Abstract
Passiflora edulis, the yellow passion fruit, is the main crop from the Passiflora genus, which comprises 525 species with its diversity center in South America. Genetic maps and a BAC (bacterial artificial chromosome) genomic library are available, but the nine chromosome pairs of similar size and morphology (2n = 18) hamper chromosome identification, leading to different proposed karyotypes. Thus, the aim of this study was to establish chromosome-specific markers for the yellow passion fruit using single-copy and repetitive sequences as probes in fluorescent in situ hybridizations (FISH) to allow chromosome identification and future integration with whole genome data. Thirty-six BAC clones harboring genes and three retrotransposons (Ty1-copy, Ty3-gypsy, and LINE) were selected. Twelve BACs exhibited a dispersed pattern similar to that revealed by retroelements, and one exhibited subtelomeric distribution. Twelve clones showed unique signals in terminal or subterminal regions of the chromosomes, allowing their genes to be anchored to six chromosome pairs that can be identified with single-copy markers. The markers developed herein will provide an important tool for genomic and evolutionary studies in the Passiflora genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Passiflora L. belongs to the family Passifloraceae Juss. ex Kunth, a member of the order Malpighiales (Judd et al. 2016). Passiflora is a large and morphologically variable genus, and it includes some 525 species distributed throughout the tropical and subtropical regions of America, Africa, and Asia (Ulmer and MacDougal 2004), with its diversity center in South America (Bernacci et al. 2015). The Passiflora species, popularly known as passionflowers or passion fruits, are typically tendril-bearing vines with a non-pedunculate inflorescence and one or two sessile, pentamerous flowers, many of which are highly prized for their exotic, unusual shape. Recent phylogenetic studies divide the genus into four main subgenera: Astrophea (57 species), Decaloba (220), Deidamioides (13), and Passiflora (240) (Feuillet and MacDougal 2003; Hansen et al. 2006), proposing Tryphostemmatoides (1) (Muschner et al. 2012) and Tetrapathea (3) (Krosnick et al. 2009) as new subgenera. Passiflora edulis Sims, the sour passion fruit in the Passiflora subgenus, is self-incompatible, with perfect flowers, and widely used for producing juice and in cosmetics and herbal products. It is also consumed as fresh fruit (Rudnicki et al. 2007).
Genome sizes (1C = 1,049.4 ± 547.7 Mbp) and flower diameters (6.12 ± 2.75 cm) vary substantially in the genus (Yotoko et al. 2011). Based on all data available for genome size, the difference between the largest and smallest genomes has been reported to be as high as 10 times (207.4 Mbp in P. organensis, Decaloba subgenus; 2,621 Mbp in P. quadrangularis, Passiflora subgenus) (Souza et al. 2004; Yotoko et al. 2011).
Passiflora species also show a variation in chromosome size and number and are classified in four groups according to their basic chromosome numbers: x = 6, 9, 10, and 12 (Melo and Guerra 2003; Hansen et al. 2006). P. edulis, like most species in the subgenus Passiflora, is diploid with 2n = 18. Several karyotypes have been proposed for the species (Oliveira 1996; Soares-Scott 1998; Melo et al. 2001; Melo and Guerra 2003; Cuco et al. 2005; Praça et al. 2008; Viana and Souza 2012), but there is no consensus on chromosomal morphology, with reported values ranging from two pairs (1 and 8) of submetacentric chromosomes and seven pairs of metacentrics (Cuco et al. 2005) to three pairs of submetacentrics (1, 8, and 9) and six pairs of metacentrics (2 to 7) (Praça et al. 2008).
The number and locations of secondary constrictions detected in the passion fruit karyotype also vary, and they have been detected on chromosome 8 (Oliveira 1996); on the long arms of chromosomes 4 and 7 (Soares-Scott 1998); and on chromosomes 1, 2, 7, and 8 (Praça et al.2008). Active nucleolar organizer regions (NORs) were detected by silver–nitrate staining on chromosomes 8 and 9 of P. edulis, associated with their secondary constrictions (Mayeda 1997). Using CMA (chromomycin A3) and DAPI (4′,6-diamidino-2-phenlindol) banding, CMA-positive blocks were detected in the terminal region of two chromosome pairs and no DAPI-positive heterochromatin was visualized. The CMA+ blocks were restricted to 35S ribosomal DNA (rDNA) sites on the long arm of chromosomes 7 and 9 (Melo et al. 2001; Melo and Guerra 2003). However, 35S rDNA sites have also been reported on the short arm of chromosome 7 and long arm of chromosome 8 (Praça et al. 2008). According to Cuco et al. (2005), 5S rDNA was located on the long arm of chromosome 4 and according to Melo and Guerra (2003), on the long arm of chromosome 5.
Due to its commercial interest, linkage maps based on molecular markers have been constructed for the passion fruit (Carneiro et al. 2002; Lopes et al. 2006; Oliveira et al. 2008). However, no cytogenetic and contig physical maps are currently available for the species, and only partial genome sequencing data have been published (Araya et al. 2017; Munhoz et al. 2018).
