Abstract
The cytogenetic characterization of the carrot genome (Daucus carota L., 2n = 18) has been limited so far, partly because of its somatic chromosome morphology and scant of chromosome markers. Here, we integrate the carrot linkage groups with pachytene chromosomes by fluorescent in situ hybridization (FISH) mapping genetically anchored bacterial artificial chromosomes (BACs). We isolated a satellite repeat from the centromeric regions of the carrot chromosomes, which facilitated the study of the pachytene-based karyotype and demonstrated that heterochromatic domains were mainly confined to the pericentromeric regions of each chromosome. Chromosome-specific BACs were used to: (1) physically locate genetically unanchored DNA sequences, (2) reveal relationships between genetic and physical distances, and (3) address chromosome evolution in Daucus. Most carrot BACs generated distinct FISH signals in 22-chromosome Daucus species, providing evidence for syntenic chromosome segments and rearrangements among them. These results provide a foundation for further cytogenetic characterization and chromosome evolution studies in Daucus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The cultivated carrot (Daucus carota subsp. sativus) is the most economically important and extensively studied species of the Apiaceae family. It ranks among the top ten vegetable crops in the world (Rubatzky et al. 1999; Simon et al. 2008) and is an excellent source of dietary provitamin A carotenoids (Simon 2000). Carrot is a diploid outcrossing species like the majority of the approximately 20 Daucus species. However, carrot, Daucus capillifolius, Daucus sahariensis, and Daucus syrticus are the only members of the genus with 2n = 18 as most Daucus species are 2n = 20 or 22. The haploid genome size of carrot has been estimated at 473 Mbp (Arumuganathan and Earle 1991), which is similar to rice.
Several projects have been initiated to enhance genetic resources for carrot. In the last two decades, several carrot linkage maps have been developed, based on different types of mostly dominant markers (Simon et al. 2008). This effort was recently complemented by construction and end sequence evaluation of a deep-coverage bacterial artificial chromosome (BAC) library (Cavagnaro et al. 2009) and expressed sequence tags (EST) database development underway (Bowman et al. 2010). Despite these advances in the molecular analysis of the carrot genome, very limited work has been done on the cytological characterization of the carrot genome. Somatic chromosome identification in carrot has been hampered by their small sizes and similar morphology (Iovene et al. 2008a), and carrot chromosomes and linkage maps are not integrated.
The development of cytogenetic maps using a BAC fluorescent in situ hybridization (FISH) approach is a well-established procedure in plant species with small to medium genome sizes. These maps have played an important role in genome sequencing projects of several model and crop species and in comparative mapping studies (Jiang and Gill 2006; Figueroa and Bass 2010).
In this report, we describe a standard carrot karyotype and assign the carrot linkage groups to specific pachytene chromosomes by FISH mapping of 17 map-anchored BACs. We identified a tandem repeat that hybridized to the centromeric regions of all carrot chromosomes. Comparative FISH mapping of carrot BACs to two 2n = 22 different Daucus species (one from Europe and one from the Americas) established a framework of chromosomal synteny that provides insights into chromosome evolution in Daucus species with varying chromosome numbers.
Materials and methods
Plant materials and chromosome preparation
Seeds of D. carota L. var. sativus male-fertile inbred lines B2566 and B493 were germinated in Petri dishes to obtain root tips for mitotic chromosomes preparation, as previously described (Dong et al. 2000; Iovene et al. 2008a). Vernalized plants were also grown to flowering in the greenhouse to obtain meiotic samples, and immature umbels were fixed in 3:1 Carnoy’s solution. In preliminary experiments, slides were prepared by squashing the whole flower bud. However, this resulted in poor preparations with relatively few pollen mother cells dispersed among many floral somatic cells. For this reason, the anthers were isolated under a dissecting microscope and digested in an enzyme mixture as previously described (Iovene et al. 2008b). After digestion, three to five anthers were transferred to a glass slide, macerated in a drop of 3:1 methanol/acetic acid solution with fine-pointed forceps and finally “flame-dried” over an alcohol flame. For comparative FISH mapping, mitotic chromosomes of the Mediterranean Daucus crinitus (2n = 2x = 22; Ames 26417) and the American Daucus pusillus (2n = 2x = 22; PI 511864; Sáenz Laín 1981) were prepared as previously described (Iovene et al. 2008a).
