Abstract
Background
Although long non-coding RNA (lncRNA) HCP plays essential roles in human cancers, its function and mechanism in multiple myeloma (MM) have not crystallized.
Methods
HCP5 level in MM was assessed through qRT-PCR. A series of functional investigations were conducted to evaluate the influences of HCP5 on proliferation and apoptosis. Bioinformatics analysis and RIP/RNA pull-down assays were carried out to determine the relationships among HCP5, miR-128-3p, and PLAGL2. Relative protein level was determined through Western blot. A xenograft tumor model was applied for validating the roles of HCP5/miR-128-3p/PLAGL2 axis in vivo.
Results
HCP5 was significantly increased in MM. HCP5 knockdown effectively thwarted the proliferative rate and cell cycle of MM cell lines and suppressed tumor growth. HCP5 regulated PLAGL2 expression by sponging miR-128-3p. PLAGL2 overexpression effectively rescued cells from influences by sh-HCP5 on cell proliferative and apoptotic rates. Additionally, HCP5 knockdown significantly inhibited Wnt/β-catenin/cyclin D1 signaling, and these effects were eliminated by PLAGL2 overexpression.
Conclusion
Our study revealed that HCP5/miR-128-3p/PLAGL2 is closely correlated to MM development by modulating Wnt/β-catenin/cyclin D1 signaling.
Graphical abstract
HCP5 promoted cell proliferation and tumor formation of MM cells by activating the Wnt/β‐catenin/CCND1 signaling pathway by sponging miR-128-3p to increase PLAGL2 expression

Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple myeloma (MM) represents a common hematological tumor with a higher incidence and mortality followed by non-Hodgkin lymphoma (Gupta et al. 2020). Investigation of 36 cancers in 185 countries identified that MM accounts for approximately 1% of all malignancies and at least 13% of all hematological cancers (Bray et al. 2018). Although extensive advantages have been made recently in conventional chemotherapy and stem cell transplantation (Detappe et al. 2018), the 5-year survival rate of MM patients remains poor. Therefore, an in-depth understanding of the potential mechanistic interplays implicated in MM at the genomic/transcriptomic levels is still necessary.
Long chain non-coding RNAs (lncRNAs) with lengths > 200 nucleotides play essential roles in oncogenesis in humans, such as MM (Amodio et al. 2017; Garzon et al. 2010). Recently, lncRNAs have been intimately linked to multiple physiological processes in human cancers (Adams et al. 2017). For example, PVT1 promotes prostate cancer growth in vivo (Yang et al. 2017). CDC6 promotes breast cancer progression, including proliferation, invasion, and migration, by sponging miR-215 (Kong et al. 2019). LncRNA HLA complex P5 (HCP5) functions as a tumorigenic factor in a series of human tumors, including cervical cancer (Yu et al. 2018), follicular thyroid carcinoma (Liang et al. 2018), breast cancer (Wang et al. 2019), and prostate cancer (Hu and Lu 2020). However, its function in MM has not been reported.
MicroRNAs (miRNAs) can repress mRNA expression by interfering mRNA stability or protein translation by binding to the 3′ untranslated region (3′ UTR) of target mRNAs (Lu and Rothenberg 2018). Past investigations recognized that aberrantly expressed miRNAs are frequently observed in various human tumors including MM (Wang et al. 2015). For instance, MiR-145-3p exacerbates autophagy and enhances chemo-resistance sensitivity of bortezomib in MM by directly targeting HDAC4 (Wu et al. 2020). Increasing investigations indicated miR-128-3p is significantly downregulated and plays important roles in various cancers, including glioblastoma (Zhang et al. 2009), lung cancer (Weiss et al. 2008), and acute lymphocytic leukemia (She et al. 2014).