Bacterial artificial chromosomes (BACs) contain genomic inserts with average sizes of approximately 100 kb (Men et al. 2001; Wu et al. 2004) and are important tools for genomic analysis and physical map construction (Peterson et al. 2000). With this in mind, a BAC library consisting of approximately 83,000 clones with an average insert size of 108 kb was constructed for passion fruit (accession ‘IAPAR-123’), providing approximately sixfold coverage of its genome (Santos et al. 2014) estimated at about 1,230 Mbp by flow cytometric analysis (Yotoko et al. 2011). Recently, over 100 large inserts were completely sequenced resulting in a prediction of around 1,900 genes; the richness of repetitive elements, such as transposable elements (TEs) and microsatellite sequences (SSRs), has also been evaluated (Munhoz et al. 2018).
In this scenario, the aim of this study was to construct a BAC-FISH-based idiogram for the passion fruit using P. edulis BAC clones (Santos et al. 2014; Munhoz et al. 2018), integrating the locations of single-copy sequences and the distribution of major repeats in a reference karyotype.
Material and methods
Plant material and chromosome preparation
Root tips obtained from germinated seeds derived from the cross-hybridization between ‘IAPAR-123’ × ‘IAPAR-06’ were pretreated with 2 mM 8-hydroxyquinoline for 4.5–18 h at 10 °C, fixed in ethanol–acetic acid (3:1 v/v), and stored in fixative at − 20 °C for up to several weeks. The root tips were digested using a solution containing 2% cellulase and 20% pectinase (w/v) for 90 min at 37 °C and chromosomes prepared by air drying (De Carvalho and Saraiva 1993). Slides were selected after staining with 2 μg/ml DAPI in 50% glycerol (Cabral et al. 2006).
Library screening for isolation and validation of clones
BACs were selected with sets of four to five genes (see Munhoz et al. 2015; Munhoz et al. 2018) using the same strategy previously employed for other sets of genes (Santos et al. 2014). Positive clones were confirmed by the polymerase chain reaction (PCR) using gene-specific primers (Suppl. Table 2). Bacterial colonies were used as templates for the amplification of each gene using pairs of primers designed with Primer3 software (Rozen and Skaletsky 2000). The reactions contained 1× PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTP, 0.3 μM each primer, and 1 U Taq DNA polymerase enzyme in a volume of 16 μl. Amplifications were performed in an Applied Biosystems Veriti thermal cycler using the following program: initial denaturation at 94 °C for 5 min, followed by 30 cycles at 94 °C for 40 s, 60 °C for 40 s, and 72 °C for 1 min; and final extension at 72 °C for 8 min. Amplification products (5 μl aliquots) were electrophoresed on 1% (w/v) agarose gels and visualized under UV light, and a 100-bp GeneRuler DNA ladder (Fermentas) used as a molecular marker.
Dot blot analysis for detection of BACs containing repetitive DNA
Denatured DNA from selected BAC colonies was dotted onto a Nylon membrane (Hybond-N+; GE) and subjected to hybridization with the total genomic DNA of P. edulis as the probe, which in turn was extracted essentially according to Doyle and Doyle (1987). The probe was labeled with digoxigenin-dUTP using the DIG High-Prime DNA labeling kit (Roche Diagnostics). The membrane was hybridized overnight with probe DNA in DIG Hyb hybridization buffer (Roche Diagnostics) at 37 °C. After hybridization, membranes were washed twice in 2× saline–sodium citrate (SSC) buffer and 0.1% sodium dodecyl sulfate (SDS) for 5 to 15 min, and in 0.5× SSC buffer and 0.1% SDS for 15 min at 68 °C. Detection was performed using anti-DIG alkaline phosphatase conjugate (Roche Diagnostics) and CDP-Star (Roche Diagnostics) chemiluminescent substrate, according to the manufacturer’s instructions. Signals were captured on an X-ray ECL film (GE).
DNA probes
Both repetitive and single-copy probes were used for FISH. To detect repetitive DNA, three partial sequences of retrotransposons from different families characterized from BACs (Munhoz et al. 2018) were used as probes. These sequences were amplified using specific primers designed using Geneious pro v.7.1.8 (Suppl. Table 1) and under the thermal conditions described above. Primers were designed to amplify the RNAse-H domain from a LTR/Gypsy element, the RT domain for the element LINE/L1, and an undetermined fragment of the LTR/Copia element. The D2 probe, a 500-bp fragment containing 5S rDNA from Lotus japonicus (Pedrosa et al. 2002), and a 25-28S, 5.8S, and 18S rDNA clone (pTa71) from Triticum aestivum (Gerlach and Bedbrook 1979), labeled with Cy3-dUTP (5-amino-propargyl-2′-deoxyuridine 5′- triphosphate coupled to red cyanine fluorescent dye; GE) or digoxigenin-11-dUTP (Roche), were used to localize 5S and 35S rDNA sites, respectively.