BAC library screening, probes, and FISH
Most BAC clones used for FISH mapping were identified by screening two carrot BAC libraries (Cavagnaro et al. 2009) with mapped sequences using a PCR-based approach (Table 1). A total of 17 mapped markers were used for screening. Of these, 15 markers were included in the carrot reference linkage map, constructed previously using an F2 population derived from a cross between the dark orange inbred B493 and a wild white-rooted carrot Queen Anne’s Lace (QAL) (Santos and Simon 2002, 2004; Grzebelus et al. 2007; Just et al. 2007; Cavagnaro et al. 2009, 2010). These markers were distributed across all nine linkage groups (Grzebelus et al. 2007; Just et al. 2007; Cavagnaro et al. 2009, 2010) and included eight STS markers (based on putative carotenoid biosynthetic gene sequences), four simple sequence repeats (SSRs), one sequence-characterized amplified region (SCAR), and two PCR markers (for Rs and DCOR sequences; Table 1). In addition, two other markers from different carrot linkage maps were used to screen the BAC library and develop potential FISH probes. These markers were a SCAR (Q1/800) linked to the nematode resistance locus Mj-1 (Boiteux et al. 2000), and an RFLP (DCG321) linked to a phenotypic locus conditioning purple root pigmentation (P1; Vivek and Simon 1999). Because their linkage was established in genetic background unrelated to the B493 × QAL cross, and since no common markers among these linkage groups were available, the linkage relationships between these two markers and those previously mentioned were initially unknown. Finally, four BAC clones not genetically anchored to any linkage map were also included. Of these non-anchored clones, BACs 124B06 and 238G06 were selected because their ends contained gene sequences (invertase and phenyl alanine ammonia-lyase) potentially associated with important carrot root traits (i.e., reduced sugar content and anthocyanin pigmentation, respectively); another clone (019A16) was identified in the library using the primers for isopentenyl pyrophosphate isomerase gene, which could not be mapped by Just et al. (2007) due to lack of polymorphism in the B493 × QAL F2; and BAC clone 112G15 was randomly selected. Other DNA clones were pTa71 and pTa794 containing the 18S–25S rDNA unit and the 5S rDNA unit, respectively (Gerlach and Bedrock 1979; Gerlach and Dyer 1980). Overall, a total of 23 FISH probes were used for physical mapping in carrot, and—from these—a subset of 16 and 13 carrot BAC probes were used for comparative FISH mapping in D. crinitus and D. pusillus, respectively.
BAC DNA was isolated using the QIAGEN plasmid midi-kit and labeled with either biotin-16-UTP or digoxigenin-11-dUTP (Roche Diagnostic, Indianapolis, IN) by standard nick translation reaction. The FISH procedure was applied to both mitotic and meiotic chromosomes according to published protocols (Dong et al. 2000; Iovene et al. 2008b). Most BAC probes required small or moderate amounts of carrot-sheared genomic DNA in the hybridization mixture to reduce background signal (approximately a 20:1 ratio of sheared genomic DNA/BAC probe was used). A few BACs (for example, BAC 002B20, 009K15, and 098O13) required a larger amount of blocking DNA in the hybridization mixture (ratio, ~100:1) to produce a clear signal. To obtain more consistent measurements, high-quality pachytene chromosomes were used for repeated probing up to four times using the procedure described by Cheng et al. (2001a). Alternatively, FISH cocktails consisting of up to ten different BAC probes were applied. The carrot cot-1 DNA fraction was prepared according to Zwick et al. (1997) and used as a FISH probe along with BAC 004H08 (a clone containing a satellite repeat associated with all carrot centromeres; see below).
Biotin- and digoxigenin-labeled probes were detected by fluorescein isothiocyanate-conjugated anti-biotin antibody and rhodamine-conjugated anti-digoxigenin antibody (Roche Diagnostic), respectively. Chromosomes were counterstained by 4′, 6-diamidino-2-phenylindole (DAPI) in antifade solution VectorShield (Vector Laboratories, Burlingame, CA).
Cytological measurements and analysis
All images were captured using a SenSys CCD camera attached to an Olympus BX60 epifluorescence microscope, and the IPLab Spectrum v3.1 software (Signal Analytics, Vienna, VA) on a Macintosh computer. Final image adjustments were done with Adobe Photoshop software v6.0. Measurements were made on digital images using IPLab software. For karyotype description, all chromosomes in 24 pachytene cells were measured. The position of each BAC clone on a specific chromosome is given as a relative distance in percent, calculated as the distance of the FISH signal from the end of the short-arm relative to the total length (in micrometer) of the chromosome.
DNA sequencing and assembly
BAC 004H08 was sequenced by shotgun Sanger DNA sequencing essentially as described in Zhu et al. (2008) with the modifications of shotgun insert size range of 2–6 kbp and assembly with TIGR Assembler (Sutton et al. 1995). The low pass sequence resulted in 35 pieces and the Phase I BAC sequence is available in GenBank under the accession number HM565961.
Results and discussion
Identification of a satellite repeat associated with the centromeres of D. carota chromosomes
BAC clone 004H08 was initially identified by screening the library for the phytoene synthase 1 gene (PSY1; Cavagnaro et al. 2009). FISH analysis revealed that BAC 004H08 hybridized to the centromeric regions of all nine pairs of the carrot chromosomes and not elsewhere. The intensity of the FISH signals differed among chromosomes and overlapped with the FISH signals of the carrot cot-1 DNA fraction, suggesting that this BAC contains a dominant repeat associated with the centromere (Supplementary Fig. S1). The partial sequence of the BAC clone 004H08 revealed a tandem repeat with a typical length of ~159 bp, which we named Cent-Dc. This monomer is composed of a variable number of subrepeats, typically four (Table 2, Supplementary File S1). These subrepeats vary slightly in sequence, each of which has a period size of 39–40 bp. Cent-Dc has 91% similarity to DCRE20, a carrot-specific repeat element previously identified in the BAC end sequences of the same library (Cavagnaro et al. 2009). Considering a period size of 40 bp, these authors estimated the carrot genome to contain > 27,000 copies of DCRE20 (Cavagnaro et al. 2009).
