Abstract
The distribution of telomeric repeats was analyzed by fluorescence in situ hybridization in 15 species of arvicoline rodents, included in three different genera: Chionomys, Arvicola, and Microtus. The results demonstrated that in most or the analyzed species, telomeric sequences are present, in addition to normal telomeres localization, as large blocks in pericentromeric regions. The number, localization, and degree of amplification of telomeric sequences blocks varied with the karyotype and the morphology of the chromosomes. Also, in some cases telomeric amplification at non-pericentromeric regions is described. The interstitial telomeric sequences are evolutionary modern and have rapidly colonized and spread in pericentromeric regions of chromosomes by different mechanisms and probably independently in each species. Additionally, we colocalized telomeric repeats and the satellite DNA Msat-160 (also located in pericentromeric regions) in three species and cloned telomeric repeats in one of them. Finally, we discuss about the possible origin and implication of telomeric repeats in the high rate of karyotypic evolution reported for this rodent group.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Telomeric sequences are widely conserved among vertebrates and consist of extended arrays of the TTAGGG hexamer (Meyne et al. 1989; Zakian 1995). These sequences are normally found at the ends of chromosomes (telomeres); however, distribution at intrachromosomal sites (ITSs) have been observed in a variety of species (Meyne et al. 1990; Nanda and Schmid 1994; Vermeesch et al. 1996; Garagna et al. 1997; Metcalfe et al. 1997, 1998; Ono and Yoshida 1997; Slijepcevic et al. 1996; Bolzán and Bianchi 2006). The most common non-telomeric positions correspond to pericentromeric regions where ITSs could expand in large arrays up to hundred of kilobases within or at the margins of constitutive heterochromatin (het-ITSs) (Meyne et al. 1990; Nanda and Schmid 1994; Garagna et al. 1997). Other short stretches of interstitial telomeric sequences (s-ITSs) are variable located at internal sites of chromosomes and presumable present in all vertebrate species (for a review, see Ruiz-Herrera et al. 2008).
It is assumed that bulk of interstitial telomeric repeats could be generated by mechanisms such as mutation, unequal crossing-over, transposition, or amplification from intrachromosomal short telomeric sequences (Wiley et al. 1992; Vermeesch et al. 1996; Garagna et al. 1997; Sharma and Sharma 1998). Also, ITSs are considered remnants of chromosomal rearrangements, such as, inversions, centric, or tandem fusions, that occurred during the species’ karyotype evolution (Lee et al. 1993; Fagundes et al. 1997; Pellegrino et al. 1999). Recent studies provided evidences that s-ITSs could be produced during the process of DNA double strand breaks repair. These sequences could be originated by either telomerase activity or direct insertion of a double strand telomeric fragment into a chromosomal DNA break (Nergadze et al. 2004, 2007).
The function, if any, of telomeric interstitial repeats have not been elucidated yet, but most probably they do not work as functional telomeres (Meyne et al. 1990). However, they have been cytogenetically associated with chromosomal reorganizations, fragile sites, and recombination hotspots (Ruiz-Herrera et al. 2005a, b). Previous cytogenetic studies revealed that ITSs often corresponded with preferential sites of breakage and fragile sites, providing evidence of their potential role in genomic and chromosomal instability (Slijepcevic et al. 1996; Ruiz-Herrera et al. 2005a). Also, these sequences have been associated with recombination hotspots regions, which indicated a possible implication in recombination events (Ashley and Ward 1993; Day et al. 1998; Pluta and Zakian 1989; Nanda et al. 2002).
Among placental mammals, rodents seem to present the highest rate of karyotypic evolution (Wilson et al. 1975). One remarkable group is the subfamily Arvicolinae, which has undergone a huge evolutionary radiation accompanied with a rapid karyotypic change. Chromosomal variation in the Arvicolinae ranges from 2n = 17 in Ellobius lutescens (Vogel et al. 1998) and Microtus oregoni (Modi 1987b) to 2n = 64 in M. longicaudus (Modi 1987b). Comparative G-bandig and, more recently, chromosome painting analyses demonstrated that chromosomal fusions and fissions are frequent chromosomal rearrangements implicated on the karyotypic evolution of this group (Burgos et al. 1988a; Li et al. 2006; Sitnikova et al. 2007; Lemskaya et al. 2010).
The genus Microtus includes about 62 extant species and represents a notable example of rapid mammalian radiation (Musser and Carleton 2005; Shenbrot and Krasnov 2005), and karyotypic variability (Maruyama and Imai 1981). Sitnikova et al. (2007) had estimated speciation rates 60–100 times higher in Microtus than in other mammals, with most sibling species showing major differences in karyotypes than in morphological characters. Lemskaya et al. (2010) estimated that during evolution of species from Microtus genus, approximately six rearrangements occurred per million of years, which is an extreme high rate of chromosomal evolution compared with other rodent groups, where the postulated rate is one chromosomal fusion/fission event per million of years (Schibler et al. 1998). In addition, sex chromosomes are prone to change as consequence of a heterochromatin enlargement trend, which gives rise in some species to giant sex chromosomes containing heterochromatic blocks of variable length and composition (Modi 1987a; Burgos et al. 1988b; Marchal et al. 2003, 2004; Nanda et al. 1988; Kalscheuer et al. 1996).
The karyotype of Arvicolinae species are well studied by conventional and molecular cytogenetic methods (Modi 1987a; Burgos et al. 1988a, b; Modi 1993; Mayorov et al. 1996; Neitzel et al. 1998; Singh et al. 2000; Mazurok et al. 2001; Modi et al. 2003; Marchal et al. 2004; Sitnikova et al. 2007; Lemskaya et al. 2010; Acosta et al. 2008, 2009, 2010). However, few of these analyses described chromosomal distribution of telomeric sequences (Gornung et al. 2011; Liu and Fredga 1999). Here, we analyzed distribution of telomeric sequences in 15 species belonging to three different genera of Arvicolinae. This comparison was attended to determine if ITSs occurred in these species associated with chromosomal rearrangements. Interestingly, in eight species we found a large bulk of het-ITSs arranged at pericentric regions of different bi-armed and acrocentric chromosomes. Also, we analyzed and compared co-distribution of telomeric sequences and a satellite DNA, Msat-160, in pericentromeric heterochromatin of three species. Finally, cloning and sequence analyses in Microtus (Terricola) thomasi atticus show that canonical and non-canonical telomeric repeats are intermixed in the genome.
