Abstract
Rodents are a very diverse group with large chromosome variability. One of the most species rich linage in the Neotropics is the Sigmodontinae. Among them, the tribe Abrotrichini was recently defined and its taxonomy and phylogeny were mostly elucidated through molecular and morphological evidence. Meanwhile, chromosome data were only secondarily used because of fragmentary information. In this contribution, we conduct a chromosome characterization of Abrothrix hirta, A. olivacea, A. andina, and Paynomys macronyx, review the cytogenetic background of the tribe, and contrast it with molecular data. Chromosomes were analyzed by conventional and differential techniques. All Abrothrix species presented 2n = 52/FNa = 56, with a high similarity in the banding patterns reflecting a conserved karyotype, which does not coincide with its high molecular variability. In turn, P. macronyx have 2n = 54/FNa = 58–59, varying due to a heteromorphic pair of autosomes. In addition, in this last species, different morphologies of the X chromosome and the presence of B chromosomes were detected. Heterochromatin was involved in these variants. The telomeric probe in P. macronyx marks terminal regions of all chromosomes. B chromosomes generated strong telomeric signals. The Ag-NORs banding revealed the same patterns in Abrothrix and Paynomys. Cytogenetic data support phylogenetic relationships previously proposed and suggest that the specious genus Abrothrix could have retained the ancestral karyotype of the subfamily. In the tribe, the relatively conserved chromosome complement contrasts with its high molecular variability. This indicates decoupling between the rates of chromosomal and molecular divergence, as observed in other rodent lineages. In abrotrichines, chromosome evolution was slower.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The subfamily Sigmodontinae is one of diversified and widely distributed group of rodents in South America. The Abrotrichini, initially referred as the “Andean Clade”, is one of the tribes included in this Neotropical subfamily. This tribe has its geographic distribution in the central and southern Andes in Argentina, Bolivia, Chile, and Peru, and several studies recover it as a monophyletic lineage (Smith and Patton 1999; D’Elía 2003; Cañón et al. 2014; Teta et al. 2016). The species of this major lineage evidence an important ecomorphologic diversification, including the occurrence of terrestrial and fossorial forms, this last being an uncommon lifestyle among sigmodontines (Patton et al. 2015).
The tribe Abrothrichini was traditionally composed by the genera Abrothrix, Chelemys, Geoxus, Notiomys, and Pearsonomys; most of which were previously assigned to the tribe Akodontini (Reig 1987). Molecular analyses, based on mitochondrial and nuclear sequences, have placed those genera in a separate clade with high support and consistency (Smith and Patton 1999; D’Elía 2003). More recently, Teta et al. (2016) found some inconsistent phylogenetic relationships among this group and proposed a new classification for the tribe, including Abrothrix (with four subgenera [Abrothrix, Angelomys, Chroeomys, and Pegamys]), Paynomys (to include Chelemys macronyx), Chelemys (now restricted to C. megalonyx), Geoxus (including Pearsonomys), and Notiomys. In this revision, the tribe was divided into two subtribes: Abrotrichina that groups all Abrothrix species and Notiomyina for all other genera (Teta et al. 2016).
At the chromosomal level, it was suggested that this lineage shares a similar 2n = 52 karyotype (Reig 1987; Liascovich et al. 1989; Spotorno et al. 1990), which was considered a synapomorphic character for the entire tribe (Smith and Patton 1999). However, during several decades, the cytogenetic data was limited to some species of the tribe, particularly those of the genus Abrothrix (Reig 1987; Gallardo 1982; Pearson 1984; Liascovich et al. 1989; Spotorno et al. 1990). Additional data from Geoxus (= Pearsonomys) annectens and Paynomys (= Chelemys) macronyx, based in conventional cytogenetic techniques, suggest that the abrotrichines possess a higher chromosomal variation than previously suspected (Ojeda et al. 2005; D’Elía et al. 2006).
The aim of our study is: a) to present new karyotype information of the Abrothrix and Paynomys species, b) to review chromosome data in members of the tribe Abrotrichini, and c) to contrast and discuss chromosome data with molecular information.
Materials and methods
The specimens analyzed were collected in different localities of west-central Argentina (Supplementary Fig. 1 and Supplementary Table 1), and handled followed ASM guidelines for the use of wild mammals in research (Sikes et al. 2016). Fifteen individuals of Abrothrix [A. hirta (N = 2), A. olivacea (N = 8), and A. andina (N = 5)] and nine of Paynomys macronyx were studied using different cytogenetic techniques. Sampling localities are shown in Supplementary Fig. 1 and Supplementary Table 1. The studied specimens, including skins, skeletons, and cellular suspensions are housed at the Mammals Collection of the Instituto Argentino de Investigaciones de Zonas Áridas IADIZA, CCT-CONICET, Mendoza (Supplementary Table 1).