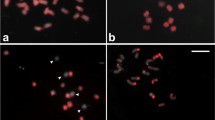
A 1,816 bp subclone (named K11) containing approximately ten Cent-Dc repeats was labeled and hybridized to mitotic and meiotic chromosomes of carrot (Fig. 1a–c). At metaphase I, FISH signals were consistently detected in one discrete region only at the most poleward position of each bivalent chromosome, suggesting that Cent-Dc is located at the chromosomal region associated with the kinetochore complex (Fig. 1b). Centromeric repeats have been isolated in a number of plant species (Jiang et al. 2003). The Cent-Dc repeat has mononucleosomic size that is typical for previously characterized centromeric satellite repeats. The structure and the cytological location of Cent-Dc suggest this repeat is likely associated with the functional centromeres of carrot chromosomes. Therefore, this subclone was used to anchor the centromere position of the carrot pachytene chromosomes (see below, Fig. 1c).
FISH mapping of Cent-Dc repeat to the centromeric regions of all carrot chromosomes and distribution of euchromatin and heterochromatin in the carrot genome. a Carrot somatic chromosomes probed with the Cent-Dc repeat (red signal). b Cent-Dc detected on carrot chromosomes at anaphase I and c at pachythene. d DAPI-stained chromosomes in (c) are converted to a black-and-white image to enhance the visualization of distribution of euchromatin and heterochromatin along the pachytene chromosomes. Chromosomes are numbered according their decreasing length. Bar scale = 5 μm
A pachytene chromosome-based karyotype of carrot
Pachytene chromosome-based karyotypes are superior to those based on somatic metaphase chromosomes and have been developed in a number of species with small to medium size genomes (Arabidopsis thaliana: Fransz et al. 1998; cucumber: Koo et al. 2005; Lotus japonicus: Ohmido et al. 2010; Medicago truncatula: Kulikova et al. 2001; potato: Iovene et al. 2008b; Tang et al. 2009; rice: Cheng et al. 2001b; sorghum: Islam-Faridi et al. 2002; Kim et al. 2005).
The somatic chromosomes of carrot are only 2–4 μm in length and morphologically similar (Sharma and Ghosh 1954; Schrader et al. 2003; Iovene et al. 2008a), thus they are difficult to identify. We developed a pachytene chromosome-based karyotype of carrot from inbred line B2566. The chromosome length and arm ratio data were collected from all chromosomes in 24 pollen mother cells (Table 3). The pachytene chromosomes are ordered from 1 to 9 according to their descending length. On average, the length of the carrot pachytene complement is ~193 ± 18 μm, which is only seven times longer than its mitotic counterpart. In plant species with a wide range of genome sizes, including Arabidopsis (Fransz et al. 1998), cotton (Wang et al. 2009), cucumber (Koo et al. 2005), maize (Wang et al. 2006), M. truncatula (Kulikova et al. 2001), potato (Tang et al. 2009), and rice (Cheng et al. 2001b), the pachytene chromosomes are approximately 15–50 times longer than the somatic metaphase chromosomes. The relatively short length of the carrot pachytene chromosomes may decrease the spatial resolution of FISH mapping in this species compared with other plant taxa. However, preparation of super-stretched pachytene chromosomes as demonstrated in maize (Koo and Jiang 2009) may overcome this limitation.
Carrot pachytene chromosomes could be individually recognized based on chromosome length, arm ratio and the DAPI staining pattern. In general, heterochromatic regions represented a small fraction of all the chromosomes and were mainly located in the pericentric regions (Figs. 1d and 2). Centromeric regions of carrot pachytene chromosomes did not have obvious primary constrictions and distinct differential staining as has been observed in the pachytene chromosomes of tomato and maize (Barton 1950; Dempsey 1994). Thus, Cent-Dc was useful to highlight the centromeric regions of each pachytene chromosome (Fig. 1) allowing us to perform more accurate measurements. Overall, the arm ratios of the carrot pachytene chromosomes ranged from 1.2 to 10.6 (although for most chromosomes, it ranged from 1.2 to 4.9; Table 3) compared with ~1 to ~4 for mitotic chromosomes (Iovene et al. 2008a). This discrepancy may reflect an unequal condensation between the chromosome arms. Based on the position of the centromere, carrot pachytene chromosomes were divided into the following three groups:
Ideogram of the distribution of heterochromatin on the pachytene chromosomes of the carrot line B2566. Solid/shaded thickenings represent the heterochromatic regions. Shaded thickenings indicate regions that were less stained by DAPI than the regions marked by solid thickenings. Hatched thickenings indicate DAPI-stained regions that were not consistently observed in each pollen mother cell and/or were sometimes separated into multiple smaller regions. Open circles represent the location of the centromeres, based on the hybridization to Cent-Dc. The dashed line on the top of chromosome 4 represents the 18S–25S rDNA; the dotted oval on the long arm of chromosome 8 represents the 5S rDNA. The relative length of each chromosome and the arm ratio were drawn based on data presented in Table 3
A group of three chromosomes (1, 6, and 8) with subterminal centromeres: pairwise t tests indicated that chromosomes 1, 6, and 8 varied significantly in length (P < 0.01). Thus, these chromosomes could be distinguished based on their length and DAPI staining pattern. In addition to being the longest chromosome, chromosome 1 had a small heterochromatic knob that was quite consistently detected on the long arm, at ~39% from the end of the short arm (based on 18 measurements out of the 24 pollen mother cells observed). This structure is potentially diagnostic to distinguish between chromosomes 1 and 6.