Materials and methods
Species analyzed and chromosome preparations
For this study, we used chromosome preparations from 15 species of arvicoline rodents, included in three different genera: Chionomys, Arvicola, and Microtus. The complete list of species is included in Table 1. Chromosome preparations were obtained either from permanent fibroblast-cell lines belonging to one male specimen, according to Neitzel et al. (1998), or from bone marrow cells of one male specimen following the method of Burgos et al. (1986) (Table 1).
FISH
Chromosomal localization of telomeric sequences was analyzed on 15 species. The telomeric probe labeled with biotin was produced following the method of Ijdo et al. (1991), a modified PCR which required (TTAGGG)5 and (CCCTAA)5 as primers and no template. Fluorescence in situ hybridization (FISH) was performed as previously described (Fernández et al. 2001) and required three rounds of signal amplification with avidin-FITC/antiavidin system (Oncor) in species with normal telomeric sequences distribution at telomeres. However, only two rounds were applied in species with accumulation of telomeric repeats in order to avoid the strong signals that overlap the chromosome and do not allowed to see the telomere signals.
Co-localization of telomeric sequences and Msat-160 satellite, another repeat previously described in pericentromeric regions of several arvicolids (Modi 1992; Fernández et al. 2001; Acosta et al. 2010), was analyzed on Arvicola terrestris, Microtus cabrerae, and Microtus (T) thomasi thomasi chromosomes. In this two-color FISH, we employed simultaneously a telomeric probe, direct labeled by PCR with SpectrumOrange (Abbot), and a probe for Msat-160 DNA either direct labeled with SpectrumGreen (Abbot) or biotin labeled according to (Fernández et al. 2001; Acosta et al. 2010). In A. terrestris and M. cabrerae, we used the SpectrumGreen labeled probe for Msat-160 DNA co-localization while in M. (T) thomasi thomasi, we employed the biotin-labeled probe and one round of signal amplification for detection of this satellite DNA. FISH protocol was similar to that previously described in Fernández et al. (2001) and Marchal et al. (2004). FISH images were captured using a fluorescence microscope (Olympus BX51) equipped with a CCD camera (Olympus DP70).
Cloning and analysis of telomeric sequences
In our laboratory, we have been performing several experiments in order to characterize satellite DNA composition in M. (T) thomasi atticus. Thus, after restriction of genomic DNA with one enzyme, AluI, a prominent band was observed and cloned in pGEM-T easy vector (Promega) following previously described methods (Sánchez et al. 1996; Acosta et al. 2009). Sequencing analyses of the obtained clones revealed that four of them (Mtho-2-2-AluI-att-25, 29, 43, and 51), unexpectedly, contained canonical and non-canonical telomeric repeats.
Pairwise sequence alignments and multiple alignments were carried out with the program CLUSTAL W 1.6 (Thompson et al. 1994).
Results
Chromosome distribution of telomeric sequences
Genus Chionomys
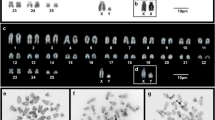
From this genus, we only analyzed the species Chionomys nivalis, where telomeric sequences were located at normal terminal position of all chromosomes, without detection of interstitial signals in any of them (Fig. 1a).
In situ hybridisation with the telomeric probe in male chromosomes of the following species: a Chionomys nivalis, b Arvicola sapidus, c A. terrestris, d Microtus arvalis, e M. guentheri, f M. oeconomous, g M. cabrerae, h M. agrestis, i M. rossiaemeridionalis, j M. (Terricola) brachycercus niethammericus, k M. (T) thomasi thomasi, and l M. (T) thomasi atticus. Insert in l the same X chromosome of the metaphase but only DAPI stained. Scale bars indicate 5 μm
Genus Arvicola
In addition to normal terminal positions, ITSs were detected in both species from this genus. Most ITSs were organized as large blocks at pericentromeric regions of the chromosomes, although differences in their number and size existed (Fig. 1b, c).
In Arvicola sapidus (2n = 40), 13 pairs of autosomes either bi-armed or acrocentric showed het-ITSs at pericentric regions. In A. terrestris (2n = 36), they were detected only on pericentromeric regions of three pairs of biarmed autosomes while several others presented faint telomeric signals at the same region (Fig. 1b, c). Concerning sex chromosomes, the submetacentric X chromosome of both species contained het-ITSs also on pericentromeric heterochromatin. In A. sapidus, an additional interstitial band was observed on Xp (Fig. 1b, c). On the Y chromosome of A. sapidus, no ITSs signals were detected, while on A. terrestris three big het-ITSs bands were detected on the heterochromatic Yq arm (Fig. 1b, c).
Genus Microtus
Subgenus Microtus
In this subgenus, only two species (Microtus arvalis and Microtus guentheri) of the six analyzed did not contain blocks of telomeric repeats at pericentric regions. Curiously, in both species some chromosomes seemed to have more copies of telomeric repeats at one of the ends, as evidenced the more intense hybridization signals (Fig. 1d, e).
FISH analyses in the other four species (Microtus oeconomous, M. cabrerae, Microtus agrestis, and Microtus rossiaemeridionalis) showed het-ITSs in pericentromeric regions of most chromosomes, in addition to normal telomere signals (Fig. 1f–i). In relation to sex chromosomes, only the biarmed X chromosomes of M. oeconomous and M. cabrerae presented large het-ITSs signals on the pericentromeric regions (Fig. 1f, g), while no ITSs accumulation was observed on the Y chromosome of all the analyzed species (Fig. 1f–i). Furthermore, no telomeric repeats were detected in our analyses on the enlarged heterochromatic blocks existing on the X and Y chromosomes of three species (M. cabrerae, M. agrestis, and M. rossiaemeridionalis) (Fig. 1g–i).
Subgenus Terricola
In four species from this group Microtus (T) brachycercus niethammericus, M. (T) duodecimcostatus, M. (T) lusitanicus, and M. (T) savii savii, we observed only the normal terminal localization for telomeric sequences (one representative example, M. (T) brachycercus niethammericus, is presented in Fig. 1j). However, the karyotypes of M. (T) thomasi thomasi (Fig. 1k) and M. (T) thomasi atticus (Fig. 1l) presented both pericentromeric het-ITSs arrays on most autosomes. Notably, the pattern of pericentric distribution was slightly different, as ITSs occupied the DAPI positive bands in M. (T) thomasi atticus while in M. (T) thomasi thomasi localized surrounding the DAPI positive band. Additionally, interstitial faint signals in some other autosomal regions were also detected in both species.