Mitotic and meiotic chromosome preparations were obtained from bone marrow and testes, respectively, using standard techniques (Ford and Hamerton 1956; Evans et al. 1964). Chromosomes were stained with Giemsa (pH = 6.8). Fundamental numbers (FNa) refer only to autosomes (Patton 1967). The distribution of constitutive heterochromatin (CH, C-bands) was determined using the method of Sumner (1972). The technique of Schweizer (1980) was used for DAPI staining. Ag-NORs staining was performed with the technique proposed by Howell and Black (1980). Fluorescent in situ hybridization (FISH) was performed with a Cy3-conjugated PNA pan-telomeric probe [Cy3-(CCCTAA)3] obtained from PNABio Inc. (CA, USA), according to the protocol provided by the supplier, as previously described (Lanzone et al. 2015). The FISH technique was only performed in P. macronyx. Photomicrographs were obtained using an Olympus BX 50 photomicroscope, with a Sony Exwave HAD digital camera. Fluorescence microscopy was performed on a Nikon Eclipse 50i epifluorescence microscope equipped with an HBO 100 mercury lamp, a Nikon high-resolution digital color camera (DS-Ri-U3), and filters for DAPI and Cy3 (Chroma Technology Corp., Rockingham, VT, USA).
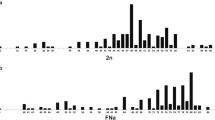
In addition, we carried out an extensive review of the literature and compiled chromosomal information of abrotrichines species (Supplementary Table 2). We described the frequencies and distribution of the diploid numbers (2n) and FNa in the tribe to investigate variability in both parameters, and contrasted the chromosomal data with the phylogeny previously proposed by other authors (Cañón et al. 2014; Teta et al. 2016).
Finally, we performed basic molecular analyses of cytochrome b (cytb), Adh, βfbg, and IRBP sequences to investigate the degree of divergence in the tribe in these molecular markers. For this approach, we used some sequences of the same dataset employed by Teta et al. (2016) available in GenBank. We calculated the genetic distances for all pairwise comparisons with the MEGA 6.0 software (Tamura et al. 2013). The selected model was K2P since all DNA regions displayed Tv/Ts bias and showed relatively low genetic variability, besides that it is the most used model in molecular analyses (Supplementary Table 3).
Results
In this paper we present, for the first time, the chromosome complements of members of the tribe Abrotrichini from San Juan (A. andina) and Mendoza (A. hirta, A. olivacea, and P. macronyx) provinces in Argentina, analyzed with conventional and differential cytogenetic techniques (Supplementary Tables 1 and 2). All specimens of Abrothrix studied, including samples of two different subgenera [A. (Abrothrix) hirta, A. (Angelomys) olivacea, and A. (Angelomys) andina] had the same 2n = 52 and FNa = 56 (Fig. 1). These complements consisted of 25 autosomes and a pair of sex chromosomes XX/XY. Among the autosomes, 22 pairs were acrocentric and three pairs were small submetacentrics. The subtelocentric X was one of the largest chromosomes in the complement, while the Y was one of the smallest chromosomes, with very small short arms, often difficult to identify. In A. andina, the Y chromosome was larger than in the other species (Fig. 1d). In all specimens, DAPI and C-bands were similar (Fig. 1c–g). Small pericentromeric C-bands in all autosomes and in the X chromosome were observed (Fig. 1e–g). One small biarmed pair exhibited a small interstitial C mark. The Y chromosome of all specimens was heterochromatic. However, in A. andina, a larger amount of CH and a DAPI neutral pericentromeric region, absent in other specimens, were detected (Fig. 1c–g).