It has been reported that pleomorphic adenoma gene like-2 (PLAGL2) promotes colorectal cancer proliferation by activating β-catenin to elevate cyclin D1 expression (Zhou et al. 2018). Moreover, cumulative evidences also demonstrate the importance of Wnt signaling in MM cells and its close relationship with cellular processes, including proliferation, differentiation, and migration (Spaan et al. 2018). In addition, PLAGL2 was also reported to act as a direct downstream gene of miRNAs such as miR-449a and miR-214-3p to affect cancer progression (Xu et al. 2018; Zhou et al. 2020). However, the roles of PLAGL2, especially the lncRNA-miRNA-PLAGL2 axis in MM are not explored.
In this study, we found that HCP5 was upregulated in MM and revealed that HCP5 promoted the progression of MM by triggering the Wnt/β‐catenin signaling pathway via targeting miR-128-3p/PLAGL2, suggesting that HCP5 is a potential target as novel MM therapies.
Materials and methods
Bioinformatics analysis
Raw expression profiles of MM (GSE125361 and GSE125363) were obtained from the Gene Expression Omnibus (GEO) database. The Limma package of R language was employed to analyze differential expression in two datasets using |log2FC|> 1.0 and p < 0.05 as the significant parameters.
Cell culture
Human MM cell lines (NCI-H929, U266, RPMI-8266, and MM.1S) and the normal plasma cell line (nPC) were procured from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were grown in RPMI-1640 supplemented with 10% FBS (Gibco, Life Technologies) in a 37 °C incubator with addition of 5% CO2.
Clinical specimens
A total of 86 MM bone marrow tissues together with 14 non-diseased control tissues were obtained from the Department of Hematology, the First Affiliated Hospital of Anhui Medical University between 2017 and 2019. All individuals signed the informed consent.
Cell transfection
Lentiviruses containing the short hairpin for HCP5 (sh-HCP5: 5ʹ-GCTGATGAGTAGGACATTT-3′) or negative control (sh-NC), HCP5 overexpression vector pcDNA3.1-HCP5 (HCP5 in short), PLAGL2 overexpression vector pcDNA3.1-PLAGL2 (PLAGL2 in short), and the corresponding negative control (empty vector pcDNA3.1) were synthesized and purchased from GenePharma (Shanghai, China). In addition, the small interfering RNA targeting PLAGL2 (si-PLAGL2), si-NC, miR-128-3p mimics/inhibitor, and their relevant control molecules (miR-NC and inhibitor NC) were developed by RiboBio™ (Guangzhou, China) and transiently transfected into NCI-H929 and MM.1S cells using Lipofectamine 2000® (Invitrogen).
Luciferase reporter assay
Bioinformatics tools in microRNA.org were employed to predict the possible binding regions between miR-128-3p and HCP5, and several tools, including PicTar, targetscan, miRDB, miRWalk, and microT-CDS, were applied to predict the binding sites between miR-128-3p and 3ʹ-UTR of PLAGL2. To determine their relationships, wild-type HCP5 sequence (WT-HCP5) or PLAGL2 3ʹ-UTR (WT-PLAGL2) and mutated HCP5 sequence (MUT-HCP5) or PLAGL2 3ʹ-UTR (MUT-PLAGL2) were generated and cloned into pmirGLO® luciferase vector (Promega™, USA). Each plasmid was co-transfected with either miR-128-3p mimics or miR-NC into NCI-H929 and MM.1S cells using Lipofectamine 2000®. Two days later, relative luciferase activity was determined using a dual-luciferase reporter system.
CCK-8 assay
Briefly, 2 × 103 cells per well were seeded onto 96-well plates and grown for 24, 48, 72, and 96 h. Consequently, cells were incubated with 10 μL of CCK8 solution (Cell Counting Kit-8, Solarbio, Beijing, China) for 2 h. Absorbance values at 450 nm were determined using a microplate reader (BioTek™, VT, USA).
Colony formation assay
Approximately 1000 transfected cells were seeded into 6-well plates and grown over 2 weeks. Cells were fixed with 10% formaldehyde followed by staining using 0.1% crystal violet. Finally, all cell colonies were observed and counted under a light microscope.
EdU staining assay
Cellular proliferative rate was determined using an EdU Staining Proliferation Kit® (Abcam™). In brief, cells were incubated with 100 μL EdU medium (50 μM) for 3 h, and stained with 10 μg/mL Hoechst 33,342 for 15 min. Ultimately, EdU positive cell populations were calculated under an Olympus fluorescence microscope.