Differences in het-ITSs accumulation were also observed on the X chromosomes of both species. Both shared an interstitial band on the euchromatic Xq arm but, in addition, the pericentric region and the Xp arm of M. (T) thomasi atticus was heavily comprised of telomeric repeats. Finally, the Y chromosome was depleted of ITSs in both species.
Colocalization of telomeric sequences and Msat-160 satellite DNA
In three species from different genera, we have also performed a simultaneous co-localization of het-ITSs and Msat-160, a satellite DNA broadly distributed in arvicolids (Modi 1992; Fernández et al. 2001; Acosta et al. 2010). This comparison is especially interesting in order to characterize more in detail pericentromeric organization and structure in these species. In order to obtain a precise mapping of both sequences and avoid magnification of FISH signals by antibody incubations, we employed direct labeled probes. The telomeric probe correctly mapped pericentric het-ITSs but produced no signals or very faint ones on telomeric positions.
In A. terrestris, Msat-160 and telomeric sequences did not co-localize at the same pericentromic regions and both were found accumulated in different chromosomes (Fig. 2a–c). However, in M. cabrerae and M. (T) thomasi thomasi most acrocentric chromosomes included both sequences on two contiguous regions at pericentromeric heterochromatin: het-ITSs more internally arranged, closed to the euchromatin, and Msat-160 more terminal accumulated, closed to the telomere border, with an overlapping region of variable length (Fig. 2d–i). In biarmed chromosomes of M. cabrerae, both probes produced a three-banding pattern on the pericentromeric region, with het-ITSs accumulated in two external bands and intermixed with Msat-160 sequences in a third internal band (Fig. 2f). Also in both species, some chromosomes showed a unique predominant signal at pericentromeric heterochromatin with either Msat-160 or telomeric probes.
Double in situ hybridisation with the telomeric (red) and Msat-160 satellite DNA (green) probes in male chromosomes of A. terrestris (a–c), M. cabrerae (d–f), and M. (T) thomasi thomasi (g–i). Left columns, telomeric probe; middle columns, Msat-160 probe; right columns, both probes merged. Scale bars indicate 5 μm
Cloned telomeric sequences
The four sequenced clones from M. (T) thomasi atticus contained insert sizes of 264 (Mtho-2.2-AluI-att-25 and 29), 406 (Mtho-2.2-AluI-att-43), and 429 bp (Mtho-2.2-AluI-att-51). Clones 25 and 29, both with equal sequences, were 82.57% and 83.33% identical with clones 43 and 51, respectively, while clones 43 and 51 were 85.22% identical. When aligned with a telomeric consensus (TTAGGG) n array of the same length the identity percentages were 86.74% (clones 25 and 29), 89.65% (clone 43), and 94.17% (clone 51) (Fig. 3). Clones 25 and 29 included 44 telomere repeat units, 12 of them canonical and 32 non-canonical. Most common change was T→G substitution in the second base of the telomeric hexanucleotide (TTAGGG to TGAGGG), occurring in 29 repeat units. Clone 43 contained 65 telomere repeats, 34 canonical and 31 non-canonical. Most common change was also substitution of T by G in the second position of the hexamere, detected in 18 repeat units. Finally, clone 51, with 71 telomeric unit repeats, presented more frequently canonical (47 repeats) than no canonical sequences (24 repeats). In this case, nucleotide changes were randomly distributed, without a particular substitution more commonly repeated.
Discussion
The analyses presented here demonstrated that het-ITS, defined as large blocks of telomeric sequences arranged in pericentromeric regions, are a common feature in arvicolid rodents. However, their number, localization, and degree of amplification varied with the karyotype analyzed and the morphology of the chromosome. Also, we found some examples of het-ITSs amplification at non-pericentromeric regions. Those examples were exclusively observed on sex chromosomes, in particular on the heterochromatic Xq arm of M. (T) thomasi atticus and along the heterochromatic Y chromosome of A. terrestris. Interestingly, the enlarged heterochromatic blocks of the giant sex chromosomes existing in three of the analyzed species (M. cabrerae, M. agrestis, and M. rossiaemeridionalis) were depleted of telomeric repeats. Finally, in this group of species we also detected faint bands composed of short stretches of ITSs (s-ITS) at the euchromatic regions of some chromosomes.
Sequencing data from whole genome projects indicate that probably all genomes contain a variable number of s-ITSs, composed of few telomeric repeats, and not detectable on FISH analyses (Ruiz-Herrera et al. 2008). Previous cytogenetic studies have demonstrated that het-ITSs occur also in many genomes arranged on large blocks at pericentromeric regions of the chromosomes. Examples have been described in primates, rodents, marsupials, carnivores, cetartiodactyla, perissodactyla, chiroptera, amphibians, reptiles, fishes, and birds (for a revision, see Ruiz-Herrera et al. 2008).
Het-ITSs and arvicolid phylogeny
From our analyses, we could hypothesized that ancestral condition in arvicolid rodents was distribution of telomeric sequences only at telomeres. Such pattern is the observed one in C. nivalis karyotype (2n = 54) (Lemskaya et al. 2010), considered very primitive within these group due to the acrocentric constitution of all chromosomes, with the exception of the X chromosome.
It is well known that Arvicolid genera and subgenera experienced a recent and rapid radiation during a short period of time. For example, origin of the genus Arvicola is dated 1.6 Myr ago (Montoya et al. 2001), the split between Chionomys and Microtus lineages is supposed to occurred 1–2.4 Myr ago (Chaline and Graf 1988; Nadachowski 1991; Janeau and Aulagnier 1997) and the origin of the subgenus Terricola is dated 0.65–0.7 Myr ago (Brunet-Lecomte 1988; Tougard et al. 2008). Our FISH analysis demonstrated that there is no correlation between presence or absence of het-ITSs and the genus or subgenus classification of the species. In fact, in the subgenus Microtus and Terricola, we have both species with het-ITSs and species without het-ITS. Hence, we can assume that variation and amplification of ITSs occurred independently in each species, as no correspondence existed with the proposed phylogeny.