Chromosomal constitution of the Abrothrix specimens analyzed in this work. a Karyotype of a male of A. olivacea with conventional staining. b In frame are sex chromosomes of one female. c DAPI bands in chromosomes of A. hirta. d In frame are the sex chromosomes of a male of A. andina. e–g C-bands in Abrothrix: eA. olivacea, fA. hirta, gA. andina
Individuals of Paynomys macronyx shared a diploid number of 2n = 54, with 26 pairs of autosomes and the sex pair XX/XY (Fig. 2). Among autosomes, pair one was subtelocentric and pairs 25 and 26 were small submetacentrics. The remaining autosomes were acrocentric and varied in size from large to small. In some individuals (five of nine; three males and two females), pair 8 was heteromorphic, formed by an acrocentric and a metacentric chromosome. This produced a variation in FNa between 58 and 59 (Fig. 2a, c). The biarmed homolog of this heteromorphic pair had a large amount of CH (Fig. 2h). The X chromosome also presented heteromorphism due to differences in length of its short arms, and could be submetacentric (with two different morphological variants) or acrocentric (Fig. 2D–E; Supplementary Table 2). Its short arms were completely heterochromatic (Fig. 2f–h) and variable to DAPI staining (Fig. 2c–e). The Y chromosome was one of the smallest DAPI positive acrocentrics (Fig. 2c). Supernumerary microchromosomes were detected in eight of nine individuals; the number of B chromosomes varied among individuals and among cells of the same individual (Supplementary Table 1). B chromosomes were C positive and neutral to DAPI staining (Fig. 2c, f–h). In the autosomes, the CH was concentrated in the pericentromeric regions (Fig. 2f–h). In P. macronyx, telomeric FISH signals were observed only at both terminal regions of each chromosome (Fig. 3a and b). Some variation in the intensity of fluorescent signals was detected at both, intra- and inter-chromosomal level. B chromosomes showed high intensity of fluorescence with the telomeric probe (Fig. 3a). In meioses, the XY pair showed the characteristic end-to-end association (Fig. 3).
Chromosome complements of Paynomys macronyx. a Conventional staining in a cell form a specimen with different X chromosomes. b In frame is a pair of X chromosomes with other morphology from another female. c DAPI bands of a male. d–e Sex chromosomes of two different females. f–g C-bands of cells from two females with different number of B chromosomes. h C-bands in a male that carried the heteromorphic pair. Note the different number of supernumerary chromosomes among the cells
a FISH in Paynomys macronyx with the pantelomeric probe. The arrows indicate the X, the biarmed chromosome of the heteromorphic pair and two B chromosomes. Note the strong signals in the supernumerary chromosomes. Arrowheads indicate chromosomes with different intensity of fluorescent signals at both ends. b Detail of the X chromosome and the biarmed homolog of the heteromophic pair. c Meioses in P. macronyx. Detail of the sex pair in diakinesis. Xe = euchromatic arm of the X chromosomes, Xh = heterochromatic arm of the X chromosome, Y = chromosome Y. d NORs in Abrothrix hirta. e NORs in P. macronyx
The Ag-NOR technique reveled the same banding pattern in both genera. Positive marks in the short arms of two pairs of acrocentric chromosomes were observed (Fig. 3d–e). The comparison between the banding chromosome complements of both genera indicated a high degree of conservation between them; however, some differences were observed. For example, all specimens of Abrothrix had an extra pair of small biarmed chromosomes. Also, pair one of P. macronys was differentiated; the long arms appeared homologous to pair three of Abrothrix and vice versa, but not its short arms. The long arms of the X chromosomes were similar in banding pattern, but the short arms of Paynomys did not evidenced visible banding homology to any chromosome of Abrothrix (Figs. 1c and 2c).
Chromosome variability within the tribe
Few different chromosome complements were described for the tribe Abrotrichini, especially for the specious genus Abrothrix (Supplementary Table 2). All diploid numbers recorded in the tribe were even. The only autosomal polymorphism was the one described here in P. macronyx, and produced variation in the FNa. The sex chromosomes were variable. The Xs were always big in size and presented different morphologies (submetacentric, subtelocentric, or acro-telocentric). In only one species, P. macronyx, we found these variants as polymorphism. The Y chromosome could be submetacentric, subtelocentric, or acro-telocentric, but always with a small size. These morphological variations were described at intra- and inter-specific level. A constant chromosome number characterizes the Abrothrix species. In this genus, few variations in the FNa, due to differences in the number of small biarmed autosomes, were reported. These chromosome pairs are difficult to distinguish, and in some cases seem to correspond to differences in chromatin condensation among samples. A 2n = 44 was referred to Abrothrix in a simple report (Supplementary Table 2). The same occur with the mentioned diploid number of 52 for Paynomys macronyx and Geoxus valdivianus, but none of these chromosome complements were showed to corroborate the information (Supplementary Table 2).
