Abstract
The role of long non-coding RNAs (lncRNAs) in the pathogenesis of neurological disorders including schizophrenia has been highlighted by independent studies. In the present study, we compared peripheral blood expression of seven lncRNAs between schizophrenic patients and sex- and age-matched controls using quantitative real-time PCR technique. FAS-AS1, PVT1 and TUG1 were significantly down-regulated in schizophrenic patients compared with healthy individuals (P = 0.007, 0.003 and 0.001, respectively). The association between FAS-AS1 expression and schizophrenia was significant in male subjects aged more than 50 but not in other subgroups. GAS5, NEAT1 and OIP5-AS1 expressions were not significantly different between patients and controls (P = 0.523, 0.739 and 0.267, respectively). The associations between GAS5, NEAT1 and OIP5-AS1 expressions and schizophrenia were significant in female subjects but not in male subjects. THRIL was up-regulated in schizophrenic patients compared with healthy subjects. Based on the results of bootstraped median regression, and after controlling for the effects of age and sex, the difference in its expression between cases and controls was significant (P = 0.014), while the interaction between group and sex was not significant. The expression of lncRNAs was not correlated with age in any study subgroups. In addition, we found sex-based pairwise correlations between PVT1 expression and expression levels of OIP5-AS1, THRIL and NEAT1. We also demonstrated high sensitivity and specificity of GAS5 for diagnosis of schizophrenia in female patients. The current study provides further evidence for the participation of lncRNAs in the pathogenesis of schizophrenia. Future studies are needed to confirm the suitability of lncRNAs as peripheral biomarkers for this psychiatric disorder.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Schizophrenia with a median lifetime prevalence of 4 per 1000 individuals is regarded as one of the most profoundly disabling psychiatric disorders. It is characterized by early adulthood onset of hallucinations, delusions, disorganized communication, decreased motivation, and reduced affect (Saha et al. 2005). The contributions of several genetic, developmental and environmental factors in the pathogenesis of schizophrenia necessitate the design of studies to explore the molecular underlying mechanisms of the disease onset and progression. The biomarker-finding investigations have been hindered by the extraordinarily complex nature of the disorder and the dissimilarities in patient populations (Schwarz and Bahn 2008). Long non-coding RNAs (lncRNAs) are among putative biomarkers for a wide range of diseases including cancer and neuropsychiatric disorders (Nikpayam et al. 2017; Taheri et al. 2018; Zuo et al. 2016). Few studies have addressed the role of lncRNAs in schizophrenia. For instance, Merelo et al. have listed a few lncRNAs whose expressions were associated with schizophrenia, among them being those with aberrant expression in schizophrenic patients as well as lncRNAs that regulate proteins or mRNAs identified to be dysregulated in schizophrenia (Merelo et al. 2015). More recently, Chen et al. have compared the expression of lncRNAs in peripheral blood mononuclear cells (PBMCs) of schizophrenic patients and healthy individuals and reported 125 differentially expressed lncRNAs in schizophrenic patients compared with healthy subjects, including lncRNAs that were associated with the response to antipsychotic treatment (Lai et al. 2016). Others have demonstrated associations between expression of some lncRNAs such as the antisense transcript HAR1, C6UAS and LINC00271 and schizophrenia (Amann-Zalcenstein et al. 2006; Morelli et al. 2000; Tamura et al. 2007). Apart from these studies, the role of lncRNAs in the pathogenesis of schizophrenia has not been fully explored. In the present study, we compared expressions of FAS-AS1, PVT1, TUG1, OIP5-AS1, THRIL, NEAT1 and GAS5 lncRNAs between schizophrenic patients and healthy subjects. The rationales for the selection of these lncRNAs were their involvement in the altered pathways in schizophrenia (Djordjevic et al. 2012; O’Brien et al. 2008; Topol et al. 2016) or their role in neuronal protection (Chen et al. 2016). We performed the current study to assess whether these lncRNAs are distinctively expressed in the peripheral blood of schizophrenic patients rather than in their brains. The results may open a different era to explore possible epigenetic mechanisms involved in the pathogenesis of this psychiatric disorder and to discover biomarkers for it.