A group of two chromosomes (2 and 4) with terminal centromeres had similar lengths and were distinguishable based on the heterochromatin patterns on their short arms. The short arm of chromosome 2 is mostly heterochromatic (Fig. 2); whereas the short arm of chromosome 4 is occupied by the Nucleolar Organizer Region (NOR).
A group of four chromosomes (3, 5, 7, and 9) with nearly median centromeres had significantly different lengths as indicated by t tests (P < 0.01), allowing them to be distinguished on this basis.
Integration of linkage map with pachytene chromosomes in carrot
The assignment of linkage groups to specific chromosomes using a BAC-FISH approach is a well-established procedure in plant species with small and medium size genomes (Dong et al. 2000; Cheng et al. 2001b; Kulikova et al. 2001; Islam-Faridi et al. 2002; Kim et al. 2005; Zhang et al. 2005; Walling et al. 2006; Pedrosa-Harand et al. 2009; Tang et al. 2009; Fonsêca et al. 2010). Integration of linkage maps with pachytene chromosomes has been useful in BAC-by-BAC-based genome sequencing efforts, to validate contig order, resolve the order of tightly linked markers located in recombination-suppressed chromosomal domains, evaluate the size of regions with low map coverage or underrepresented in the BAC library, estimate the euchromatin-heterochromatin boundaries, characterize and integrate complex chromosomal regions, such as the pericentromeric regions, in the linkage maps (Jiang and Gill 2006; Figueroa and Bass 2010).
To integrate the carrot linkage maps with individual chromosomes, 15 marker-anchored BACs were hybridized to specific carrot pachytene chromosomes (Table 1, Fig. 3). The markers anchoring the BACs were mapped on the nine linkage groups of carrot with one to three markers for each linkage group. This enabled us to associate all linkage groups with chromosomes and to orientate six of the nine carrot linkage groups (with the exception of LG 3, LG 6, and LG 7, for which a single anchored BAC was used) according to the short/long arm of the corresponding chromosomes. Using the convention that the short chromosome arm is oriented north and the long arm south, the north/south orientation of the current carrot genetic linkage groups 1 and 5 match with the short/long arm; however, the orientation of linkage groups 2, 4, 8, and 9 should be reversed relative to the order of molecular markers published. Furthermore, all linkage groups should be renamed according to the chromosome numbering.
A standard carrot karyotype using the chromosome-specific BACs (red and green signals) listed in Table 1. Only BAC clone 064A04 on the short arm of chromosome 8 is not shown in this image. BAC names are indicated for each chromosome. BAC names reported in bold were used in the probe cocktail in Fig. 4. Arrows indicate the centromeric regions based upon hybridization to Cent-Dc
FISH mapping of additional DNA sequences
The developed chromosome-specific carrot BACs provided us a tool to localize additional DNA sequences that were either not mapped or mapped in genetic backgrounds unrelated to the B493 × QAL population used to construct the reference linkage map (Santos and Simon 2002, 2004; Grzebelus et al. 2007; Just et al. 2007; Cavagnaro et al. 2010). Thus, we were able to assign chromosomal positions to the rDNA gene clusters, and to six BAC probes (four unanchored and two mapped in unrelated genetic backgrounds) relative to the chromosome-specific BACs described above (Table 1, Figs. 2 and 3).
For example, two BACs (BAC 002B20 and BAC 032K14) were selected using markers mapped in two different unrelated populations, thus the genetic relation between these and the previous markers was unknown. FISH analysis of these two clones revealed that BAC 002B20, harboring a SCAR marker linked to the nematode resistance locus Mj-1 (Boiteux et al. 2000), is located on the long arm of chromosome 8 (LG 9), distal to BAC 009K15 and the 5S rDNA; whereas BAC 032K14, carrying a RFLP marker linked to P1, a phenotypic locus conditioning purple root pigmentation (Vivek and Simon 1999), is located on the long arm of chromosome 3 (LG 8) (Fig. 3). The same chromosome bears 067N21, a carrot BAC containing sequences homologous to the cauliflower Or gene (Lu et al. 2006; Cavagnaro et al. 2009).
Altogether, we have mapped 21 BACs on the nine carrot pachytene chromosomes. Using two-color FISH, a subset of these clones consisting of ten BACs hybridizing eight chromosomes provided a probe cocktail to reliably identify carrot pachytene chromosomes (Fig. 4).
Utilization of a probe cocktail consisting of ten chromosome-specific carrot BACs for straight forward chromosome identification in carrot. a Carrot pachytene chromosomes probed with two BACs specific to chromosomes 6 (red signal) and 8 (green signal). b The same preparation probed with a cocktail of eight BACs specific to six chromosomes. c The DAPI-stained chromosomes are converted to a black-and-white image. Arrows point to the centromeric region of each chromosome. Bar scale = 5 μm
Relationship between genetic and chromosomal distances in carrot
Cytological, genetic, and molecular studies have demonstrated that recombination events are not evenly distributed along the chromosomes (for a review, see Mézard 2006; Li et al. 2007), and significant discrepancies in relative genetic and physical distances have been reported in a number of plant species. Such discrepancies limit the use of linkage maps to guide genome sequence assembly or for map-based cloning. Pachytene FISH provides an efficient tool to visualize such disparities (Cheng et al. 2001a).