Molecular divergence within the tribe
Molecular divergences based on cytochrome b sequences were very high among all the studied species of the tribe (Supplementary Table 3), whereas the nuclear markers showed lower levels of variability. The variation in the mitochondrial marker was in general an order of magnitude greater than in nuclear sequences. The number of substitutions found in the βFBG was superior to the one observed in the Adh; the IRBP presented the lowest variation. There was a taxonomic scaling in the genetic distances in all molecular markers analyzed. But Abrotrichina, which include all its diversity in the unique genus Abrothrix, displayed similar values of maximum divergence than Notiomyina, in which the comparisons encompassed three to four genera depending on the marker (Supplementary Table 3). The only exception was the βFBG, where the values of maximum divergence were similar to the maximum observed for the entire tribe.
Discussion
Earlier taxonomic assessments, mostly based on morphological evidences, placed Abrotrichini as part of Akodontini, with whom they share several morphological traits (see Reig 1987). Molecular data provided by recent studies enable the novel diagnosis of the tribe, which is strongly supported in all phylogenetic analyzes (Smith and Patton 1999; D’Elía 2003; Parada et al. 2013; Cañón et al. 2014), and is characterized by deep genetic divergences between species (see Supplementary Table 3). Cytogenetic evidence indicate a conserved 2n = 52 and FNa = 56 with minor variations in the FNa for the genus Abrothrix, including representatives of the four known subgenera (Bianchi et al. 1971; Gallardo 1982; Rodriguez et al. 1983; Patterson et al. 1984; Liascovich et al. 1989; Spotorno et al. 1990; Feijoo et al. 2010; present work). The morphology of the Y chromosome varies at both intra- and inter-specific level, as in others genera of sigmodontine rodents (Lanzone et al. 2016). The occurrence of a population of Abrothrix cf. A. olivacea in Comodoro Rivadavia (Chubut, Argentina) with 2n = 44 (Rodriguez and Theiler 2007) needs further confirmation. This 2n resembles the chromosome complements of some species of Akodon, a genus morphologically similar to Abrothrix, with which it can be easily confused (Teta et al. 2016 and references therein).
Due to its extensive occurrence, some authors suggest that the 2n = 52 represents a synapomorphic character for the tribe Abrotrichini (Smith and Patton 1999). The revision of the literature indicates that this 2n is shared by all Abrothrix species (subtribe Abrotrichina; sensu Teta et al. 2016), but not for its sister clade Notiomyina (Teta et al. 2016). This 2n is also shared by some akodontines such as Thaptomys and Brucepattersonius (Lanzone et al. 2018 and references therein), as well as by some species from other tribes, such as the Oryzomyini Nectomys rattus (Maia et al. 1984; Bonvicino et al. 1996), and the Phyllotini Eligmodontia moreni (Lanzone et al. 2016), among others. Evidences based on the chromosome painting technique, combined with molecular phylogeny, indicate that 2n = 52 is the plesiomorphic condition for the subfamily Sigmodontinae (Swier et al. 2009). This suggests that the chromosome complement of Abrothrix species could be very similar to that of the ancestral complement of the subfamily, which reinforces the hypothesis of chromosome stability within this genus.
There is no chromosomal data for some of the species within the Notiomyina clade (see Supplementary Table 2). However, Paynomys and Geoxus display chromosomal differentiation in the diploid and fundamental numbers, complementing and confirming previous observations (Ojeda et al. 2005; D’Elía et al. 2006; present work). Considering the 2n = 52 ancestral for the tribe, these species show an increase in the 2n and FNa, which can be distinctive for the subtribe. Phylogenetic analyzes, based on molecular and morphological characters, showed that Pearsonomys and Geoxus are congeneric forms (Smith and Patton 1999; D’Elía et al. 2006; Teta et al. 2016). This relationship is also supported by chromosome data, because both taxa share very similar chromosome complements (D’Elía et al. 2006; Supplementary Table 2). Pearson (1984) described a 2n = 52 for Geoxus valdivianus, indicating that this 2n was identical- or almost- to that of P. macronyx. However, none of these chromosome complements were exhibited to corroborate the information and may represent errors in the literature.
To accommodate the taxonomy with the evolutionary history of the taxa recovered in the phylogenetic trees, Teta et al. (2016) named a new genus, Paynomys, to include Chelemys macronyx, which in turn do not form a monophyletic group with C. megalonyx (the type species of the genus Chelemys). Unfortunately, the karyotype of C. megalonyx is unknown. On the other hand, P. macronyx not only differs from all other abrotrichines in terms of their 2n and FN, but also differs in the presence of B chromosomes, which varies at intra- and inter-population levels (Ojeda et al. 2005 and data cited there; present work). Other chromosome characteristics, apparently unique in Paynomys within abrotrichines, are the polymorphism in the X chromosome and the occurrence of a medium sized chromosome almost completely heterochromatic. All these differential chromosomal characteristics support the inclusion of P. macronyx in its own genus.