Materials and Methods
Patients
The current case–control study was conducted on blood samples obtained from 50 unrelated schizophrenic patients who were referred to the psychology department of Hamadan University of Medical Sciences and 50 age- and sex-matched healthy subjects. Patients were evaluated based on the Diagnostic and Statistical Manual of Mental Disorders, fifth edition (DSM-V) criteria for schizophrenia (Association 2013). All patients were under treatment with Clozapine™ as the single antipsychotic drug (Rahimi et al. 2017). Patients had no substance abuse or cigarette smoking. Individuals recruited in the control group were evaluated through a structured psychiatric interview to rule out the existence of any psychiatric disorder. Individuals suffered from any concomitant conditions such as malignancy, recent or persistent infectious disorder, auto-immune disease, nerve muscle coupling disorders and pregnancy were excluded from the study. The study protocol was approved by the Ethical Committee of Hamadan University of Medical Sciences (IR.UMSHA.REC.1396.928). Informed written consent was obtained from all study participants (including patients and parents of some patients).
Expression Analysis
Experiments were performed on total RNA extracted from whole venous blood of study participants using the Hybrid-RTM blood RNA extraction Kit (Geneall Biotechnology, South Korea). After evaluation of the quality and quantity of extracted RNA using Nanodrop equipment (Thermo Scientific, MA, USA), cDNA was synthesized using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems). Expressions of lncRNAs were compared between cases and controls using TaqMan Universal PCR Master Mix (Takara Bio, Shiga, Japan) on the rotor gene 6000 Corbett Real-Time PCR System. The HPRT1 gene was used as the reference gene. The nucleotide sequences of primers and probes as well as amplicon length are set out in Table 1.
Statistical Analysis
Relative expressions of lncRNAs were compared between schizophrenic patients and healthy subjects using the frequentist method. Pairwise correlations between transcript levels of lncRNAs were evaluated in each set of samples using the Spearman rank order correlation test. Correlations between transcript levels of lncRNAs and the age of study participants were assessed using the same method. The effects of possible confounding variables such as age and sex were estimated through the application of Quantile regression. P values less than 0.05 were regarded as significant. The receiver operating characteristic (ROC) curve was depicted to appraise the properness of transcript levels of lncRNAs for defining disease status.
Results
General Features of Cases and Controls
Table 2 shows the general features of cases and controls. No significant difference was found between the age and sex ratios of cases and controls (P values of 0.65 and 0.68).
Relative Expression of lncRNAs in Schizophrenic Patients Compared with Healthy Subjects
Relative expressions of lncRNAs were compared between total schizophrenic patients and healthy subjects as well as sex- and age-based subgroups of patients and controls (Table 3).
Subsequently, the Quantile regression method was used for controlling the effects of age and sex (Table 4). FAS-AS1, PVT1 and TUG1 were significantly down-regulated in schizophrenic patients compared with healthy individuals (P = 0.007, 0.003 and 0.001, respectively). The interaction between group and sex was significant for FAS-AS1 but not for PVT1 and TUG1 which shows a sex-based difference in the expression of FAS-AS1 between cases and controls. The association between FAS-AS1 expression and schizophrenia was significant in male subjects aged more than 50 but not in other subgroups. GAS5, NEAT1 and OIP5-AS1 expressions were not significantly different between patients and controls (P = 0.523, 0.739 and 0.267, respectively). However, the interactions between group and sex were significant for all three lncRNAs which shows difference in the expression of these lncRNAs between cases and controls based on the sex of study participants. The associations between GAS5, NEAT1 and OIP5-AS1 expressions and schizophrenia were significant in female subjects but not male subjects. THRIL was up-regulated in schizophrenic patients compared with healthy subjects. Based on the results of Bootstraped median regression and after controlling the effects of age and sex, the difference in its expression between cases and controls was significant (P = 0.014) while the interaction between group and sex was not significant.