Cell cycle analysis
Cell cycle analysis was performed using propidium iodide (PI) staining as described previously (Pawlyn et al. 2017). In brief, 5 × 105 transfected cells were stained using 10 μL of PI staining solution for 20 min. Subsequently, cell cycle distribution was evaluated using flow cytometry (FACSCalibur®, BD Biosciences™).
RNA isolation and qRT-PCR
Total RNA was isolated using TRIzol® (Invitrogen™, Carlsbad, CA, USA). All cDNAs were generated using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems™). QRT-PCR assay was conducted on ABI 7500® Real-Time PCR System (Applied Biosystems™). The relative expression was determined using the 2−ΔΔCt method with U6 and β-actin as the internal/normalization control genes. The primers used in qRT-PCR assays were HCP5 forward 5ʹ-GACTCTCCTACTGGTGCT TGGT-3ʹ and reverse 5ʹ-CACTGCCTGGTGAGCCTGTT-3ʹ; PLAGL2 forward 5ʹ-GTGCCAAGGAGAAGAAGCAC-3′ and reverse 5ʹ-CTAACATGTCCACGGGCTC T-3′; β-actin forward 5ʹ-GTCACCGGAGTCCATCACGAT-3′ and reverse 5ʹ-TCACC AACTGGGACGACATG-3′; miR-128-3p forward 5ʹ-GGTCACAGTGAACCGGTC-3′ and reverse 5ʹ-GTGCAGGGTCCGAGGT-3′; and U6 forward 5′-ATCCGGCAGAT GGCTGTTGAC-3′ and reverse 5′-GGCCGGTACACCATTCCGATTC-3′.
Western blot
Total proteins were collected using RIPA lysis buffer, and the proteomic levels were determined using BCA Protein Assay Kits (Tiangen™, Beijing, China). Equivalent proteomic loads were separated on 12% SDS-PAGE and transferred onto PVDF membranes that were consequently incubated with primary antibodies against cyclin D1 (1:500, SC-8396, Santa Cruz, USA), non-phospho-(active) β-catenin (Ser33/37/Thr41) (D13A1) (1:1000, #8814, CST, USA), PLAGL2 (1:800, ab139509, Abcam, UK), and β-actin (1:1000, ab8227, Abcam, UK) overnight at 4 °C. All protein bands were observed using the ECL Kit (Millipore).
RNA immunoprecipitation (RIP) assay
RIP assays were conducted by a Magna RIP RNA-Binding Protein Immunoprecipitation Kit® (Millipore™, Darmstadt, Germany). Briefly, MM cells were ruptured, and the post-lysis supernatants were incubated with anti-Ago2 (Millipore™, USA) or mouse IgG (Invitrogen™, USA) conjugated magnetic beads overnight at 4 °C. Finally, the co-precipitated RNAs were isolated and subjected to qRT-PCR assay.
RNA pull-down assay
MM cells were transfected with 50 nM mutated (MUT)-biotinylated or wild-type (WT)-biotinylated miR-128-3p (GenePharma, China). The cell lysates were incubated with pre-coated M-280 streptavidin beads (Sigma Aldrich™) for 4 h at 4 °C. After washing with PBS twice and cold lysis solution twice, the beads-bound RNAs were purified and subjected to qRT-PCR assays.
TOP/FOP flash assay
This assay was conducted as described previously (Chen et al., 2019). In brief, approximately 5000 cells were transfected with 100 ng TOP/FOP flash reporter plasmids (Millipore™, MA, USA), 100 ng expression vector (vector or HCP5), or sh-RNA (sh-NC or sh-HCP5). After 2 days, the luciferase activity was determined using a Dual-Luciferase Kit (Promega™, USA).