The karyotypes of Paynomys and Geoxus are morphologically very similar, supporting their close relationship. Geoxus annectens and P. macronyx share a pair one subtelocentric and a submetacentric X chromosome (Ojeda et al. 2005; D’Elía et al. 2006; present work). In all Abrothrix species, pair one is acrocentric and the X is subtelocentric. Thus, chromosome characteristics support the differentiation of both major clades within Abrotrichini.
Species within the tribe Abrotrichini have low chromosome variability. Several taxa share the same complement, and the ranges of variation in the 2n and FNa are narrow. In addition, species diverging in the 2n and FNa were very similar in DAPI, C, and Ag-NOR banding pattern, which indicate conservation in most chromosomes pairs. However, the divergence among cytochrome b sequences within the abrotrichines is high, compared with that of most rodent species (Baker and Bradley 2006), indicating uncoupling between the rates of chromosomal and mitochondrial evolution. In the case of abrotrichines, chromosome evolution appears to be slower than molecular one in the mitochondrial genome. This incongruence between cytogenetic and molecular evidence was also observed in some other rodents, where chromosome evolution appeared accelerated compared with a low molecular divergence (Buschiazzo et al. 2018). On the other hand, molecular divergences in the nuclear sequences were low, as observed in other rodents (D’Elía 2003; Steppan et al. 2007), regardless of its chromosome variability. The βFBG was the marker with the highest number of substitutions, which was also observed in other taxa (Henson and Bradley 2009; Machado et al. 2014). However, there are different combinations of molecular markers used in different works, especially from the nuclear genome, which prevent broader comparisons among sigmodontines.
Chromosomes in P. macronyx
This species is very peculiar among rodents, since it possess three chromosome variations that are uncommon to this group. Paynomys macronyx has B chromosomes, heterochromatic variations of the X chromosome, and a heteromorphic pair of autosomes. However, FISH signals were observed only at the ends of all chromosomes. Some variations in the intensity of signals in some autosomal pairs were observed, but not in the chromosomes that present polymorphic variants. This suggests that telomeric sequences are not related to the chromosomal modifications detected in Paynomys. Strict telomeric signals are also common in some sigmodontines displaying chromosome rearrangements (Lanzone et al. 2015). However, in the B chromosomes, signals were very strong, suggesting that they are composed in a large proportion of telomeric sequences. Additional telomeric repeats were also recorded in several B chromosomes of other mammals (Vujošević et al. 2018).
Supernumerary chromosomes are infrequent in mammals, with most descriptions in the literature belonging to rodents from the superfamily Muroidea (Palestis et al. 2004; Vujošević and Blagojević 2004; Vujošević et al. 2018). In general, these chromosomes are heterochromatic, as observed in P. macronyx, but they are found in low frequencies within populations (Palestis et al. 2004; Vujošević and Blagojević 2004). Some exceptions are that of Trinomys iheringi, where dot-like B chromosomes were observed in all individuals (Fagundes et al. 2004), plus that of some species of Nectomys in which several variants of supernumerary chromosomes were found in high frequency in some populations (Maia et al. 1984). Also, in two Apodemus species, A. peninsulae and A. flavicollis, Bs were found in almost all populations, in frequencies reaching 100 percent in the former species (Kartavtseva and Roslik 2004; Wójcik et al. 2004). In P. macronyx, these chromosomes were also dot-like and unstable, producing mosaicism in somatic cells in all specimens. In a review of Brazilian rodents carrying B chromosomes, it was reported that Akodon montensis, N. squamipes, Oligoryzomys flavescens, Proechimys sp., and T. iheringi presented mosaicism; but except for Trinomys, in all the other species this mitotic instability was observed only in some individuals (Fagundes et al. 2004; Silva and Yonenaga-Yassuda 2004). The high frequency of these chromosomes in P. macronyx and their instability suggest that they are in an early evolutionary stage of accumulation (Camacho et al. 2000), at least in the studied region.
Another unusual variation observed in P. macronyx is the polymorphism of the X chromosome (Fredga 1970, 1988; Paresque et al. 2007). These variations are due to differences in the length of its short arms, which are heterochromatic, as in N. squamipes (Maia et al. 1984) and O. nigripes (Paresque et al. 2007). Also, the occurrence of a mostly heterochromatic medium sized autosome is a rare cytogenetic feature. Intriguingly, a similar case was described in the phyllotine Phyllotis xanthopygus, from a population of the same geographic region as that of P. macronyx described here (Labaroni et al. 2014).