Correlations Between Expression of lncRNAs and Age of Study Participants
No significant correlation was found between expression of lncRNAs and age of study participants in any study subgroup (Table 5).
Pairwise Correlation Between Expression Levels of lncRNAs
Significant correlations were found between expression levels of lncRNAs in almost all study subgroups independent of disease status (Table 6). Notably, GAS5 levels were inversely correlated with the expression of other lncRNAs in all study subgroups. Moreover, the pairwise correlation between PVT1 and OIP5-AS1 was sex-dependent as it was merely seen in female subjects. Conversely, PVT1 expression levels were correlated with THRIL and NEAT1 expression only in male subjects.
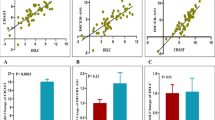
Finally, we depicted the correlation matrix showing the distribution of gene expressions in total study participants (Fig. 1).
ROC Curve Analysis
The area under the curve (AUC) values were calculated for transcript levels of all lncRNAs in sex- and age-based subgroups of study. FAS-AS1 transcript levels had 90.48% sensitivity and 66.67% specificity (AUC = 0.825, P < 0.0001) for the diagnosis of schizophrenia in male subjects aged more than 50 years (Fig. 2). GAS5 transcript levels had 100% sensitivity and 86.96% specificity (AUC = 0.93, P < 0.0001) for the diagnosis of schizophrenia in female subjects (Fig. 3). The sensitivity and specificity values were increased to 100% in female subjects aged less than 50 (Fig. 4). NEAT1 had 86.67% sensitivity and 78.2% specificity (AUC = 0.86, P < 0.0001) for the diagnosis of schizophrenia in female subjects (Fig. 5). Such values were not significantly different in between age-based subgroups. OIP5-AS1 had 100% sensitivity and 60.87% specificity (AUC = 0.87, P < 0.0001) in female subjects (Fig. 6) and the AUC value increased in female subjects aged less than 50. THRIL had 86.67% sensitivity and 78.26% specificity (AUC = 0.817, P < 0.0001) in female subjects with no difference between age-based subgroups (Fig. 7). TUG1 had 71.43% sensitivity and 85.19% specificity (AUC = 0.832, P < 0.0001) in male subjects (Fig. 8). Finally, PVT1 had 76.47% sensitivity and 85.19% specificity (AUC = 0.83, P < 0.0001) in male subjects (Fig. 9).
Discussion
In spite of extensive efforts in the identification of biomarkers for schizophrenia, the results of these studies have not yet been translated into clinical application due to the unsuitability of brain biopsies and the expensiveness of neuroimaging methods. Therefore, blood-based biomarkers are considered effective alternatives in this regard (Lai et al. 2016). Based on the fundamental roles of lncRNAs in the regulation of gene expression in brain tissue (Andersen and Lim 2018), and their observed dysregulation in neuropsychiatric disorders (Wang et al. 2015), lncRNAs are putative biomarkers for this kind of disorder. Moreover, as lncRNAs are not translated into proteins, their transcript levels are more confidently associated with their function compared with other types of transcripts. In the present study, we compared the expression of seven lncRNAs between schizophrenic patients and healthy subjects and found significant down-regulation of FAS-AS1, PVT1 and TUG1 in patients. We also detected higher levels of THRIL in patients compared with healthy individuals.