Although there were relatively few genetically anchored BACs placed on each chromosome, the available FISH data shed light on the relationship between genetic and physical distances on carrot chromosomes. First, all pairs of linked molecular markers were also physically linked (Table 1, Figs. 3 and 5, Supplementary Fig. S2). Second, the order of three genes on the map of linkage group 2 was consistent with the physical order of three corresponding BACs on chromosome 5. Third, comparing the ends of specific linkage groups to the ends of the corresponding chromosomes, all of the BACs used as anchor markers to map the end of a linkage group were indeed located at the chromosome ends (Table 1, Fig. 5, Supplementary Fig. S2), suggesting a good coverage for these chromosome arms. For example, BAC 012D08 was located ~4% away from the end of the long arm of chromosome 2 based on physical distance. This BAC clone was anchored at 0.0 cM on the map of LG 4 of the carrot inbred line B493 (Cavagnaro et al. 2010).
Relationships between BAC-FISH signals on pachytene chromosomes 2, 5, and 8 and the corresponding genetic linkage groups 4, 2, and 9 in carrot. For each chromosome, BACs are placed according to their relative distance from the end of the short arm. Linkage groups and genetic distances (in cM) refers to the D. carota B493 map (Santos and Simon 2002; Just et al. 2007; Cavagnaro et al. 2010). Dashed lines connecting cytogenetic and genetic maps are drawn only between the map-anchored BAC clones and the corresponding genetic markers. Solid circles represent BAC clones, open circles represent centromeres. The orientation of the linkage groups was inverted to agree with the conventional north/south orientation of a linkage group according to the chromosome arm
Discrepancies between the genetic and physical distance of anchored BAC clones were observed from the available data set. For example, BAC clone 064A04 was physically detected ~11% away from the end of the short arm of chromosome 8 (corresponding to LG 9). The corresponding anchor marker, GSSR107 was mapped at 76 cM, which is a relative genetic distance from the end of the linkage group 9 of about 52%. The discrepancy indicated a genetic map distance five times longer than the physical map in this region. Another disproportion on chromosome 8 (LG 9) was detected between BACs 064A04 and 009K15, which are located close to flanking boundaries of the pericentromeric region on the short and long arm of the chromosome, respectively. These two BACs span 20% of the physical length of chromosome 8 but genetically they are separated by only 4 cM (Table 1, Fig. 5). The drop in recombination rate in the pericentromeric regions is a genetic feature common to all eukaryotes. However the size of these recombination-suppressed regions varies among species. In rice, which has a genome size similar to carrot, and in Arabidopsis, these regions contain few megabases of DNA (Yan et al. 2005, 2006), but in large grass genomes up to tens of megabases are involved (Mézard 2006).
Comparative FISH mapping in Daucus species with 2n=22
The genus Daucus contains about 20 species, mainly diploid with n = 9, 10, and 11. It has been hypothesized that the basic chromosome number in the Apioideae is x = 11 because this is the most common base number in this subfamily of the Apiaceae (Moore 1971). According to this hypothesis, n = 9 and 10 should be considered derived karyotypes. However, phylogenetic analysis of combined morphological and the internal transcribed spacer region of the 18S–25S nuclear rDNA sequence data has suggested that Daucus is not monophyletic (Lee and Downie 1999; Lee et al. 2001). Those studies did not include all the species and subspecies, thus the current classification of the genus Daucus is unresolved.
We performed comparative FISH mapping using a subset of carrot BAC probes in D. crinitus and D. pusillus (both 2n = 2x = 22) which are both in section Daucus of the genus (Sáenz Laín 1981). The probes used (16 for D. crinitus and 13 for D. pusillus) included BACs mapped to carrot chromosomes 3 (LG 8), 4 (LG 6; used only in D. crinitus), 5 (LG 2), 7 (LG 5), and 8 (LG 9). All the clones (with the exception of BACs 098O13 and 009K15) generated distinct FISH signals in both species. Overall, our analysis suggested that D. crinitus and D. pusillus chromosomes shared synteny (i.e., no evidence for translocation) with carrot chromosomes 7 and 8; whereas carrot chromosomes 3 and 5 had been involved in more extensive reshuffling due to putative translocations (Fig. 6, Supplementary Figs. S3, S4, and S5). In particular, the four clones mapped to carrot chromosome 3 (067N21, 025M09, 032K14, and 238G06) hybridized to two different chromosome pairs in D. crinitus (Fig. 6b, Supplementary Fig. S3a–c). Moreover, the relative order of BACs 025M09 and 032K14 was inverted in D. crinitus compared with carrot (Supplemental Fig. S3d). These FISH data suggest that carrot chromosome 3 has been involved in at least one translocation and one inversion. A similar hybridization pattern was detected in D. pusillus (not shown). However, due to the lack of plant material, BAC 025M09 was not used in D. pusillus, thus the aforementioned inversion could not be confirmed in this species. Since BAC clone 238G06 (mapped on carrot chromosome 3) was detected on the NOR chromosome in D. crinitus (Fig. 6b), we wanted to investigate whether this chromosome is partially syntenic to the NOR chromosome in carrot (carrot chromosome 4). Clones 019A16 and 069N01, both physically linked to the NOR on carrot chromosome 4, were also linked in D. crinitus, but they were not located on the NOR chromosome and thus, they were not linked to clone 238G06 (Fig. 6c).