Furthermore, significant variation in het-ITSs pattern was observed also within closely related species or subspecies form this group. That occurred for the Arvicola species analyzed, which split 0.25 Myr ago (Centeno-Cuadros et al. 2009), and also when comparing the subspecies of M. (T) thomasi, both recently evolved (0.015–0.01 Myr) after the last glacial period (Mitsainas et al. 2009). Also, central Europe and Turkey populations of A. terrestris presented a very small Y chromosome while the specimens analyzed here have a big Y chromosome with a long arm containing three het-ITSs bands (Zima and Kral 1984; Gözcelioğlu et al. 2006). Thus, rapid amplification and variation of het-ITSs might be product of the evolutionary dynamics noticed in arvicolid genome for repeated DNA. Thus, several studies have described a complex pattern of amplification, accumulation and variation for different repeated DNA sequences (satellite DNA, non satellite DNA, retroelements, and pseudogenes) in heterochromatic regions (either pericentromeric or not) of Arvicolinae karyotypes (Modi 1993; Marchal et al. 2004, 2008; Acosta et al. 2008; 2010).
Our sequencing data of some telomeric arrays are also in agreement with a rapid evolutionary drift for these repeated DNA. Thus, we detected both canonical and non-canonical telomeric sequences, consequence of mutations arising at many different positions. The number of different nucleotides may be correlated with the time after the insertions, as was demonstrated in primates and rodent species where the more ancient insertion presented more changes and the recent ones less (Nergadze et al. 2007). Such novel ITSs sequences probably are being homogenized and expanded in concerted evolution pattern, an evolutionary path common in satellite DNAs.
Numerous studies have showed that block of telomeric repeats are unstable regions which represent chromosomal fusion and fission, tandem fusion, or inversion points (Ashley and Ward 1993; Slijepcevic et al. 1996; Pluta and Zakian 1989; Nanda et al. 2002; Farré et al. 2009). In addition, het-ITSs have been related with either induced or spontaneous chromosomal rearrangement, breakage, or fragile sites, and with recombination hotspots at the genome (Ruiz-Herrera et al. 2008; López-Fernández et al. 2006; Álvarez et al. 1993; Fernández et al. 1995; Bertoni et al. 1996; Slijepcevic et al. 1996). There are even evidences of their direct implication in genomic instability and chromosomal pathologies (Kilburn et al. 2001).
Het-ITSs might have played a significant role in karyotypic variation and evolution of Arvicolinae species, one of the highest described in mammals (Wilson et al. 1975; Maruyama and Imai 1981; Acosta et al. 2010). However, het-ITSs are not correlated with the rearrangements occurred during the karyotypic evolution of the analyzed species. For example, the karyotypes of the two Arvicola species differ only by two Robertsonian fusions but the accumulation and distribution of het-ITS in both karyotypes is very different, as A. sapidus show big blocks of telomeric repeats in most pericentric regions while in A. terrestris they are observed only in very few chromosomes. Similarly, in Microtus genus there are species where het-ITS are accumulated in most pericentric regions while other species lack het-ITS, and such distribution is independent of the chromosomal reorganizations identified in previous studies (Burgos et al. 1988a; Lemskaya et al. 2010).
Het-ITSs: spreading mechanisms in Arvicolid chromosomes
The pattern of co-localization observed here for het-ITSs and satellite DNA Mast-160 is of valuable information to discern het-ITSs presence in arvicolid species. That co-distribution was significantly different in A. terrestris compared with M. cabrerae and M. (T) thomasi thomasi. Hence in A. terrestris het-ITSs and MSAT-160 are independently arrayed, composing the pericentromeric regions of different biarmed chromosomes. Most probably, the origin for these telomeric blocks is chromosomal fusions of acrocentric or telocentric chromosomes, with reduced loss of terminal telomeric sequences, with subsequent amplification and spread along pericentromeric heterochromatin (Fig. 4a). In fact, such rearrangements might have frequently occurred in the common ancestor of Arvicola species and would explain the origin of the Arvicola karyotypes, with 2n = 36 or 2n = 40 (Díaz de la Guardia and Pretel 1979), from the supposed ancestral arvicolid karyotype, with 2n = 54 and all chromosomes acrocentric (Martínková et al. 2004). Further amplification/deletions processes occurring independently at novel pericentromeric regions in each Arvicola species would give rise to the observed differences in number and amount of het-ITSs in both species.
Diagram showing possible evolutionary origins of het-ITSs in the chromosomes of A. terrestris (a), M. cabrerae, and M. (T) thomasi thomasi (b, c). Chr fusion chromosomal fusion, ITS ampl interstitial telomeric sequences amplification, Tel ampl telomeric sequences amplification, Per inv pericentromeric inversion, Tel lost telomere lost. Red color represents telomeric sequences and green represents Msat-160 satellite DNA sequences
Alternatively, it cannot be discarded that het-ITSs appeared first in Arvicola karyotypes after enlargement of terminal telomeric sequences in some acrocentric chromosomes, which further were implicated in fusion events giving rise to bi-armed chromosomes with pericentromeric telomere repeats (Fig. 4a). Finally, occurrence of biarmed chromosomes without pericentromeric het-ITSs in Arvicola species could be explained by total reduction of telomeric sequences during the process of chromosomal fusion.
In M. cabrerae and M. (T) thomasi thomasi, ITSs and satellite DNA co-distribution displayed a scenario more complicated to elucidate. One intricate pattern is observed in the centromeric heterochromatin of some acrocentric autosomes, composed by telomeric repeats on internal part, closed to euchromatin, and Msat-160 satellite more terminal to the centromere, with a variable region of overlapping. Such organization could appear if telomeric repeats are first amplified at terminal end of the chromosome and subsequently transferred to inner parts, as consequence of pericentric inversions (Fig. 4b). Alternatively, pericentric inversions in acrocentric chromosomes could also originated short ITSs at the inner part of chromosomes, nearby to Msat-160, and could become later enlarged by common mechanisms of repeated DNA amplification.
Either proposed mechanism is unlikely to have occurred simultaneously in most acrocentric chromosomes of both species. Instead, once het-ITSs appeared/evolved in one or few chromosomes, common mechanisms producing amplification and homogenization of repeated DNA might amplify, transfer and extend them through the karyotype. A similar path was postulated to explain telomeric repeat accumulation in lemur species (Go et al. 2000). Interestingly, in our FISH analyses we regularly observed physical associations of pericentromeric regions in some metaphases, and patched patterns of Msat-160 and telomeric signals suggestive of heterochromatic association in interphase nucleus (data not shown). Such heterochromatic association has been also described in several mammalian species, and it is considered part of the mechanisms responsible of repeated DNA dispersion and homogenization between chromosomes (Go et al. 2000).