In P. macronyx, the CH content was higher than in the studied species of Abrothrix. Not only the pericentromeric blocks were bigger, but also larger amounts of CH in the X chromosome and in one autosome were detected. The occurrence of a large variability in the amount of CH in the X chromosomes, in addition to the presence of B chromosomes in high frequency, was also detected in N. squamipes (Maia et al. 1984). This suggests some relationship between the accumulation of CH in the standard complement, especially in the variable short arms of the X, and the presence of supernumerary chromosomes in these species. The relation between sex chromosomes and the origin of supernumerary chromosomes was demonstrated in some species (Karamysheva et al. 2002; Silva and Yonenaga-Yassuda 2004; Vujošević and Blagojević 2004; Rajičić et al. 2017), and seems to be the case for P. macronyx.
References
Baker RJ, Bradley RD (2006) Speciation in mammals and the genetic species concept. J Mammal 87:643–662
Bianchi NO, Reig A, Molina OJ, Dulout FN (1971) Cytogenetics of the South American akodont rodents (Cricetidae). I A progress report of Argentinian and Venezuelan forms. Evolution 25:724–736
Bonvicino CR, D’Andrea PS, Cerqueira R, Seuánez HN (1996) The chromosomes of Nectomys (Rodentia, Cricetidae) with 2n=52, 2n=56, and interspecific hybrids (2n = 54). Cytogenet Cell Genet 73:190–193
Buschiazzo LM, Caraballo DA, Cálcena E, Longarzo ML, Labaroni CA, Ferro JM, Rossi MS, Bolzán AD, Lanzone C (2018) Integrative analysis of chromosome banding, telomere localization and molecular genetics in the highly variable Ctenomys of the Corrientes group (Rodentia; Ctenomyidae). Genetica 146:403–414
Camacho JPM, Sharbel TF, Beukeboom LW (2000) B chromosome evolution. Phil Trans Royal Sc B: Biol Sc 355:163–178
Cañón C, Mir D, Pardiñas UFJ, Lessa EP, D’Elía G (2014) A multilocus perspective on the phylogenetic relationships and diversification of rodents of the tribe Abrotrichini (Cricetidae: Sigmodontinae). Zool Sc 43:443–454
D’Elía G (2003) Phylogenetics of Sigmodontinae (Rodentia, Muroidea, Cricetidae), with special reference to the Akodon group, and with additional comments on historical biogeography. Cladistics 19:307–323
D’Elía G, Ojeda A, Mondaca F, Gallardo MH (2006) New data of the long-clawed mouse Pearsonomys annectens (Cricetidae, Sigmodontinae) and additional comments on the distinctiveness of Pearsonomys. Mamm Biol 71:39–51
Evans EP, Breckon G, Ford CE (1964) An air-drying method for meiotic preparations from mammalian testes. Cytogenetics 3:289–294
Fagundes V, Camacho J, Yonenaga-Yassuda Y (2004) Are the dot-like chromosomes in Trinomys iheringi (Rodentia, Echimyidae) B chromosomes? Cytogenet Genome Res 106:159–164
Feijoo M, D’Elía G, Pardiñas UFJ, Lessa EP (2010) Systematics of the southern Patagonian-Fueguian endemic Abrothrix lanosus (Rodentia: Sigmodontinae): phylogenetic position, karyotypic and morphological data. Mamm Biol 75:122–137
Ford CE, Hamerton JL (1956) A colchicine, hypotonic citrate, squash sequence for mammalian chromosomes. St Technol 31:247–251
Fredga K (1970) Unusual sex chromosome inheritance in mammals. Phil Trans Royal Sc B: Biol Sc 259:15–36
Fredga K (1988) Aberrant chromosomal sex-determining mechanisms in mammals, with special references to species with XY females. Phil Trans Royal Sc B: Biol Sci 322:83–95
Gallardo MH (1982) Chromosomal homology in southern Akodon. Experientia 38:1485–1487