Three groups of BAC clones were used as FISH probes: (1) 20 BACs selected with genes by library screening and dot blot (by Santos et al. 2014 and present work); (2) three partially sequenced clones (BES) considered conserved (rearranged and collinear) after microsynteny analysis against complete genomes of Arabidopsis thaliana, Vitis vinifera, and Populus trichocarpa (Santos et al. 2014); (3) 13 fully sequenced clones (Munhoz et al. 2018) selected based on high gene content and low repeat content (Table 1). Details of the BAC sequences are given in Munhoz et al. (2018) and were deposited at https://genomevolution.org/coge/GenomeInfo.pl?gid=52053. BAC DNA was extracted using the Qiagen Plasmid Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s specifications, with adaptations for use with BACs. All selected sequences were labeled by nick translation (Invitrogen or Roche Diagnostics) with Cy3-dUTP or Alexa Fluor® 488-5-dUTP (Thermo Fisher).
Fluorescence in situ hybridization
The FISH procedure applied to mitotic chromosomes was essentially the same as previously described (Fonsêca et al. 2010). The hybridization mix consisted of 50% (v/v) formamide, 10% (w/v) dextran sulfate, 2× SSC, and 5-10 ng/μl probe. The slides were denatured for 5 min at 75 °C and hybridized for up to 40 h at 37 °C. A final stringency of 76% was obtained. P. edulis total genomic DNA in 10- to 100-fold excess was added to the hybridization mix when necessary to block repetitive sequences. Digoxigenin-labeled probes were detected with 0.4 μl (1:100) sheep anti-digoxigenin conjugated with fluorescein isothiocyanate (FITC; Roche Diagnostics) and amplified with 0.2 μl (1:200) anti-sheep-FITC (Dako) in 1% (w/v) BSA. Re-hybridization of slides for localization of different DNA sequences in the same cell was performed following Heslop-Harrison et al. (1992).
Microscopical data analysis
Photographs were taken in an epifluorescence Leica DMLB microscope using Leica Las-AF software. For final processing, images were artificially pseudo-colored using Adobe Photoshop version 10.0 and adjusted for brightness and contrast only. To construct the idiograms, five cells identified with specific BAC markers were measured using the Adobe Photoshop Ruler tool. Chromosomes were identified and classified according to the length and arm ratio following Guerra (1986). To measure the positions of the signals of each BAC along the chromosome, 14 to 20 chromatids were selected and measured as described by Fonsêca et al. (2010).
Results
Distribution of repetitive sequences
Three retrotransposon sequences, identified as the most abundant by a previous BAC-end analysis (Santos et al. 2014), were used as probes to investigate their chromosomal distribution. The L1-1Cpa, a LINE (long interspersed nuclear element, Fig. 1a), and Gypsy-22, a Ty3-gypsy LTR retrotransposon (Fig. 1b), showed a dispersed and uniform distribution throughout the genome. By contrast, the Copy-13 element, a Ty1-copia LTR retrotransposon, revealed a dispersed but not entirely uniform distribution, with four small chromosomes showing weaker labeling (Fig. 1c).
Fluorescent in situ hybridization to P. edulis chromosomes. a LINE (long interspersed nuclear element) and bTy3-gypsy LTR retrotransposon, showing a dispersed and uniform distribution. cTy1-copia LTR retrotransposon showing a dispersed but not entirely uniform distribution (arrow heads indicate four unlabeled small chromosomes). d BAC Pe55J16 (red) showing a subtelomeric pattern on most chromosome ends. Bar = 5 μm
Mapping BAC clones
BAC clones were selected using three complementary approaches. First, membrane screening of the BAC library with four sets of gene probes (four or five target genes within each probe, 17 genes in total, Munhoz et al. 2018) allowed 33 positive BAC clones to be identified (Suppl. Table 1). All 33 positive BAC clones were subjected to PCR validation using specific primers to recognize the respective gene sequences with bands of the predicted lengths. Eighteen clones corresponding to nine genes were validated by PCR: CHI-B (Pe20N03; Pe108P03 clones), LHCB (Pe86H07; Pe95P11; Pe103L12), CYS3 (Pe61D03; Pe70N03; Pe88O02; Pe96N23), CESA (Pe25C22; Pe65F07; Pe108C16), COX2 (Pe27O13), KTI (Pe55J16; Pe58C13), GOX (Pe64C12), NDHJ (Pe85L08), and STP13 (Pe24G19) (Table S1). None of the clones matched the ARF2, EDA-2, GLR, KIN, or LOX2 genes. No amplification products were observed with primers for GLN2, GSH1, and S-ACP-DES.
In order to select the BAC clones with the lowest repetitive content for each gene region, the eighteen validated clones were subjected to dot blot using total genomic DNA as probe, as well as three clones selected in a second approach, based on BES information (Santos et al. 2014). Ten clones previously validated for the ACO1, CYCD1, EMB2765, ERS2, G3PD, LOX, MIPS, and NDID genes (Santos et al. 2014) were also used for FISH in the present work. Altogether, 20 BACs selected with gene probes and showing lower amounts of repetitive DNA were used as FISH probes (Table 1).