Schematic drawings of selected chromosomes of carrot, D. crinitus and D. pusillus summarizing the results of the comparative FISH mapping of carrot-specific-chromosome BACs (solid circles). Open circles represent carrot centromeres. Striped circles (in a, b, and e) indicate the approximate location of the centromeric regions in D. crinitus and D. pusillus. a The linkage and relative order of the three clones mapped to carrot chromosome 7 are conserved in both D. crinitus and D. pusillus. B–c Carrot chromosome 3 has been involved in at least one translocation and one inversion relative to D. crinitus and D. pusillus. BAC 238G06 is located on the NOR chromosome in D. crinitus, but not in carrot. Two BAC clones located on the NOR chromosome in carrot hybridized to a different chromosome pair (not bearing the 18S–25S rDNA) in D. crinitus. d The linkage between two clones specific to carrot chromosome 5 is disrupted in both D. crinitus and D. pusillus. e BAC clones and 5S rDNA located on chromosome 8 in carrot are also linked in both D. crinitus and D. pusillus. Their relative order is different among the three species and might be the result of multiple inversions
FISH data suggested the involvement of carrot chromosome 5 in another putative translocation, since the two clones located on carrot chromosome 5 hybridized to two different chromosome pairs in both D. crinitus and D. pusillus (Fig. 6d, Supplementary Figs. S4a, b). Identification of additional clones associated with carrot chromosome 5 (LG 2) may provide conclusive evidence that this is, perhaps, a key chromosome associated with variation in chromosome number in Daucus.
A previous survey on the cytological location of the ribosomal gene cluster in several Daucus species indicated that the 5S rDNA is located interstitially on the long arm of one chromosome pair in all the species analyzed with the exception of D. crinitus (Iovene et al. 2008a). In this species, the 5S rDNA locus was located distally on the short arm of one chromosome pair. Ribosomal DNA may move without the involvement of chromosomal rearrangements, and without altering the colinearity of other markers (Dubcovsky and Dvořák 1995). Thus, we used the BACs located on carrot chromosome 8 to assess synteny between carrot, D. crinitus and D. pusillus, and to test the hyposthesis that a chromosome rearrangement accounted for the repositioning of the 5S rDNA in Daucus. We found that 5S rDNA and BACs 002B20, 064A04, and 124B06 were also linked in both D. crinitus and D. pusillus. However, their relative order was different in any pairwise comparison among these three species (Fig. 6e, Supplementary Fig. S5) and compatible with the involvement of one or more inversions. To draw more certain conclusions, analysis of pachytene chromosomes of D. pusillus and D. crinitus, as well as of other 22- and 20-chromosome Daucus and the use of additional BAC probes, is highly desirable. Moreover, analysis of other Daucus and Daucus-related species and a better phylogenetic framework are also necessary to understand the direction of the chromosomal rearrangements.
The most common comparative FISH approach is mapping a common set of DNA clones in related species. Recent applications of this approach in plant species have yielded exciting results, including the first evidence for a centromere repositioning event in plants (Han et al. 2009) and the discovery of chromosomal inversions among Solanum species that diverged up to 12 Ma (Lou et al. 2010). The results obtained in our study further demonstrate the power of comparative FISH mapping to study synteny and chromosome rearrangements among related plant species lacking saturated genetic linkage maps and the availability of appropriate mapping populations (Lou et al. 2010; Lysak et al. 2006; Mandakova and Lysak 2008).
This study begins to reveal the translocations, inversions, and fissions/fusions that shaped the evolution of the genome for several species of Daucus. Whole genome sequencing of 18-, 20-, and 22-chromosome Daucus will be important to confirm and expand upon these observations.
In conclusion, we have described a standard carrot karyotype and established the relationship between carrot linkage groups and specific pachytene chromosomes using a set of map-anchored BACs. We have demonstrated that comparative FISH mapping is a useful approach to study synteny among Daucus species. The power of this strategy will be fully exploited once a more accurate phylogeny of the genus is established.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- FISH:
-
Fluorescent in situ hybridization
- LG:
-
Linkage group
- STS:
-
Sequence-tagged site marker
- SSR:
-
Simple sequence repeats
- SCAR:
-
Sequence-characterized amplified region
References
Arumuganathan K, Earle ED (1991) Nuclear DNA content of some important plant species. Plant Mol Biol Rep 9:208–218
Barton DW (1950) Pachytene morphology of the tomato chromosome complement. Am J Bot 37:639–643
Boiteux LS, Belter JG, Roberts PA, Simon PW (2000) RAPD linkage map of the genomic region encompassing the root-knot nematode (Meloidogyne javanica) resistance locus in carrot. Theor Appl Genet 100:439–446
Bowman M, Senalik D, Matvienko M, Van Deynze A, Simon PW (2010) Developing genomic resources for the Apiaceae. In: Plant & Animal Genomes XVIII Conference, San Diego, CA, 9–13 January 2010. P037.
