI 1K22CA127519-01A1 and National Institute of Environmental Health Sciences Center grants ES09145 and ES007784.
Conflicts of interest
Authors have no conflicts of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kushiro, K., Núñez, N.P. Ob/ob serum promotes a mesenchymal cell phenotype in B16BL6 melanoma cells. Clin Exp Metastasis 28, 877–886 (2011). https://doi.org/10.1007/s10585-011-9418-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-011-9418-4