A third approach was to select, from fully sequenced BAC clones (Munhoz et al. 2018), the ones containing the highest number of annotated genes and the lowest number of annotated transposable elements in their inserts. Some of the fully sequenced BACs without transposable annotation were also evaluated by dot blot, and a total of 13 clones were selected after this third approach. A total of 36 clones, 20 selected with gene probes, three selected after BES sequencing, and 13 fully sequenced, were used for BAC-FISH (Table 1). Of the total of 36 BACs, 12 (33.3%) showed single-copy signals (Figs. 2 and S1), 12 (33.3%) showed repetitive signals dispersed across all chromosomes (data not shown), and 1 (2.8%) showed a subtelomeric pattern (Fig. 1d). This clone contained repetitive DNA, based on dot blot analysis. The remaining 11 (30.6%) showed no FISH signal.
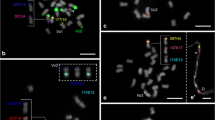
Fluorescent in situ hybridization to P. edulis chromosomes. a BAC Pe93G04 (blue) and Pe164K17 (red); b Pe93G04 (red), Pe216B22 (blue), and Pe214H11 (yellow); c Pe216B22 (red) and Pe79I13 (inset for one homolog in blue); d Pe164M13 (red) and 35S rDNA (green); e Pe125I23 (red), Pe215I08 (blue), Pe214H11 (yellow), and 5S rDNA (pink). Chromosomes were counterstained with DAPI, are visualized in gray, and are indicated by number. BACs were labeled with Cy3-dUTP and pseudocolored. Bar = 5 μm
Most single-copy BACs showed terminal or subterminal signals on short or long chromosome arms. Chromosomes varied from 2.21 to 3.19 μm and were ordered and named by decreasing size. Two of them (pairs 2 and 7) were submetacentric and the others metacentric. It was possible to establish markers for six chromosome pairs. Chromosome 1 (previously named chromosome 3 by Santos et al. 2014) is metacentric and harbors the sequence present in BACs Pe134H15 (Fig. S1a), Pe93G04, and Pe164K17 (Figs. 2a, b and 3), all located in the distal region of the long arm. Chromosome 2 (submetacentric) harbors the sequence present in Pe216B22 and Pe79I13 located respectively in the distal region of the short and long arms (Figs. 2b, c and 3). Chromosome 3 (metacentric) harbors the sequence present in Pe214H11 located in the distal region of the short arm (Figs. 2b, e and 3). On the other hand, Pe125I23 and Pe215I08 were mapped on the short arm of chromosome 5 (metacentric), which also showed the 5S rDNA site in the subterminal region of the long arm (Figs. 2e and 3). Pe173B16 and Pe195F4 were mapped in the distal region of the short arm of chromosome 7 (submetacentric), which also contains a 35S rDNA site in the terminal region of the long arm (Figs. S1c–e and 3). Chromosome 8 (metacentric) shows a second 35S rDNA site in the terminal region of the short arm and harbors Pe164M13 (Figs. 2d, 3, and S1c) and Pe164A12, both located on the long arm (Figs. S1b and 3). Chromosomes 4, 6, and 9 showed no chromosome-specific BAC marker (Fig. 4).
Physical localization of BAC clones for P. edulis mitotic chromosomes counterstained with DAPI (in gray). Single-copy clones (pseudocolored in yellow, blue, and red) are ordered according to their localization along the short and long arms (from top to bottom) of chromosomes from different cells. The 5S (pink) and 35S rDNA (green) rDNA loci are also shown. Bar = 5 μm
Discussion
In the present study, 36 BACs containing Passiflora genes or gene regions were used as probes for the construction of an idiogram of the sour passion fruit by BAC-FISH. Twelve of these clones showed single-copy signals and could be mapped, allowing six of the chromosomes to be identified. The advantages of FISH for physical mapping (also known as a cytogenetic mapping) include the visualization of the location of the cloned DNA along the chromosome, assignment of mapped sequences to chromosome domains (such as subtelomeres or pericentromeres), and the potential to construct a physical map at a relatively low cost (Jiang and Gill 2006). In addition, new clones can be successively incorporated into the map to increase density. Furthermore, chromosome-specific markers are useful for chromosome identification, redefining pre-existing karyotypes as described here in several species of agronomic interest, such as rice (Cheng et al. 2001), cotton (Wang et al. 2008), potato (Dong et al. 2000), common bean (Fonsêca et al. 2010), and sugar beet (Paesold et al. 2012).
The established karyotype of passion fruit enabled us to recognize chromosomes containing different genes, including the 5S and 35S rDNA sites. The number and positions of secondary constrictions, which correspond to active 35S rDNA sites, have been controversial in passion fruit. Oliveira (1996) reported only one secondary constriction located on chromosome 8, while Mayeda (1997) reported two unspecified pairs of chromosomes with satellites. Soares-Scott (1998) also reported the presence of two secondary constrictions, one on chromosome 4 and one on chromosome 7, both on the long arms. In a subsequent study, at least three secondary constrictions were reported, but the author did not specify their positions (Soares-Scott et al. 2005). The presence of secondary constrictions was recognized in chromosomes 8 and 9 (Cuco et al. 2005), and Melo and Guerra (2003) localized 35 rDNA sites on the long arm of chromosomes 7 and 9. The small size of P. edulis chromosomes (~ 2.5 μm), the degree of chromosome compaction at metaphase, and the difficulty in obtaining consistent chromosome spreads of good quality hampered the correct assignment of chromosome morphology to specific chromosomes.