An alternative hypothesis based on amplification of pre-existing short ITSs might be considered to explain the co-distribution pattern of ITSs and satellite DNA observed in M. cabrerae and M. (T) thomasi thomasi. Thus, short-ITSs are variably inserted at different intra-chromosomal regions in mammalian genomes (Ruiz-Herrera et al. 2008). Such insertions are proposed to occur during reparation of DNA double strand breaks, by direct insertion of a blunt-end telomeric sequence or directly though a mechanism that required telomerase intervention (Nergadze et al. 2004, 2007). Hence, s-ITS could be originated at pericentromeric regions of arvicolid chromosomes during DNA repair process; afterwards, s-ITS could give stretched into het-ITSs through any of the mechanisms expanding tandem repeats at heterochromatin. It would be of interest to determine if het-ITSs are hot-spots for spontaneous or induced chromosome breakage in these species, as demonstrated in Chinese hamster (Simi et al. 1998; Bolzán and Bianchi 2006).
Finally, in some bi-armed chromosomes from M. cabrerae and M. (T) thomasi thomasi, telomeric repeats are arranged in pericentric external regions, separated by an inner Msat-satellite band. This pattern is most likely originated by centric fusions of acrocentric chromosomes containing het-ITSs and Msat-satellite repeats on pericentric and inner centromeric regions respectively (Fig. 4c). Those fusions might occur following breakage within Msat-160 satellite DNA and elimination of normal telomeric sequences. A similar process is documented in wild mice in which Robertsonian rearrangements generated fusions through minor satellite DNA and subsequent elimination of telomeric sequences (Garagna et al. 1995, 2001).
Het-ITSs and heterochromatin
Accordingly to the organization and distribution shown in arvicolid chromosomes, het-ITSs constituted a new component of satellite DNA in these species. In arvicolids, different families of satellites have been separately localized at centromeric regions. Our data of co-distribution for het-ITSs and Msat-160 satellites on metaphase chromosomes are of interest to understand the organization and structure of centromeric heterochromatin. Both repeats occupied adjacent regions in some chromosomes, with a variable overlapping region visible in some of them. In Chinese hamster, het-ITSs repeats and the satellite CH5 are also located in contiguous compartments at centromeric heterochromatin, but the molecular organization differed from the observed here, being the telomeric sequences in the middle of two blocks of satellite DNA (Faravelli et al. 2002). In other species where het-ITSs have been mapped they localized within or at the margins of constitutive heterochromatin (Meyne et al. 1990; Vermeesch et al. 1996; Ono and Yoshida 1997; Go et al. 2000), however data about co-localization with other repeats are still scarce.
A remarkable question to address for het-ITSs blocks is their chromatin structure. Thus, interstitial telomere repeats could share common structural features with the telomeric chromatin sited at the end of chromosomes, as a higher nucleosome density compared with non-telomeric chromatin (Revaud et al. 2009) and similar epigenetic modifications. Concerning last ones, both terminal and interstitial telomeric sequences were found associated with the telomeric proteins TRF1 and TRF2 in CHO cells and also in cells from a human patient showing aberrant ITSs (Mignon-Ravix et al. 2002; Smogorzewska et al. 2000). Moreover, functional assays on CHO cells showed that TRF1 and histone H3 trimethylated, a typical repressive chromatin modification, did not co-localize (Svetlova et al. 2007). In the same study, it was also detected transcription of some unique sequences adjacent to het-ITSs blocks, indicating that TTAGGG repeats do not work as general transcription silencers. Thus, het-ITSs chromatin may lack typical hallmarks of constitutive or facultative heterochromatin. Further epigenetic and functional analyses in arvicolid species and other organisms are required to better clarify the functional significance of het-ITSs chromatin.
In conclusion, we demonstrated here that ITSs are a common feature of arvicolid karyotypes. They are evolutionary modern and rapidly colonized and spread on chromosomes via different mechanisms and probably independently in each species. These telomeric sequences are probably implicated, together with other repeated sequences, in the high rate of karyotypic evolution reported for this rodent group. However, no correlation seemed to exist between chromosomal rearrangements and het-ITS distribution in these species. Future molecular analysis involving sequence cloning and FISH co-localization in metaphase chromosomes and extended fibers are important to establish the relation of ITSs and other repeated sequences, which could aide to understand the origin and possible implication of these sequences in chromosomal and karyotypic evolution in arvicolid.
Abbreviations
- ITS:
-
Interstitial telomeric sequences
- PCR:
-
Polymerase chain reaction
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- Myr:
-
Million years
- TRF1:
-
Telomeric repeat binding factor 1
- TRF2:
-
Telomeric repeat binding factor 2
- CHO:
-
Chinese hamster ovary
References
Acosta MJ, Marchal JA, Fernandez-Espartero CH, Bullejos M, Sanchez A (2008) Retroelements (LINEs and SINEs) in vole genomes: differential distribution in the constitutive heterochromatin. Chromosome Res 16:949–959
Acosta MJ, Marchal JA, Mitsainas GP, Rovatsos MT, Fernandez-Espartero CH, Giagia-Athanasopoulou EB, Sanchez A (2009) A new pericentromeric repeated DNA sequence in Microtus thomasi. Cytogenet Genome Res 124:27–36
Acosta MJ, Marchal JA, Fernández-Espartero C, Romero-Fernández I, Rovatsos MT, Giagia-Athanasopoulou EB, Gornung E, Castiglia R, Sánchez A (2010) Characterization of the satellite DNA Msat-160 from species of Terricola (Microtus) and Arvicola (Rodentia, Arvicolinae). Genetica 138:1085–1098