The lncRNA FAS-AS1 is transcribed from the opposite strand of the Fas gene and participates in the Fas alternative splicing process. Ectopic expression of this lncRNA has decreased the sFAS isoform while increased the mFAS, thus triggering FasL-induced apoptosis. An inverse correlation has been found between expression levels of FAS-AS1 and production of sFas (Sehgal et al. 2014). A previous study has reported elevated serum levels of both sFas and FasL in schizophrenic patients compared with healthy subjects. Such a rise in apoptotic markers was independent of the disease characteristics, antipsychotic treatment, genetics basis, the first commencement of the disorder, disease duration and cigarette smoking (Djordjevic et al. 2012). The observed down-regulation of FAS-AS1 in the schizophrenic patients in the current study is in line with the previously reported up-regulation of sFas in these patients and implies a role for this lncRNA in epigenetic regulation of apoptosis in these patients. As the Quantile regression model showed a significant association between FAS-AS1 expression and schizophrenia in male subjects aged more than 50 (n = 15), but not in other subgroups, we evaluated the diagnostic power of this lncRNA in the mentioned study subgroup. The results of ROC curve analysis showed 90.48% sensitivity and 66.67% specificity of FAS-AS1 expression for the diagnosis of schizophrenia in male subjects aged more than 50 years (AUC = 0.825, P < 0.0001). The observed down-regulation of PVT1 in schizophrenic patients in the current study is consistent with the previously reported role for this lncRNA in neuronal protection (Chen et al. 2016). Li et al. have shown that PVT1-mediated autophagy may shield hippocampal neurons from synaptic plasticity damage and apoptosis, and subsequently remediate cognitive deficiency in diabetic mice (Chen et al. 2016). We also detected down-regulation of TUG1 in schizophrenic patients compared with controls. The surge in expression of TUG1 is necessary for retinal development (Young et al. 2005). TUG1 has also been shown to interact with miR-9 and seclude it directly (Chen et al. 2017). On the other hand, abnormal levels and function of miR-9 have been regarded as one of the numerous elements that participate in the pathogenesis of schizophrenia (Topol et al. 2016). Consequently, future studies are needed to explore the effects of TUG1 down-regulation on miR-9 expression in peripheral blood of schizophrenic patients and reveal whether such dysregulation contributes to the disease course.
Although GAS5, NEAT1 and OIP5-AS1 expressions were not significantly different between patients and controls, the interactions between group and sex were significant for all three lncRNAs which show differences in the expression of these lncRNAs between cases and controls based on the sex of the study participants. As we detected a significant association between GAS5 expression and schizophrenia in female subjects, we evaluated its diagnostic power in this subgroup of study participants. GAS5 transcript levels had 100% sensitivity and 86.96% specificity (AUC = 0.93, P < 0.0001) for dthe iagnosis of schizophrenia in female subjects. The sensitivity and specificity values were increased to 100% in female subjects aged less than 50 (n = 10). These results show the appropriateness of GAS5 expression levels for the diagnosis of schizophrenia in female subjects. Finally, we found up-regulation of THRIL in schizophrenic patients compared with healthy subjects. This lncRNA regulates expression of tumor necrosis factor (TNF) in human monocytes through interactions with HNRNPL (Li et al. 2014). A previous study has shown an association between increased TNF-alpha levels and acute exacerbations of schizophrenia (O’Brien et al. 2008). However, a more recent study reported elevated plasma TNF pathway markers in schizophrenic patients without an equivalent upsurge in blood cell gene expression (Hoseth et al. 2017). Consequently, we hypothesize that THRIL is involved in the pathogenesis of schizophrenia possibly through epigenetic regulation of TNF pathway. Functional studies are needed to appraise this hypothesis.
Although we did not find any difference in the expression of NEAT1 between cases and controls, this lncRNA has previously been shown to be up-regulated in other neurological disorders such as Huntington’s disease (Johnson 2012). Moreover, its up-regulation has been documented during the early stage of amyotrophic lateral sclerosis (Nishimoto et al. 2013). A recent study has shown lower levels of NEAT1 in the peripheral blood of untreated schizophrenic patients compared with healthy subjects, but the expression of this lncRNAs in treated patients was almost similar to healthy subjects (Li et al. 2018). As the patients in our cohort were all under treatment with clozapine, the similar expression levels of NEAT1 between cases and controls can be attributed to the effect of this drug.