In situ identification of chromosomes with single-copy sequences enabled us to better compare the relative sizes of each individual chromosomal pair, confirming, for example, that chromosomes 7 and 8 carry 35S rDNA sites respectively on their long and short arms and that they are larger than chromosome 9. The 5S rDNA site was detected on the long arm of chromosome 5, corresponding to the position described by Melo and Guerra (2003) and Santos et al. (2014).
BAC clones may contain a large number of repetitive sequences. Thus, by using these clones in FISH, the probability of obtaining signals on several or all chromosomes is high, making it difficult to locate single-copy sequences (Jiang and Gill 2006). Thus, this method is more appropriate for the physical mapping of species with smaller genomes (Dong et al. 2000; Jiang and Gill 2006). In the case of P. edulis (1C = 1,230 Mbp, Yotoko et al. 2011), a considerable proportion of single-copy BACs (~ 33%) could be selected, compared for instance with BACs containing RFLP markers, in which 52% of common bean single-copy sequences were selected from a genome of only 600 Mb in size (Fonsêca et al. 2010). This can be attributed to previous selection of BACs from the P. edulis genomic library for high gene and low repeat contents (see Munhoz et al. 2018).
Nevertheless, hybridization with 13 BACs resulted in strong dispersed or subtelomeric patterns on most chromosomes, indicating that they contain moderate-to-high proportion of repetitive sequences even when gene-rich. The efficiency of BAC-FISH is indeed expected to be low for medium-sized genomes (Koumbaris and Bass 2003). Therefore, the assembly of the first Passiflora genome in the future will be important to obtain additional markers for the three remaining chromosomes. Recently, three satellite DNAs were described for P. edulis using the RepeatExplorer pipeline (Pamponét et al. 2019). One of them (PeSat_3) showed a subtelomeric pattern, but different from that observed by the BAC hybridization, indicating that the subtelomeric BAC may contain a different repeat or other repeats together with PeSat_3 in its insert. A similar subtelomeric pattern was observed in common bean when using BACs harboring the subtelomeric satellite DNA khipu (Fonsêca et al. 2010).
Unlike common bean (Fonsêca et al. 2010), CMA/DAPI chromosome banding in Passiflora did not detect pericentromeric or subtelomeric blocks (Melo et al. 2001), suggesting a difference in heterochromatin composition. Nevertheless, other BACs might show a pericentromeric distribution, especially if not selected for genes or gene-rich regions. In fact, all single-copy BACs showed a terminal or subterminal distribution on the chromosome arms, suggesting that these regions are gene-rich, while the proximal regions are gene-poor and consist of dispersed repetitive sequences (Dhar et al. 2009; Stack et al. 2009). The three P. edulis retrotransposons analyzed by FISH showed a dispersed distribution along the chromosomes, in agreement with the results reported by Costa (2018), who classified most of the P. edulis transposable elements as retrotransposons. Pamponét et al. (2019) observed that the most abundant retrotransposons were classified as Ty3/gypsy (representing 33.33% of the genome), followed by Ty1/Copy (16.89%). Both showed a dispersed distribution, except for Cluster 6, a Ty3/Gypsy/Chromovirus, which was pericentromeric. LINEs are not as abundant and a possible explanation for the intense, dispersed distribution observed with the LINE probe is that the RT domain was used, which could possibly give cross-hybridization with other elements. The distribution of retrotransposons dispersed along the chromosomes, as observed in P. edulis, is also a common feature of plant genomes of similar sizes (Cheng and Murata 2003; Nagaki et al. 2004), and it corroborates the findings of Bennetzen and Wang (2014) who proposed that most TEs predominate in recombination-poor regions.
Our results confirm the importance of using BAC-FISH for chromosome identification and characterization, enabling us to develop chromosome-specific markers for P. edulis, the most important agronomic Passiflora species. Because all mapped BACs contain genes, it will be possible to integrate genome sequences directly into chromosomes when it becomes available. In addition, with the availability of chromosome-specific BACs, comparative mapping between species from the four subgenera of Passiflora will be possible, including those with different basic chromosome numbers (x) and ploidy levels (n), thanks to the high degree of conservation of nucleotide sequences in gene-rich regions observed in closely related taxa (Pedrosa et al. 2002; Lysak et al. 2006).
Conclusions
BAC-FISH enabled us to define chromosome-specific markers for six chromosome pairs of P. edulis, the sour passion fruit. The predominant terminal distributions of the genes studied herein suggest that gene-rich regions are possibly restricted to distal chromosome regions in the Passiflora genome. The main retrotransposon families show a dispersed distribution, non-uniform in the case of the Ty1-copia element. Other repetitive sequences have subtelomeric distribution. The markers established will allow the genomic sequence, once obtained, to be anchored to chromosomes, as well as comparative mapping between related species.