Álvarez L, Evans JW, Wilks R, Lucas JN, Brown JM, Giaccia AJ (1993) Chromosomal radiosensitivity at intrachromosomal telomeric sites. Genes Chromosomes Cancer 8:8–14
Ashley T, Ward DC (1993) A “hot spot” of recombination coincides with an interstitial telomeric sequence in the Armenian hamster. Cytogenet Cell Genet 62:169–171
Bertoni L, Attolini C, Faravelli M, Simi S, Giulotto E (1996) Intrachromosomal telomere-like DNA sequences in Chinese hamster. Mamm Genome 7:853–855
Bolzán AD, Bianchi MS (2006) Telomeres, interstitial telomeric repeat sequences, and chromosomal aberrations. Mutat Res 612:189–214
Brunet-Lecomte P (1988) Les campagnols souterrains (Terricola, Arvicolinae, Rodentia) actuels et fossiles d’Europe occidentale. D.Phil. thesis, University of Burgundy
Burgos M, Jiménez R, Díaz de la Guardia R (1986) A rapid, simple and reliable combined method for G-banding mammalian and human chromosomes. Stain Technol 61:257–260
Burgos M, Jiménez R, Díaz de la Guardia R (1988a) Comparative study of G- and C-banded chromosomes of five species of Microtidae: a chromosomal evolution analysis. Genome 30:540–546
Burgos M, Jiménez R, Olmos DM, Diaz de la Guardia R (1988b) Heterogeneous heterochromatin and size variation in the sex chromosomes of Microtus cabrerae. Cytogenet Cell Genet 47:75–79
Centeno-Cuadros A, Delibes M, Godoy JA (2009) Dating the divergence between Southern and European water voles using molecular coalescent-based methods. J Zool 279:404–409
Chaline J, Graf JD (1988) Phylogeny of the arvicolidae (Rodentia): biochemical and paleontological evidence. J Mamm 69:22–23
Day JP, Limoli CL, Morgan WF (1998) Recombination involving interstitial telomere repeat-like sequences promotes chromosomal instability in Chinese hamster cells. Carcinogenesis 19:259–265
Díaz de la Guardia R, Pretel A (1979) Comparative study of the karyotypes of two species of water vole: Arvicola sapidus and Arvicola terrestris (Rodentia, Microtinae). Caryologia 32:183–188
Fagundes V, Vianna-Morgante AM, Yonenaga-Yassuda Y (1997) Telomeric sequences localization and G-banding patterns in the identification of a polymorphic chromosomal rearrangement in the rodent Akodon cursor (2n = 14,15 and 16). Chromosome Res 5:228–232
Faravelli M, Azzalin CM, Bertoni L, Chernova O, Attolini C, Mondello C, Giulotto E (2002) Molecular organization of internal telomeric sequences in Chinese hamster chromosomes. Gene 283:11–16
Farré M, Ponsà M, Bosch M (2009) Interstitial telomeric sequences (ITSs) are not located at the exact evolutionary breakpoints in primates. Cytogenet Genome Res 124:128–131
Fernández JL, Gosálvez J, Goyanes V (1995) High frequency of mutagen-induced chromatid exchanges at interstitial telomere-like DNA sequence blocks of Chinese hamster cells. Chromosome Res 3:281–284
Fernández R, Barragán MJ, Bullejos M, Marchal JA, Martínez S, Díaz de la Guardia R, Sánchez A (2001) Molecular and cytogenetic characterization of highly repeated DNA sequences in the vole Microtus cabrerae. Heredity 87:637–646
Garagna S, Broccoli D, Redi CA, Searle JB, Cooke HJ, Capanna E (1995) Robertsonian metacentrics of the house mouse lose telomeric sequences but retain some minor satellite DNA in the pericentromeric area. Chromosoma 103:685–692
Garagna S, Ronchetti E, Mascheretti S, Crovella S, Formenti D, Rumpler Y, Manfredi Romanini MG (1997) Non-telomeric chromosome localization of (TTAGGG)n repeats in the genus Eulemur. Chromosome Res 5:487–491
Garagna S, Marziliano N, Zuccotti M, Searle JB, Capanna E, Redi CA (2001) Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc Natl Acad Sci USA 98:171–175
Go Y, Rakotoarisoa G, Kawamoto Y, Randrianjafy A, Koyama N, Hirai H (2000) PRINS analysis of the telomeric sequence in seven lemurs. Chromosome Res 8:57–65
Gornung E, Castiglia R, Rovatsos M, Marchal JA, Díaz de la Guardia-Quiles R, Sanchez A (2011) Comparative cytogenetic study of two sister species of Iberian ground voles, Microtus (Terricola) duodecimcostatus and M. (T.) lusitanicus (Rodentia, Cricetidae). Cytogenet Genome Res 132:144–150
Gözcelioğlu B, Colak E, Colak R (2006) Karyotype of Arvicola terrestris (Mammalia: Rodentia) in Turkish Thrace. Pak J Biol Sci 9:2387–2388
Ijdo JW, Wells RA, Baldini A, Reeders ST (1991) Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res 19:4780
Janeau G, Aulagnier S (1997) Snow vole—Chionomys nivalis (Martins, 1842). J Mt Ecol 4:1–11
Kalscheuer V, Singh AP, Nanda I, Sperling K, Neitzel H (1996) Evolution of the gonosomal heterochromatin of Microtus agrestis: rapid amplification of a large, multimeric, repeat unit containing a 3.0-kb (GATA)11-positive, middle repetitive element. Cytogenet Cell Genet 73:171–178
Kilburn AE, Shea MJ, Sargent RG, Wilson JH (2001) Insertion of a telomere repeat sequence into a mammalian gene causes chromosome instability. Mol Cell Biol 21:126–135
Lee C, Sasi R, Lin CC (1993) Interstitial localization of telomeric DNA sequences in the Indian muntjac chromosomes: further evidence for tandem chromosome fusions in the karyotypic evolution of the Asian muntjacs. Cytogenet Cell Genet 63:156–159
Lemskaya NA, Romanenko SA, Golenishchev FN, Rubtsova NV, Sablina OV, Serdukova NA, O’Brien PC, Fu B, Yiğit N, Ferguson-Smith MA, Yang F, Graphodatsky AS (2010) Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). III. Karyotype relationships of ten Microtus species. Chromosome Res 18:459–471
Li T, Wang J, Su W, Yang F (2006) Chromosomal mechanisms underlying the karyotype evolution of the oriental voles (Muridae, Eothenomys). Cytogenet Genome Res 114:50–55
Liu WS, Fredga K (1999) Telomeric (TTAGGG)n sequences are associated with nucleolus organizer regions (NORs) in the wood lemming. Chromosome Res 7:235–240