OIP5-AS1, as the other lncRNA with similar expression level in cases and controls, has previously been shown to act as a chief regulator of neurogenesis during development (Ulitsky et al. 2011). Although we did not find any difference in their peripheral expression between cases and controls, these lncRNAs might affect some aspects of schizophrenia in brain tissue.
As we detected no significant correlation between the expression of lncRNAs and the age of study participants in any study subgroup, we propose these lncRNAs as age-independent disease markers. This independence implies that they are not affected by various phenotypic deficits during the disease.
Significant correlations were found between expression levels of lncRNAs in almost all study subgroups independent of disease status, which implies that the interactions between these lncRNAs are not influenced by schizophrenia. In spite of other positive pairwise correlations, GAS5 levels were inversely correlated with the expression of other lncRNAs in all the study subgroups. The significance of such a pattern of correlation should be explored in future studies. Moreover, the pairwise correlation between PVT1 and OIP5-AS1 was oinly seen in female subjects. Conversely, PVT1 expression levels were correlated with THRIL and NEAT1 expression only in male subjects. Such sex-based correlations might show the influence of sex hormones on their expressions.
In brief, in the current study, we demonstrated dysregulation of lncRNAs in peripheral blood of schizophrenic patients and the suitability of their expression levels as diagnostic markers in certain subgroups of patients. While PVT1, FAS-AS1 and TUG1 were suitable markers in male subjects, NEAT1, OIP5-AS1, THRIL and GAS5 were more suitable for the diagnosis of the disease in female subjects. However, our study had some limitations, including small sample size and the potential effects of the medication. Moreover, the study groups were not matched in terms of education. Future studies are needed to confirm the diagnostic power of lncRNAs as peripheral biomarkers for this psychiatric disorder. Moreover, assessment of the expression of these lncRNAs in a group of drug-naïve patients would help to rule out the effects of clozapine in changing the expression of these genes.
References
Amann-Zalcenstein D et al (2006) AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia European. J Hum Genet 14:1111
Andersen RE, Lim DA (2018) Forging our understanding of lncRNAs in the brain Cell Tissue Res 371:55–71. https://doi.org/10.1007/s00441-017-2711-z
Association D-AP (2013) Diagnostic and statistical manual of mental disorders Arlington. American Psychiatric Publishing, Washington
Chen S et al (2016) Aberrant expression of long non-coding RNAs in schizophrenia patients. Med Sci Monit 22:3340–3351
Chen SC et al (2017) LncRNA TUG1 sponges microRNA-9 to promote neurons apoptosis by up-regulated Bcl2l11 under ischemia. Biochem Bioph Res Co 485:167–173. https://doi.org/10.1016/j.bbrc.2017.02.043
Djordjevic VV, Ristic T, Lazarevic D, Cosic V, Vlahovic P, Djordjevic VB (2012) Schizophrenia is associated with increased levels of serum Fas and FasL. Clin Chem Lab Med 50:1049–1054 https://doi.org/10.1515/cclm-2011-0684
Hoseth EZ et al (2017) A study of TNF pathway activation in schizophrenia and bipolar disorder in plasma and brain. Tissue Schizophr Bull 43:881–890. https://doi.org/10.1093/schbul/sbw183
Johnson R (2012) Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis 46:245–254. https://doi.org/10.1016/j.nbd.2011.12.006
Lai CY, Scarr E, Udawela M, Everall I, Chen WJ, Dean B (2016) Biomarkers in schizophrenia: A focus on blood based diagnostics and theranostics. World J Psychiatr 6:102–117. https://doi.org/10.5498/wjp.v6.i1.102