Change history
19 May 2020
The article “Identification of passion fruit (<Emphasis Type="Italic">Passiflora edulis</Emphasis>) chromosomes using BAC-FISH,” written by Mariela A. Sader, Yhanndra Dias, Zirlane P. da Costa, Carla Munhoz, Helen Penha, Hélène Bergès, Maria Lucia C. Vieira, and Andrea Pedrosa-Harand, was originally published with incorrect values in Figure 4.
Abbreviations
- BACs:
-
Bacterial artificial chromosomes
- FISH:
-
Fluorescent in situ hybridizations
- NORs:
-
Active nucleolar organizer regions
- CMA:
-
Chromomycin A3
- DAPI:
-
4′,6-Diamidino-2-phenlindol
- rDNA:
-
Ribosomal DNA
- TEs:
-
Transposable elements
- SSRs:
-
Microsatellite sequences
- PCR:
-
Polymerase chain reaction
- SSC:
-
Saline–sodium citrate buffer
- SDS:
-
Sodium dodecyl sulfate
- Cy3-dUTP:
-
5-Amino-propargyl-2′-deoxyuridine 5′-triphosphate coupled to red cyanine fluorescent dye
- Alexa:
-
Alexa Fluor 488
- FITC:
-
Fluorescein isothiocyanate
- BSA:
-
Bovine serum albumin
- LINE:
-
Long interspersed nuclear element
References
Araya S, Martins AM, Junqueira NT, Costa AM, Faleiro FG, Ferreira ME (2017) Microsatellite marker development by partial sequencing of the sour passion fruit genome (Passiflora edulis Sims). BMC Genomics 18:549. https://doi.org/10.1186/s12864-019-5576-6
Bennetzen JL, Wang H (2014) The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu Rev Plant Biol 65:505–530. https://doi.org/10.1146/annurev-arplant-050213-035811
Bernacci LC, Cervi AC, Milward-de-Azevedo MA, Nunes TS, Imig DC, Mezzonato AC (2015) Passifloraceae in Lista de Espécies da Flora do Brasil. Jardim Botânico do Rio de Janeiro. Disponível em: <http://floradobrasil.jbrj.gov.br/jabot/floradobrasil/FB12506>. Accessed August 2017
Cabral JS, Felix LP, Guerra M (2006) Heterochromatin diversity and its co-localization with 5S and 45S rDNA sites in chromosomes of four Maxillaria species (Orchidaceae). Genet Mol Biol 29:659–664. https://doi.org/10.1590/S1415-47572006000400015
Carneiro MS, Camargo LEA, Coelho ASG, Vencovsky R, Júnior RPL, Stenzel NMC, Vieira MLC (2002) RAPD-based genetic linkage maps of yellow passion fruit (Passiflora edulis Sims. f. flavicarpa Deg.). Genome 45:670–678. https://doi.org/10.1139/g02-035
Cheng ZJ, Murata M (2003) A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics 164:665–672
Cheng Z, Buell CR, Wing RA, Gu M, Jiang J (2001) Toward a cytological characterization of the rice genome. Genome Res 11:2133–2141. https://doi.org/10.1101/gr.194601
Costa ZP (2018) Estudos genômicos em Passiflora edulis (Passifloraceae). Tese, Escola Superior de Agricultura “Luiz de Queiroz”, Universidade de São Paulo
Cuco SM, Vieira MLC, Mondin M, Aguiar-Perecin MLR (2005) Comparative karyotype analysis of three Passiflora L. species and cytogenetic characterization of somatic hybrids. Caryologia 58:220–228. https://doi.org/10.1080/00087114.2005.10589454
De Carvalho CR, Saraiva LS (1993) An air drying technique for maize chromosomes without enzymatic maceration. Biotech Histochem 68:142–145. https://doi.org/10.3109/10520299309104684
Dhar MK, Fuchs J, Houben A (2009) Distribution of eu and heterochromatin in Plantago ovata. Cytogenet Genome Res 125:235–240. https://doi.org/10.1159/000230007
Dong F, Song J, Naess SK, Helgeson JP, Gebhardt C, Jiang J (2000) Development and applications of a set of chromosome-specific cytogenetic DNA markers in potato. Theor Appl Genet 101:1001–1007. https://doi.org/10.1007/s001220051573
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Feuillet C, MacDougal JM (2003) A new infrageneric classification of Passiflora L. (Passifloraceae). Passiflora 13:34–38
Fonsêca A, Ferreira J, dos Santos TRB, Mosiolek M, Bellucci E, Kami J, Gepts P, Geffroy V, Schweizer D, dos Santos KGB, Pedrosa-Harand A (2010) Cytogenetic map of common bean (Phaseolus vulgaris L.). Chromosom Res 18:487–502. https://doi.org/10.1007/s10577-010-9129-8
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Guerra M (1986) Reviewing the chromosome nomenclature of Levan et al. Braz J Genet 9:741–743
Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK (2006) Phylogenetic relationships and chromosome number evolution in Passiflora. Syst Bot 31:138–150. https://doi.org/10.1600/036364406775971769
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8:372–373. https://doi.org/10.1016/0168-9525(92)90287-E
Jiang J, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49:1057–1068. https://doi.org/10.1139/g06-076