Animal model
NCI-H929 and MM.1S cell populations were transfected with sh-HCP5 or co-transfected with negative control sh-NC and empty vector, or sh-HCP5 and PLAGL2, and then subcutaneously injected into nude mice (6 mice in each group) with 1.2 × 107 cells per mouse. Tumor volume was evaluated using the equation: 0.5 × length × width2. On the 28th day, mice were sacrificed, and the tumor xenografts from three groups were excised, weighed, imaged, and further analyzed.
Fluorescence in situ hybridization (FISH) assay
FISH assay was conducted using Servicebio lncRNA FISH Probe Mix® (Red) (G3016-3, Servicebio™, Wuhan, China) as previously described (Cao et al. 2020). Briefly, the tumor samples were paraffin-coated and sliced to 4-μm-thick sections. The sections were incubated with a hybridization solution containing 8 ng/μL HCP5 lncRNA probes at 37 °C overnight, scrutinized, and imaged using fluorescence microscopy (Nikon™, Tokyo, Japan).
Immunohistochemistry (IHC) assay
Xenograft tumors from three groups were fixed in 10% formaldehyde, paraffin-coated, sliced as 5-µm-thick sections, and consequently stained using H&E at room temperature to evaluate the cytopathological features as previously described (Parks et al. 2019). In addition, sections were incubated with anti-Ki-67 antibody (1:400, ab15580, Abcam™) at 4 °C overnight to assess proliferation as previously reported (Gu et al. 2017).
Statistical analysis
All datasets were expressed as mean ± SD using SPSS v.20.0 (SPSS, Inc., Chicago, IL, USA). Differences in quantitative data between two groups were tested by Student’s t test. p < 0.05 was deemed to confer statistical significance.
Results
HCP5 was significantly upregulated in MM
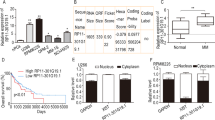
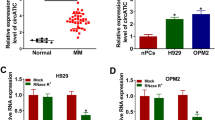
We firstly analyzed t HCP5 expression in a publicly available expression profile of plasma cells involved in MM and found that HCP5 was highly upregulated in plasma cells of MM cases than healthy donors in GSE125361 (p < 0.001; Fig. 1A). To validate its upregulation, HCP5 levels in 86 MM tissues and 14 healthy tissues were analyzed using qRT-PCR. The results indicated that typically, HCP5 expression in MM tissues was markedly elevated (p < 0.001; Fig. 1B). Furthermore, HCP5 level in clinical MM cases was intimately correlated to ISS staging, and its level was the highest at the advanced stage (p < 0.05; Fig. 1C). The 86 MM cases were divided into two groups with the median HCP5 expression as the cutoff. The patients with high HCP5 level had a poorer progress-free survival (p < 0.05; Fig. 1D) and overall survival (p < 0.001; Fig. 1E). In addition, HCP5 was also highly upregulated in five MM cell lines than nPCs (p < 0.05; Fig. 1F), indicating that HCP5 might deeply involve the development of MM.
HCP5 was significantly upregulated in MM tissues and corresponding cell lines. A Relative HCP5 expression in GSE125361 consisting of MM tissues (n = 45) and healthy tissues (n = 3). B HCP5 expression in MM tissues (n = 86) and healthy tissues (n = 14) was detected by qRT-PCR. C HCP5 expression in MM tissues of different ISS stages was evaluated by qRT-PCR (n = 86). D Progression-free curve analysis of MM patients with high (n = 43) or low (n = 43) HCP5 level. E Overall survival curve analysis of MM patients with high (n = 43) or low (n = 43) HCP5 level. F HCP5 expression in MM cell lines and normal nPC cells was detected by qRT-PCR (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001. Each experiment was repeated three times
HCP5 overexpression promoted the proliferation of MM cell lines
HCP5 knockdown or overexpression was conducted in NCI-H929 and MM.1S cells. Transfection efficiency was evaluated using qRT-PCR (p < 0.01; Fig. 2A). CCK-8 assay demonstrated that sh-HCP5 lowered the viability of these two cell lines (p < 0.01), while HCP5 overexpression promoted the viability of these two cell lines (p < 0.01) (Fig. 2B). Colony formation assay (Fig. 2C) and EdU staining (Fig. 2D) revealed that sh-HCP5 significantly decreased the proliferative ability of these two cells (p < 0.01) and HCP5 overexpression promoted the proliferative rates of these two cells (p < 0.05). In addition, flow cytometry highlighted that HCP5 downregulation inhibited cell cycle progression, and HCP5 overexpression promoted cell cycle and blocked cell cycle at G1 stage in these two cell lines (p < 0.05; Fig. 2E). These results demonstrated HCP5 overexpression promoted MM cell proliferation.