López-Fernández C, Arroyo F, Fernández JL, Gosálvez J (2006) Interstitial telomeric sequence blocks in constitutive pericentromeric heterochromatin from Pyrgomorpha conica (Orthoptera) are enriched in constitutive alkali-labile sites. Mutat Res 599:36–44
Marchal JA, Acosta MJ, Bullejos M, Diaz de la Guardia R, Sanchez A (2003) Sex chromosomes, sex determination, and sex-linked sequences in Microtidae. Cytogenet Genome Res 101:266–273
Marchal JA, Acosta MJ, Nietzel H, Sperling K, Bullejos M, Diaz de la Guardia R, Sanchez A (2004) X chromosome painting in Microtus: origin and evolution of the giant sex chromosomes. Chromosome Res 12:767–776
Marchal JA, Acosta MJ, Bullejos M, Diaz de la Guardia R, Sanchez A (2008) Origin and spread of the SRY gene on the X and Y chromosomes of the rodent Microtus cabrerae: role of L1 elements. Genomics 91:142–151
Martínková N, Nová P, Sablina OV, Graphodatsky AS, Zima J (2004) Karyotipic relationships of the Tatra vole (Microtus tatricus). Folia Zool 53:279–284
Maruyama T, Imai HT (1981) Evolutionary rate of the mammalian karyotype. J Theor Biol 90:111–121
Mayorov VI, Adkison LR, Vorobyeva NV, Khrapov EA, Kholodhov NG, Rogozin IB, Nesterova TB, Protopopov AI, Sablina OV, Graphodatsky AS, Zakian SM (1996) Organization and chromosomal localization of a B1-like containing repeat of Microtus subarvalis. Mamm Genome 7:593–597
Mazurok NA, Rubtsova NV, Isaenko AA, Pavlova ME, Slobodyanyuk SY, Nesterova TB, Zakian SM (2001) Comparative chromosome and mitochondrial DNA analyses and phylogenetic relationships within common voles (Microtus, Arvicolidae). Chromosome Res 9:107–120
Metcalfe CJ, Eldridge MD, McQuade LR, Johnston PG (1997) Mapping the distribution of the telomeric sequence (T2AG3)n in rock-wallabies, Petrogale (Marsupialia: Macropodidae), by fluorescence in situ hybridization. I. The penicillata complex. Cytogenet Cell Genet 78:74–80
Metcalfe CJ, Eldridge MD, Toder R, Johnston PG (1998) Mapping the distribution of the telomeric sequence (T2AG3)n in the Macropodoidea (Marsupialia), by fluorescence in situ hybridization. I. The swamp wallaby, Wallabia bicolor. Chromosome Res 6:603–610
Meyne J, Ratliff RL, Moyzis RK (1989) Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA 86:7049–7053
Meyne J, Baker RJ, Hobart HH, Hsu TC, Ryder OA, Ward OG, Wiley JE, Wurster-Hill DH, Yates TL, Moyzis RK (1990) Distribution of non-telomeric sites of the (TTAGGG)n telomeric sequence in vertebrate chromosomes. Chromosoma 99:3–10
Mignon-Ravix C, Depetris D, Delobel B, Croquette MF, Mattei MG (2002) A human interstitial telomere associates in vivo with specific TRF2 and TIN2 proteins. Eur J Hum Genet 10:107–112
Mitsainas GP, Rovatsos MTh, Rizou E, Giagia-Athanasopoulou EB (2009) Sex chromosome variability outlines the pathway to the chromosomal evolution in Microtus thomasi (Rodentia, Arvicolinae). Biol J Linn Soc 96:685–695
Modi WS (1987a) C-Banding analyses and the evolution of heterochromatin among Arvicolid rodents. J Mammal 68:704–714
Modi WS (1987b) Phylogenetic analyses of the chromosomal banding patterns among the Nearctic Arvicolidae. Systemat Zool 36:109–136
Modi WS (1992) Nucleotide sequence and genomic organization of tandem satellite array from the rock vole Microtus chrotorrhinus (Rodentia). Mamm Genome 3:226–232
Modi WS (1993) Rapid, localized amplification of a unique satellite DNA family in the rodent Microtus chrotorrhinus. Chromosoma 102:484–490
Modi WS, Serdyukova NA, Vorobieva NV, Graphodatsky AS (2003) Chromosomal localization of six repeated DNA sequences among species of Microtus (Rodentia). Chromosome Res 11:705–713
Montoya P, Alberdi MT, Barbadillo LJ, Van Der Made J, Morales J, Murelaga X, Peñalver E, Robles F, Ruiz-Bustos A, Sánchez A, Sanchiz B, Soria D, Szyndlar Z (2001) Une faune très diversifièe du Pléistocène inférieur de la Sierra de Quibas (province de Murcia, Espagne). CR Acad Sci Paris Sci de la Terre et des planétes 332:387–393
Musser G, Carleton M (2005) Superfamily Muroidea. In: Wilson DE, Reeder DM (eds) Mammal species of the world. The Johns Hopkins University Press, Baltimore
Nadachowski A (1991) Systematics, geographic variation, and evolution of snow voles (Chionomys) based on dental characters. Acta Theriol 36:1–45
Nanda I, Schmid M (1994) Localization of the telomeric (TTAGGG)n sequence in chicken (Gallus domesticus) chromosomes. Cytogenet Cell Genet 65:190–193
Nanda I, Neitzel H, Sperling K, Studer R, Epplen JT (1988) Simple GATCA repeats characterize the X chromosomal heterochromatin of Microtus agrestis, European field vole (Rodentia, Cricetidae). Chromosoma 96:213–219
Nanda I, Schrama D, Feichtinger W, Haaf T, Schartl M, Schmid M (2002) Distribution of telomeric (TTAGGG)(n) sequences in avian chromosomes. Chromosoma 111:215–227
Neitzel H, Kalscheuer V, Henschel S, Digweed M, Sperling K (1998) Beta-heterochromatin in mammals: evidence from studies in Microtus agrestis based on the extensive accumulation of L1 and non-L1 retroposons in the heterochromatin. Cytogenet Cell Genet 80:165–172
Nergadze SG, Rocchi M, Azzalin CM, Mondello C, Giulotto E (2004) Insertion of telomeric repeats at intrachromosomal break sites during primate evolution. Genome Res 14:1704–1710
Nergadze SG, Santagostino MA, Salzano A, Mondello C, Giulotto E (2007) Contribution of telomerase RNA retrotranscription to DNA double-strand break repair during mammalian genome evolution. Genome Biol 8:R260
Ono T, Yoshida MC (1997) Differences in the chromosomal distribution of telomeric (TTAGGG)n sequences in two species of the vespertilionid bats. Chromosome Res 5:203–205
Pellegrino KC, Rodrigues MT, Yonenaga-Yassuda Y (1999) Chromosomal evolution in the Brazilian lizards of genus Leposoma (Squamata, Gymnophthalmidae) from Amazon and Atlantic rain forests: banding patterns and FISH of telomeric sequences. Hereditas 131:15–21
Pluta AF, Zakian VA (1989) Recombination occurs during telomere formation in yeast. Nature 337:429–433