Judd WS, Campbell CS, Kellogg EA, Stevens PF, Donoghue MJ (2016) Plant systematics: a phylogenetic approach, 4th ed. Sinauer Associates, Sunderland
Koumbaris GL, Bass HW (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BAC clones. Plant J 35:647–359. https://doi.org/10.1046/j.1365-313X.2003.01829.x
Krosnick SE, Ford AJ, Freudenstein JV (2009) Taxonomic revision of Passiflora subgenus Tetrapathea including the monotypic genera Hollrungia and Tetrapathea (Passifloraceae), and a new species of Passiflora. Syst Bot 34:375–385. https://doi.org/10.1600/036364409788606343
Lopes R, Lopes MTG, Carneiro M et al (2006) Linkage and mapping of resistance genes to Xanthomonas axonopodis pv. Passiflorae in yellow passion fruit. Genome 49:17–29. https://doi.org/10.1139/g05-081
Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I (2006) Mechanisms of chromosome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci 103:5224–5229. https://doi.org/10.1073/pnas.0510791103
Mayeda LY (1997) Estudo citogenético em dez taxons do gênero Passiflora L. (Passifloraceae). Dissertação, Escola Superior de Agricultura “Luiz de Queiroz”, Universidade de São Paulo
Melo NF, Guerra M (2003) Variability of the 5S and 45S rDNA sites in Passiflora L. species with distinct base chromosome numbers. Ann Bot 92:309–316. https://doi.org/10.1093/aob/mcg138
Melo NF, Cervi AC, Guerra M (2001) Karyology and cytotaxonomy of the genus Passiflora L. (Passifloraceae). Plant Syst Evol 226:69–84. https://doi.org/10.1007/s006060170074
Men AE, Meksem K, Kassem MA, Lohar D, Stiller J, Lightfoot D, Gresshoff PM (2001) A bacterial artificial chromosome library of Lotus japonicus constructed in an Agrobacterium tumefaciens-transformable vector. Mol Plant-Microbe Interact 14:422–425. https://doi.org/10.1094/MPMI.2001.14.3.422
Munhoz CF, Santos AA, Arenhart RA, Santini L, Monteiro-Vitorello CB, Vieira MLC (2015) Analysis of plant gene expression during passion fruit–Xanthomonas axonopodis interaction implicates lipoxygenase 2 in host defence. Ann Appl Biol 167:135–155. https://doi.org/10.1111/aab.12215
Munhoz CF, Costa ZP, Cauz-Santos LA, Reátegui ACE, Rodde N, Cauet S, Dornelas MC, Leroy P, Varani AM, Bergès H, Vieira MLC (2018) A gene-rich fraction analysis of the Passiflora edulis genome reveals highly conserved microsyntenic regions with two related Malpighiales species. Sci Rep 8:13024. https://doi.org/10.1038/s41598-018-31330-8
Muschner VC, Zamberlan PM, Bonatto SL, Freitas LB (2012) Phylogeny, biogeography and divergence times in Passiflora (Passifloraceae). Genet Mol Biol 35:1036–1043. https://doi.org/10.1590/S1415-47572012000600019
Nagaki K, Neumann P, Zhang D, Ouyang S, Buell CR, Cheng Z, Jiang J (2004) Structure, divergence, and distribution of the CRR centromeric retrotransposon family in rice. Mol Biol Evol 22:845–855. https://doi.org/10.1093/molbev/msi069
Oliveira AM (1996) Reprodução e citogenética de espécies de Passiflora. Tese, Universidade Estadual Paulista São José do Rio Preto
Oliveira EJ, Vieira MLC, Garcia AAF, Munhoz CF, Margarido GRA, Consoli L, Matta FP, Moraes MC, Zucchi MI, Fungaro MHP (2008) An integrated molecular map of yellow passion fruit based on simultaneous maximum-likelihood estimation of linkage and linkage phases. J Am Soc Hortic Sci 133:35–41. https://doi.org/10.21273/JASHS.133.1.35
Paesold S, Borchardt D, Schmidt T, Dechyeva D (2012) A sugar beet (Beta vulgaris L.) reference FISH karyotype for chromosome and chromosome-arm identification, integration of genetic linkage groups and analysis of major repeat family distribution. Plant J 72:600–611. https://doi.org/10.1111/j.1365-313X.2012.05102.x
Pamponét VCC, Souza MM, Silva GS, Micheli F, de Melo CAF, de Oliveira SG, Costa EA, Corrêa RX (2019) Low coverage sequencing for repetitive DNA analysis in Passiflora edulis Sims: citogenomic characterization of transposable elements and satellite DNA. BMC Genomics 20:262. https://doi.org/10.1186/s12864-019-5576-6
Pedrosa A, Sandal N, Stougaard J, Schweizer D, Bachmair A (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH (2000) Construction of plant bacterial artificial chromosome (BAC) libraries: an illustrated guide. J Agric Genomics 5:1–100
Praça MM, Carvalho CR, Francismar CM, Correa MMA (2008) Morphological aspects of Passiflora edulis f. flavicarpa chromosomes using acridine orange banding and rDNA-FISH tools. Caryologia 61:154–159. https://doi.org/10.1080/00087114.2008.10589623