HCP5 overexpression promoted proliferation of MM cell lines. NCI-H929 and MM.1S cells were transfected with sh-HCP5, HCP5 (pcDNA3.1-HCP5 overexpression plasmid), and corresponding negative controls. A Transfection efficiency was evaluated by qRT-PCR. B Cell viability was evaluated by CCK-8 assay. C, D Cell proliferation was evaluated by colony formation assay (C) and EdU staining assay (D). E Cell cycle was evaluated by flow cytometry. *p < 0.05, **p < 0.01, ***p < 0.001. Each experiment was repeated three times
HCP5 acted as a sponge for miR-128-3p
Next, we investigated the subcellular localization of HCP5 in both NCI-H929 and MM.1S cells. RNA-FISH data highlighted HCP5 to be predominantly abundant in MM cytoplasm (Fig. 3A). To determine HCP5 downstream genes, we predicted 29 potential target miRNAs by microRNA.org, and searched all 928 downregulated miRNAs in GSE125363 profile. Moreover, Venn diagram showed that there were two overlapped miRNAs (miR-299-3p and miR-128-3p) in the two prediction results (Fig. 3B), which were selected for the subsequent validation. Then, we quantitatively detected the two putative miRNAs in NCI-H929 and MM.1S cells transfected with sh-HCP5 or HCP5 overexpression vector, and qRT-PCR revealed that sh-HCP5 upregulated miR-128-3p while HCP5 overexpression decreased miR-128-3p level (p < 0.01), but had no significant effect on miR-299-3p expression (Fig. 3C). Moreover, qRT-PCR also revealed that miR-128-3p mimics obviously downregulated HCP5 (p < 0.01) and miR-128-3p inhibitor obviously upregulated HCP5 (p < 0.001) in these two cell lines (Fig. 3D). Meanwhile, miR-128-3p expression in MM tissues was downregulated (p < 0.001; Fig. 3E). In addition, Pearson’s correlation analysis indicated an inverse-proportion relationship between HCP5 and miR-128-3p expression in MM tissues (n = 86, p < 0.001; Fig. 3F). As expected, bioinformatics prediction revealed that HCP5 might be a miR-128-3p sponge (Fig. 3G). Luciferase reporter studies demonstrated miR-128-3p mimics markedly lowered the relative luciferase activity for WT-HCP5 in the two cells (p < 0.001), while remained unaffected for MUT-HCP5 (Fig. 3H). These results revealed that HCP5 might be a sponge of miR-128-3p.