Cavagnaro PF, Chung SM, Szklarczyk M et al (2009) Characterization of a deep-coverage carrot (Daucus carota L.) BAC library and initial analysis of BAC-end sequences. Mol Genet Genomics 281:273–288
Cavagnaro PF, Chung SM, Manin S, Atkins AE, Simon PW (2010) New carrot microsatellites—linkage mapping, diversity analysis and transferability to other Apiaceae. In: Plant & Animal Genomes XVIII Conference, San Diego, CA, 9–13 January 2010. P038.
Cheng Z, Presting GG, Buell CR, Wing RA, Jiang J (2001a) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157:1749–1757
Cheng Z, Buell CR, Wing RA, Gu M, Jiang J (2001b) Toward a cytological characterization of the rice genome. Genome Res 11:2133–2141
Dempsey E (1994) Traditional analysis of maize pachytene chromosomes. In: Freeling M, Walbot V (eds) The maize handbook. Springer, New York, pp 432–441
Dong F, Song J, Naess SK, Helgeson JP, Gebhardt G, Jiang J (2000) Development and applications of a set of chromosome specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007
Dubcovsky J, Dvořák J (1995) Ribosomal RNA multigene loci: Nomads of the Triticeae genomes. Genetics 140:1367–1377
Figueroa DM, Bass HW (2010) A historical and modern perspective on plant cytogenetics. Brief Funct Genomics 9:92–105
Fonsêca A, Ferreira J, Barros R, dos Santos T et al (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosome Res 18:487–502
Fransz P, Armstrong A, Alonso-Blanco C, Fischer TC, Torres-Ruiz RA, Jones G (1998) Cytogenetics for the model system Arabidopsis thaliana. Plant J 13:867–876
Gerlach WL, Bedrock JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 109:1346–1352
Gerlach WL, Dyer TA (1980) Sequence organization of the repeating units in the nucleus of wheat which contain 5S rRNA genes. Nucleic Acids Res 8:4851–4865
Grzebelus D, Jagosz B, Simon PW (2007) The DcMaster transposon display maps polymorphic insertion sites in the carrot (Daucus carota L.) genome. Gene 390:67–74
Han YH, Zhang ZH, Liu CX et al (2009) Centromere repositioning in cucurbit species: implication of the genomic impact from centromere activation and inactivation. Proc Nat Acad Sci USA 106:14937–14941
Iovene M, Grzebelus E, Carputo D, Jiang J, Simon PW (2008a) Major cytogenetic landmarks and karyotype analysis in Daucus carota and other Apiaceae. Am J Bot 95:793–804
Iovene M, Wielgus SM, Simon PW, Buell CR, Jiang J (2008b) Chromatin structure and physical mapping of chromosome 6 of potato and comparative analyses with tomato. Genetics 180:1307–1317
Islam-Faridi MN, Childs KL, Klein PE et al (2002) A molecular cytogenetic map of sorghum chromosome 1: fluorescence in situ hybridization analysis with mapped bacterial artificial chromosomes. Genetics 161:345–353
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068
Jiang J, Birchler JA, Parrott WA, Dawe RK (2003) A molecular view of plant centromeres. Trends Plant Sci 8:570–575
Just BJ, Santos CAF, Fonseca MEN, Boiteux LS, Oloizia BB, Simon PW (2007) Carotenoid biosynthesis structural genes in carrot (Daucus carota): isolation, sequence-characterization, single nucleotide polymorphism (SNP) markers and genome mapping. Theor Appl Genet 114:693–704
Kim JS, Islam-Faridi MN, Klein PE et al (2005) Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171:1963–1976
Koo D-H, Jiang J (2009) Super-stretched pachytene chromosomes for fluorescence in situ hybridization mapping and immunodetection of DNA methylation. Plant J 59:509–516
Koo D-H, Choi H-W, Cho J, Hur Y, Bang J-W (2005) A high-resolution karyotype of cucumber (Cucumis sativus L. ‘Winter Long’) revealed by C-banding, pachytene analysis, and RAPD-aided fluorescence in situ hybridization. Genome 48:534–540
Kulikova O, Gualtieri G, Geurts R et al (2001) Integration of the FISH pachytene and genetic maps of Medicago truncatula. Plant J 27:49–58
Lee B-Y, Downie SR (1999) A molecular phylogeny of Apiaceae tribe Caucalideae and related taxa: inferences based on ITS sequence data. Syst Bot 24:461–479
Lee B-Y, Levin A, Downie SR (2001) Relationships within the spiny-fruited umbellifers (Scandiceae subtribes Daucinae and Torilidinae) as assessed by phylogenetic analysis of morphological characters. Syst Bot 26:622–642
Li J, Hsia A-P, Schnable PS (2007) Recent advances in plant recombination. Curr Opin Plant Biol 10:131–135
Lou Q, Iovene M, Spooner DM, Buell CR, Jiang J (2010) Evolution of chromosome 6 of Solanum species revealed by comparative fluorescence in situ hybridization mapping. Chromosoma 119:435–442