Henson DD, Bradley RD (2009) Molecular systematics of the genus Sigmodon: results from mitochondrial and nuclear gene sequences. Can J Zool 87:211–220
Howell WN, Black DA (1980) Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a one step method. Experientia 36:1014–1015
Karamysheva T, Andreenkova O, Bochkaerev M, Borissov Y, Bogdanchikova V, Borodin V, Rubtsov V (2002) B chromosomes of korean field mouse Apodemus peninsulae (Rodentia, Murinae) analysed by microdissection and FISH. Cytogenet Genome Res 96:154–160
Kartavtseva IV, Roslik GV (2004) A complex B chromosome system in the Korean field mouse Apodemus peninsulae. Cytogenet Genome Res 106:271–278
Labaroni C, Malleret M, Novillo A, Ojeda A, Rodriguez D, Martí DA, Lanzone C (2014) Karyotypic variation in the Andean rodent Phyllotis xanthopygus (Waterhouse, 1837) (Rodentia, Cricetidae, Sigmodontinae). Comp Cytogenet 8:369–381
Lanzone C, Labaroni CA, Suárez N, Rodríguez D, Herrera ML, Bolzán AD (2015) Distribution of telomeric sequences (TTAGGG)n in rearranged chromosomes of phyllotine rodents (Cricetidae, Sigmodontinae). Cytogenet Genome Research 147:247–252
Lanzone C, Cardozo D, Sánchez DM, Martí DA, Ojeda RA (2016) Chromosomal variability and evolution in the tribe Phyllotini (Rodentia, Cricetidae, Sigmodontinae). Mammal Res 61:373–382
Lanzone C, Labaroni CA, Formoso A, Buschiazzo LM, Da Rosa F, Teta P (2018) Diversidad, sistemática y conservación de roedores en el extremo sudoccidental del Bosque Atlántico Interior. Rev Mus Arg Cs Nat 20:151–164
Liascovich RC, Bárquez RM, Reig OA (1989) A karyological and morphological reassessment of Akodon (Abrotrix) illuteus Thomas. J Mammal 70:386–391
Machado LF, Leite YLR, Christoff AU, Giugliano LG (2014) Phylogeny and biogeography of tetralophodont rodents of the tribe Oryzomyini (Cricetidae: Sigmodontinae). Zool Sc 43:119–130
Maia V, Yonenaga-Yassuda Y, Freitas JRO, Kasahara S, Suñé-Mattevi M, Oliveira LF, Galindo MA, Sbalqueiro IJ (1984) Supernumerary chromosomes in Nectomys squamipes (Cricetidae-Rodentia). Genetica 63:121–128
Ojeda AA, D’Elía G, Ojeda R (2005) Taxonomía alfa de Chelemys y Euneomys (Rodentia, Cricetidae): el número diploide de ejemplares topotípicos de C. macronyx y E. mordax. Mastozool Neotrop 12:79–82
Palestis B, Burt A, Jones R, Trivers R (2004) B chromosomes are more frequent in mammals with acrocentric karyotypes: support for the theory of centromeric drive. Proceedings of the Royal Society of London. Series B, Biol Sc 271:22–24
Parada A, Pardiñas UFJ, Salazar-Bravo J, D’Elía G, Palma RE (2013) Dating an impressive Neotropical radiation: molecular time estimates for the Sigmodontinae (Rodentia) provide insights into its historical biogeography. Mol Phylogenet Evol 66:960–968
Paresque R, Silva M, Yonenaga-Yassuda Y, Fagundes V (2007) Karyological geographic variation of Oligoryzomys nigripes Olfers, 1818 (Rodentia, Cricetidae) from Brazil. Genet Mol Biol 30:43–53
Patterson BD, Gallardo MH, Freas KE (1984) Systematics of mice of the subgenus Akodon (Rodentia:Cricetidae) in southern South America, with the description of a new species. Field Zool 23:1–16
Patton JL (1967) Chromosome studies in certain pocket mouse, genus Perognathus (Rodentia, Heteromyidae). J Mammal 48:27–37
Patton JL, Pardiñas UFJ, D’Elía G (2015) Mammals of South America. 2 Rodents. University of Chicago Press, Chicago, Illinois.