Revaud D, Mozziconacci J, Sabatier L, Desmaze C, Lavelle C (2009) Sequence-driven telomeric chromatin structure. Cell Cycle 8:1099–1100
Ruiz-Herrera A, García F, Giulotto E, Attolini C, Egozcue J, Ponsà M, Garcia M (2005a) Evolutionary breakpoints are co-localized with fragile sites and intrachromosomal telomeric sequences in primates. Cytogenet Genome Res 108:234–247
Ruiz-Herrera A, García F, Mora L, Egozcue J, Ponsà M, Garcia M (2005b) Evolutionary conserved chromosomal segments in the human karyotype are bounded by unstable chromosome bands. Cytogenet Genome Res 108:161–174
Ruiz-Herrera A, Nergadze SG, Santagostino M, Giulotto E (2008) Telomeric repeats far from the ends: mechanisms of origin and role in evolution. Cytogenet Genome Res 122:219–228
Sánchez A, Bullejos M, Burgos M, Jiménez R, Díaz de la Guardia R (1996) An alternative to blunt-end ligation for cloning DNA fragments with incompatible ends. Trends Genet 12:44
Schibler L, Vaiman D, Oustry A, Giraud-Delville C, Cribiu EP (1998) Comparative gene mapping: a fine-scale survey of chromosome rearrangements between ruminants and humans. Genome Res 8:901–915
Sharma GG, Sharma T (1998) Unusual chromosomal organization of telomeric sequences and expeditious karyotypic differentiation in the recently evolved Mus terricolor complex. Cytogenet Cell Genet 80:204–208
Shenbrot GI, Krasnov BR (2005) An Atlas of the geographic distribution of the Arvicoline rodents of the world (Rodentia, Muridae: Arvicolinae). Pensoft Publ, Sofia, p 336
Simi S, Attolini C, Giulotto E (1998) Intrachromosomal telomeric repeats and stabilization of truncated chromosomes in V79 Chinese hamster cells. Mutat Res 397:229–233
Singh A, Henschel S, Sperling K, Kalscheuer V, Neitzel H (2000) Differences in the meiotic pairing behavior of gonosomal heterochromatin between female and male Microtus agrestis: implications for the mechanism of heterochromatin amplification on the X and Y. Cytogenet Cell Genet 91:253–260
Sitnikova NA, Romanenko SA, O’Brien PC, Perelman PL, Fu B, Rubtsova NV, Serdukova NA, Golenishchev FN, Trifonov VA, Ferguson-Smith MA, Yang F, Graphodatsky AS (2007) Chromosomal evolution of Arvicolinae (Cricetidae, Rodentia). I. The genome homology of tundra vole, field vole, mouse and golden hamster revealed by comparative chromosome painting. Chromosome Res 15:447–456
Slijepcevic P, Xiao Y, Dominguez I, Natarajan AT (1996) Spontaneous and radiation-induced chromosomal breakage at interstitial telomeric sites. Chromosoma 104:596–604
Smogorzewska A, van Steensel B, Bianchi A, Oelmann S, Schaefer MR, Schnapp G, de Lange T (2000) Control of human telomere length by TRF1 and TRF2. Mol Cell Biol 20:1659–1668
Svetlova MP, Solovjeva LV, Smirnova AN, Tomilin NV (2007) Long interstitial (TTAGGG)n arrays do not colocalize with repressive chromatin modifications in Chinese hamster cells. Cell Biol Int 31:308–315
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tougard C, Montuire S, Brunet-Lecomte P, Fabre M (2008) Speciation of two allopatric Terricola species (Arvicolinae, Rodentia) from molecular and morphological data. Biol J Linn Soc 93:309–323
Vermeesch JR, De Meurichy W, Van Den Berghe H, Marynen P, Petit P (1996) Differences in the distribution and nature of the interstitial telomeric (TTAGGG)n sequences in the chromosomes of the Giraffidae, okapai (Okapia johnstoni), and giraffe (Giraffa camelopardalis): evidence for ancestral telomeres at the okapi polymorphic rob(5;26) fusion site. Cytogenet Cell Genet 72:310–315
Vogel W, Jainta S, Rau W, Geerkens C, Baumstark A, Correa-Cerro LS, Ebenhoch C, Just W (1998) Sex determination in Ellobius lutescens: the story of an enigma. Cytogenet Cell Genet 80:214–221
Wiley JE, Meyne J, Little ML, Stout JC (1992) Interstitial hybridization sites of the (TTAGGG)n telomeric sequence on the chromosomes of some North American hylid frogs. Cytogenet Cell Genet 61:55–57
Wilson AC, Bush GL, Case SM, King MC (1975) Social structuring of mammalian populations and rate of chromosomal evolution. Proc Natl Acad Sci USA 72:5061–5065
Zakian VA (1995) Telomeres: beginning to understand the end. Science 270:1601–1607
Zima J, Kral B (1984) Karyotypes of European mammals II. Acta Sci Nat Brno 18:1–62
Acknowledgments
The authors wish to express their gratitude to Dr. K. Sperling and Dr. H. Neitzel for providing the cell lines used in this work, to R. Castiglia and E Gornung for providing chromosomes of M. (T) savii savii and M. (T) brachycercus niethammericus, to the Research Technical Services from the University of Jaén for maintenance of cell cultures, and to the cooperation received from the Parque Nacional de Sierra Nevada (Granada, Spain) and the Junta de Andalucía in the capture of C. nivalis. Permission for captures were provided by the Junta de Andalucía (M. cabrerae, C. nivalis, M. (T) doudecimcostatus, and A. sapidus), the Junta de Castilla-La Mancha and Comunidad de Madrid, Ministerio das Cidades, Ordenamento do Território e Ambiente (Portugal) (M. cabrerae), Junta de Castilla-León (A. terrestris and M. (T) lusitanicus), and the Comunidad de Navarra (M. agrestis). We are also grateful to the Jerez de la Frontera Zoo for providing samples from M. cabrerae. This work was supported by the Spanish Ministerio de Ciencia y Tecnología through project number CGL2009-07754 (co-funded by the European Regional Development Fund) and by the program ‘Ayudas a grupos de investigación’ to CVI 220 group.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Fengtang Yang.
Rights and permissions
About this article
Cite this article
Rovatsos, M.T., Marchal, J.A., Romero-Fernández, I. et al. Rapid, independent, and extensive amplification of telomeric repeats in pericentromeric regions in karyotypes of arvicoline rodents. Chromosome Res 19, 869–882 (2011). https://doi.org/10.1007/s10577-011-9242-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9242-3