Lu S, Van Eck J, Zhou X et al (2006) The cauliflower Or gene encodes a DnaJ cysteine-rich domain-containing protein that mediates high levels of beta-carotene accumulation. Plant Cell 18:3594–3605
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci USA 103:5224–5229
Mandakova T, Lysak MA (2008) Chromosomal phylogeny and karyotype evolution in x = 7 crucifer species (Brassicaceae). Plant Cell 20:2559–2570
Mézard C (2006) Meiotic recombination hotspots in plants. Biochem Soc Trans 34:531–534
Moore DM (1971) Chromosome studies in the Umbelliferae. In: Heywood VH (ed) The biology and chemistry of the Umbelliferae. Academic, London, UK, pp 233–255
Ohmido N, Ishimaru A, Kato S, Sato S, Tabata S, Fukui K (2010) Integration of cytogenetic and genetic linkage maps of Lotus japonicus, a model plant for legumes. Chromosome Res 18:287–299
Pedrosa-Harand A, Kami J, Gepts P, Geffroy V, Schweizer D (2009) Cytogenetic mapping of common bean chromosomes reveals a less compartmentalized small-genome plant species. Chromosome Res 17:405–417
Rubatzky VE, Quiros CF, Simon PW (1999) Carrots and related vegetable Umbelliferae. CABI Publishing, New York
Sáenz Laín C (1981) Research on Daucus L. (Umbelliferae). An Inst Botanico AJ Cavanilles 37:481–533
Santos CAF, Simon PW (2002) QTL analyses reveal clustered loci for accumulation of major provitamin A carotenes and lycopene in carrot roots. Mol Genet Genomics 268:122–129
Santos CAF, Simon PW (2004) Merging carrot linkage groups based on conserved dominant AFLP markers in F2 populations. J Amer Soc Hort Sci 129:211–217
Schrader O, Ahne R, Fuchs J (2003) Karyoptype analysis of Daucus carota L. using Giemsa C-Banding and FISH of 5S and 18S–25S rRNA specific genes. Caryologia 56:149–154
Sharma AK, Ghosh C (1954) Cytogenetics of some of the Indian umbellifers. Genetica 27:17–44
Simon PW (2000) Domestication, historical development, and modern breeding of carrot. Plant Breed Rev 19:157–189
Simon PW, Freeman RE, Vieira JV (2008) Carrot. In: Prohens J, Nuez F (eds) Handbook of Plant Breeding, Vegetables II: Fabaceae, Liliaceae, Solanaceae, and Umbelliferae, vol 2. Springer, Berlin, pp 327–357
Sutton GG, White O, Adams MD, Kerlavage AR (1995) TIGR assembler: a new tool for assembling large shotgun sequencing projects. Genome 1:9–19
Tang X, de Boer JM, vanEck HJ, Bachem CWB, Visser RGF, de Jong H (2009) Assignment of genetic linkage maps to diploid Solanum tuberosum pachytene chromosomes by BAC-FISH technology. Chromosome Res 17:899–915
Vivek BS, Simon PW (1999) Linkage relationships among molecular markers and storage root traits of carrot (Daucus carota L. ssp. sativus). Theor Appl Genet 99:58–64
Walling JG, Shoemaker RC, Young ND, Mudge J, Jackson SA (2006) Chromosome level homeology in paleopolyploid soybean (Glycine max) revealed through integration of genetic and chromosome maps. Genetics 172:1893–1900
Wang C-JR, Harper L, Cande ZW (2006) High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18:529–544
Wang K, Yang Z, Shu C et al (2009) Higher axial-resolution and sensitivity pachytene fluorescence in situ hybridization protocol in tetraploid cotton. Chromosome Res 17:1041–1050
Yan HH, Jin W, Nagaki K et al (2005) Transcription and histone modifications in the recombination-free region spanning a rice centromere. Plant Cell 17:3227–3238
Yan HH, Ito H, Nobuta K et al (2006) Genomic and genetic characterization of rice Cen3 reveals extensive transcription and evolutionary implications of a complex centromere. Plant Cell 18:2123–2133
Zhang D, Yang Q, Bao W et al (2005) Molecular cytogenetic characterization of the Antirrhinum majus genome. Genetics 169:325–335
Zhu W, Ouyang S, Iovene M et al (2008) Analysis of 90 Mb of the potato genome reveals conservation of gene structures and order with tomato but divergence in repetitive sequence composition. BMC Genomics 9:286
Zwick MS, Hanson RE, Mcknight TD et al (1997) A rapid procedure for the isolation of Cot-1 DNA from plants. Genome 40:138–142
Acknowledgment
We thank Dr. Jiri Macas for his invaluable help in the analysis of the BAC sequences generated in this work.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Pat Heslop-Harrison.
Sequence data from this article have been deposited with the EMBL/GenBank Data Libraries under accession no. HM565961.
Rights and permissions
About this article
Cite this article
Iovene, M., Cavagnaro, P.F., Senalik, D. et al. Comparative FISH mapping of Daucus species (Apiaceae family). Chromosome Res 19, 493–506 (2011). https://doi.org/10.1007/s10577-011-9202-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9202-y