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Rudnicki M, Silveira MM, Pereira TV, Oliveira MR, Reginatto FH, Dal-Pizzol F, Moreira JCF (2007) Protective effects of Passiflora alata extract pretreatment on carbon tetrachloride induced oxidative damage in rats. Food Chem Toxicol 45:656–661. https://doi.org/10.1016/j.fct.2006.10.022
Santos AA, Penha HA, Bellec A, Munhoz C, Pedrosa-Harand A, Bergès H, Vieira ML (2014) Begin at the beginning: a BAC-end view of the passion fruit (Passiflora) genome. BMC Genomics 15:816. https://doi.org/10.1186/1471-2164-15-816
Soares-Scott, MD (1998) Caracterização citogenetica de algumas especies e hibridos interespecificos de Passiflora Dissertação, Universidade Estadual de Campinas. Disponível em: <http://www.repositorio.unicamp.br/handle/REPOSIP/317970>. Accessed 23 July 2018
Soares-Scott MD, Meletti LMM, Bernacci LC, Passos IRS (2005) Citogenética clássica e molecular em passifloras. In: Maracujá: Germoplasma e Melhoramento Genético; Faleiro, F.G., Junqueira, N.T.V., Braga, M.F., eds.; Embrapa Cerrados: Planaltina, Brazil, pp. 210–240
Souza MM, Palomino G, Pereira TNS, Pereira MG, Viana AP (2004) Flow cytometric analysis of genome size variation in some Passiflora species. Hereditas 141:31–38. https://doi.org/10.1111/j.1601-5223.2004.01739.x
Stack SM, Royer SM, Shearer LA, Chang SB, Giovannoni JJ, Westfall DH, White RA, Anderson LK (2009) Role of fluorescence in situ hybridization in sequencing the tomato genome. Cytogenet Genome Res 124:339–350. https://doi.org/10.1159/000218137
Ulmer T, MacDougal JM (2004) Passiflora: passionflowers of the world. Timber Press, Portland-Cambridge, 430
Viana AJC, Souza MM (2012) Comparative cytogenetics between the species Passiflora edulis and Passiflora cacaoensis. Plant Biol 14:820–827. https://doi.org/10.1111/j.1438-8677.2011.00557.x
Wang K, Guan B, Guo W (2008) Completely distinguishing individual A-genome chromosomes and their karyotyping analysis by multiple bacterial artificial chromosome fluorescence in-situ hybridization. Genetics 178:1117–1122. https://doi.org/10.1534/genetics.107.083576
Wu C, Sun S, Nimmakayala P, Santos FA, Meksem K, Springman R, Ding K, Lightfoot DA, Zhang HB (2004) A BAC- and BIBAC-based physical map of the soybean genome. Genome Res 14:319–326. https://doi.org/10.1101/gr.1405004
Yotoko KSC, Dornelas MC, Togni PD, Fonseca TC, Salzano FM, Bonatto SL, Freitas LB (2011) Does variation in genome sizes reflect adaptive or neutral processes? New clues from Passiflora. PLoS One 6:e18212. https://doi.org/10.1371/journal.pone.0018212
Acknowledgments
We thank Mr. Steve Simmons for proofreading the manuscript.
Funding
The authors received financial support from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). Scholarships from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) were awarded to MA Sader, ZP Costa, and A. Pedrosa-Harand. This study was supported in part by the Coordenacão de Aperfeicoamento de Pessoal de Nível Superior - Brasil (CAPES, Finance Code 001), and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP grant no. 2014/25215-2).
Author information
Authors and Affiliations
Contributions
MAS and APH conceived and designed the research. HB, HP, and MLCV contributed to BAC library, plant material, and bioinformatic data. MAS and YD conducted the experiments. CM, MAS, YD, and ZPC analyzed the data. MAS and APH wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Additional information
Responsible Editor:Jiming Jiang
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Figure S1
Fluorescent in situ hybridization to P. edulis chromosomes. (a) BAC Pe134H15 (red); (b) Pe164A12 (red) and 35S rDNA (green); (c) BAC Pe173B16 (green) and Pe164M13 (red); (d) BAC Pe195F4 (red) and 35S rDNA (green); (e) BAC Pe173B16 (red) and 35S rDNA (green). Chromosomes were counterstained with DAPI, are visualized in grey and indicated by number. BACs were labeled with Cy3-dUTP and pseudocolored. Bar = 5 μm. (PNG 8848 kb)
ESM 1
(DOCX 22 kb)
Rights and permissions
About this article
Cite this article
Sader, M.A., Dias, Y., Costa, Z.P. et al. Identification of passion fruit (Passiflora edulis) chromosomes using BAC-FISH. Chromosome Res 27, 299–311 (2019). https://doi.org/10.1007/s10577-019-09614-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-019-09614-0