Li Z et al (2014) The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci USA 111:1002–1007
Li J, Zhu L, Guan F, Yan Z, Liu D, Han W, Chen T (2018) Relationship between schizophrenia and changes in the expression of the long non-coding RNAs Meg3, Miat, Neat1 and Neat2. J Psychiatr Res 106:22–30
Merelo V, Durand D, Lescallette AR, Vrana KE, Hong LE, Faghihi MA, Bellon A (2015) Associating schizophrenia, long non-coding RNAs and neurostructural dynamics. Front Mol Neurosci 8:57
Morelli C, Magnanini C, Mungall AJ, Negrini M, Barbanti-Brodano G (2000) Cloning and characterization of two overlapping genes in a subregion at 6q21 involved in replicative senescence. and schizophrenia. Gene 252:217–225
Nikpayam E, Tasharrofi B, Sarrafzadeh S, Ghafouri-Fard S (2017) The role of long non-coding RNAs in ovarian cancer. Iran Biomed J 21:3–15. https://doi.org/10.6091/.21.1.24
Nishimoto Y et al (2013) The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain 6:31 https://doi.org/10.1186/1756-6606-6-31
O’Brien SM, Scully P, Dinan TG (2008) Increased tumor necrosis factor-alpha concentrations with interleukin-4 concentrations in exacerbations of schizophrenia. Psychiatr Res 160:256–262. https://doi.org/10.1016/j.psychres.2007.11.014
Rahimi S, Sayad A, Moslemi E, Ghafouri-Fard S, Taheri M (2017) Blood assessment of the expression levels of matrix metalloproteinase 9 (MMP9) and its natural inhibitor, TIMP1 genes in Iranian schizophrenic patients. Metab Brain Dis 32:1537–1542. https://doi.org/10.1007/s11011-017-0043-z
Saha S, Chant D, Welham J, McGrath J (2005) A systematic review of the prevalence of schizophrenia. PLoS Med 2:e141 https://doi.org/10.1371/journal.pmed.0020141
Schwarz E, Bahn S (2008) Biomarker discovery in psychiatric disorders. Electrophoresis 29:2884–2890. https://doi.org/10.1002/elps.200700710
Sehgal L et al (2014) FAS-antisense 1 lncRNA and production of soluble versus membrane Fas in B-cell lymphoma. Leukemia 28:2376–2387. https://doi.org/10.1038/leu.2014.126
Taheri M, Omrani MD, Ghafouri-Fard S (2018) Long non-coding RNA expression in bladder cancer. Biophys Rev 10:1205–1213. https://doi.org/10.1007/s12551-017-0379-y
Tamura Y, Kunugi H, Ohashi J, Hohjoh H (2007) Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol Psychiatr 12:519:593–600. https://doi.org/10.1038/sj.mp.4001965
Topol A et al (2016) Dysregulation of miRNA-9 in a subset of schizophrenia patient-derived neural progenitor cells. Cell Rep 15:1024–1036
Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147:1537–1550. https://doi.org/10.1016/j.cell.2011.11.055
Wang Y et al (2015) Genome-wide differential expression of synaptic long noncoding RNAs in autism spectrum disorder. Transl Psychiat. https://doi.org/10.1038/tp.2015.144
Young TL, Matsuda T, Cepko CL (2005) The noncoding RNA Taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 15:501–512. https://doi.org/10.1016/j.cub.2005.02.027
Zuo L et al (2016) Long non-coding RNAs in psychiatric disorders. Psychiatr Genet 26:109
Funding
This study was financially supported by Hamadan University of Medical Sciences (Grant Number 970121264).
Author information
Authors and Affiliations
Contributions
SGF wrote the manuscript. SAJ analyzed the data. MT and AK supervised the study. MRS performed the laboratory tests.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Safari, M.R., Komaki, A., Arsang-Jang, S. et al. Expression Pattern of Long Non-coding RNAs in Schizophrenic Patients. Cell Mol Neurobiol 39, 211–221 (2019). https://doi.org/10.1007/s10571-018-0640-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-018-0640-3