Pearson OP (1984) Taxonomy and natural history of some fossorial rodents of Patagonia, southern Argentina. J Zool 202:225–237
Rajičić M, Romanenko SA, Karamysheva TV, Blagojević J, Adnađević T, Budinski I, Bogdanov AS, Trifonov VA, Rubtsov NB, Vujošević M (2017) The origin of B chromosomes in yellow-necked mice (Apodemus flavicollis)-break rules but keep playing the game. PLoS One 12(3):e0172704
Reig OA (1987) An assessment of the systematics and evolution of the Akodontini, with the description of new fossil species of Akodon (Cricetidae: Sigmodontinae). Field Zool 39:347–399
Rodriguez VA, Theiler GR (2007) Micromamíferos de la Región de Comodoro Rivadavia (Chubut, Argentina). Mastozool Neotrop 14:97–100
Rodriguez M, Montoya V, Venegas W (1983) Cytogenetic analysis of some Chielan species of the genus Akodon Meyen (Rodentia, Cricetidae). Caryologia 36:129–138
Schweizer D (1980) Simultaneous fluorescent staining of R-bands and specific heterochromatic regions (DA/DAPI bands) in human chromosomes. Cytogenet Cell Genet 27:190–193
Sikes RS, and the Animal Care and Use Committee of the American Society of Mammalogists (2016) Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal 97:663–688
Silva MJJ, Yonenaga-Yassuda Y (2004) B chromosomes in Brazilian rodents. Cytogenet Genome Res 106:257–263
Smith MF, Patton JL (1999) Phylogenetic relationships and the radiation of Sigmodontine rodents in South America: evidence from Cytochromo b. J Mamm Evol 6:89–127
Spotorno AE, Zuleta CA, Cortes A (1990) Evolutionary systematics and heterocrony in Abrotrix species (Rodentia, Cricetidae). Evol Biol 4:37–62
Steppan SJ, Ramirez O, Banbury J, Huchon D, Pacheco V, Walker LI, Spotorno AE (2007) A molecular reappraisal of the systematics of the leaf-eared mice Phyllotis and their relatives. In: Kelt DA, Lessa EP, Salazar-Bravo JA, Patton JL (eds) The quintessential naturalist: honoring the life and legacy of Oliver P, vol 134. Univ California Publ Zool USA, Pearson, pp 799–826
Sumner AT (1972) A simple technique for demostrating centromeric heterochromatin. Exp Cell Res 75:304–306
Swier VJ, Bradley RD, Rens W, Elder FFB, Baker RJ (2009) Patterns of chromosomal evolution in Sigmodon, evidence from whole chromosome paints. Cytogenet Genome Res 125:54–66
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Teta P, Cañon C, Patterson BD, Pardiñas UFJ (2016) Phylogeny of the tribe Abrotrichini (Cricetidae, Sigmodontinae): integrating morphological and molecular evidence into a new classification. Cladistics 33:153–182
Vujošević M, Blagojević J (2004) B chromosomes in populations of mammals. Cytogenet Genome Res 106:247–256
Vujošević M, Rajičić M, Blagojević J (2018) B chromosomes in populations of mammals revisited. Genes 9:487
Wójcik JM, Wójcik AM, Macholán M, Piálek J, Zima J (2004) The mammalian model for population studies of B chromosomes: the wood mouse (Apodemus). Cytogenet Genome Res 106:264–270
Acknowledgments
Our thanks to Benjamín Bender for his assistance with the material deposited in the Mammal Collection of the Instituto Argentino de Zonas Aridas (CMI), IADIZA-CONICET, Mendoza.
Funding
This research has been partially funded by PIP-CONICET 1122015 0100258 CO: RAO, PT, CL; PICT 2016-0537: PT, CL, as well as by PIP No 0182, CONICET, RI: ADB and funds from the CICPBA and UNLP of Argentina.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Fig 1:
Map showing the sampling localities of the abrotrichines analyzed with chromosome techniques in this work: 1- Barreal, San Juan; 2- Las Heras, Mendoza; 3- Tunuyán, Mendoza; 4-San Carlos, Mendoza; 5- Malargüe, Mendoza; 6- San Rafael, Mendoza. (JPG 2242 kb)
Supplementary Table 1:
Species, collection number, sex and chromosome constitution of the specimens analyzed in this work. For Paynomys macronyx the different morphology of the X chromosome, the presences of the heteromorphic pair and the number of B chromosomes per cell per individual, are indicated. (XLSX 13 kb)
Supplementary Table 2:
Chromosome data in the tribe Abrothichini. In grey are chromosome complements that could not be confirmed. The species name were in accordance with the revision done by Teta et al. (2016) (XLSX 12 kb)
Supplementary Table 3:
K2P genetic distances of different DNA regions (Cyt-b; FBG; IRBP; ADH) of abrotrichines obtained from GenBank sequences. The minimum and maximum of intrageneric comparisons within Abrothrix (Abrotrichina sensu Teta et al. 2016), intergeneric comparisons within Notiomyina, and inter subtribe genetic distances (between samples from Abrotrichina and Notiomyina) are showed. (XLSX 31 kb)
Rights and permissions
About this article
Cite this article
Da Rosa, F., Ojeda, A., Novillo, A. et al. Chromosome variability and evolution in rodents of the tribe Abrotrichini (Rodentia, Cricetidae, Sigmodontinae). Mamm Res 65, 59–67 (2020). https://doi.org/10.1007/s13364-019-00463-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13364-019-00463-0