HCP5 acted as a sponge for miR-128-3p. A FISH analysis of the subcellular location of HCP5 in NCI-H929 and MM.1S cells. B The Venn diagram of target miRNAs of HCP5 predicted by microRNA.org and downregulated miRNAs in GSE125363. C NCI-H929 and MM.1S cells were transfected with sh-HCP5, HCP5 (pcDNA3.1-HCP5 overexpression plasmid), and corresponding negative controls. The expression of the two miRNA candidates was detected by qRT-PCR (n = 3). D NCI-H929 and MM.1S cells were transfected with miR-128-3p mimics, miR-128-3p inhibitor, or their negative controls. The expression of HCP5 was detected by qRT-PCR (n = 3). E The expression of miR-128-3p in MM tissues (n = 86) and healthy tissues (n = 14) was detected by qRT-PCR. F The correlation between the levels of HCP5 and miR-128-3p in MM tissues (n = 86). G Schematic illustration of WT-HCP5 and MUT-HCP5 luciferase reporter vectors. H The relative luciferase reporter activity of WT or MUT-HCP5 in two MM cells was detected by the dual-luciferase reporter system (n = 3). **p < 0.01, ***p < 0.001. Each experiment was repeated three times
PLAGL2 is targeted by miR-128-3p
Next, several predictive tools, including PicTar, targetscan, miRDB, miRWalk, and microT-CDS software, were applied. We also searched all upregulated mRNAs and overlapped the predicted target mRNAs. We found that 13 potential mRNAs might be targets of miR-128-3p (Fig. 4A). Consequently, MM.1S cells were transfected with miR-128-3p mimics or miR-NC, and qRT-PCR results highlighted only PLAGL2 level was downregulated in miR-128-3p mimics group (p < 0.001; Fig. 4B). Western blot indicated that miR-128-3p mimics obviously decreased PLAGL2 expression, and miR-128-3p inhibitor obviously increased PLAGL2 protein expression (Fig. 4C). To determine their relationship, we also constructed the luciferase reporter plasmids containing the putative binding site of PLAGL2 3ʹ-UTR (Fig. 4D). The results indicated that miR-128-3p mimics markedly reduced the relative luciferase reporter activity for WT-PLAGL2 3ʹ-UTR in the two cells (p < 0.01) and had no effect on MUT-PLAGL2 3ʹ-UTR (Fig. 4E). PLAGL2 expression in MM tissue was significantly elevated (p < 0.001; Fig. 4F). In addition, an inverse-proportion relationship existed between miR-128-3p and PLAGL2 expression in MM tissues (p < 0.001; Fig. 4G), suggesting PLAGL2 to be directly targeted by miR-128-3p.
HCP5 upregulated PLAGL2 expression by sponging miR-128-3p. A Schematic illustration to search the target mRNAs of miR-128-3p predicted by online systems (PicTar, targetscan, miRDB, miRWalk, and microT-CDS) and the upregulated mRNAs in GSE125361 database. B MM.1S cells were transfected with miR-128-3p mimics or miR-NC, and the expression of 13 mRNA candidates was evaluated by qRT-PCR (n = 3). C NCI-H929 and MM.1S cells were transfected with miR-128-3p mimics, miR-128-3p inhibitor, or their control. PLAGL2 protein expression was detected by western blot (n = 3). D Schematic illustration of WT and MUT PLAGL2 3ʹ-UTR luciferase reporter vectors. E The relative luciferase reporter activity of WT or MUT PLAGL2 3ʹ-UTR in two MM cell lines was detected by the dual-luciferase reporter system (n = 3). F PLAGL2 expression in MM tissues (n = 86) and healthy tissues (n = 14) was evaluated by qRT-PCR. G The correlation between miR-128-3p and PLAGL2 in MM tissues (n = 86). H The direct interaction among HCP5, miR-128-3p, and PLAGL2 was determined by RIP assay (n = 3). I The enrichment of HCP5 and PLAGL2 in MM.1S cells by using biotin-labeled miR-128-3p probe or biotin-NC probe (n = 3). J, K The correlation between HCP5 and PLAGL2 in 86 MM tissues (J) and GSE125361 dataset (K). L NCI-H929 and MM.1S cells were transfected with sh-HCP5, miR-128-3p inhibitor, or their controls. PLAGL2 protein expression was evaluated by western blot. *p < 0.05, **p < 0.01, ***p < 0.001. Each experiment was repeated three times
HCP5 upregulated PLAGL2 expression by sponging miR-128-3p
We further investigated the relationship among HCP5, miR-128-3p, and PLAGL2 in MM.1S cells using RIP/RNA pull-down assays. RIP assay revealed that HCP5, miR-128-3p, and PLAGL2 were all efficiently enriched by anti-Ago2 (p < 0.001; Fig. 4H). Meanwhile, RNA pull-down assay highlighted HCP5 and PLAGL2 were highly enriched in biotin-labeled miR-128-3p probe (p < 0.001; Fig. 4I). Furthermore, a directly proportional relationship between PLAGL2 and HCP5 expressions was identified in 86 MM tissue samples (p < 0.05; Fig. 4J) and GSE125361 dataset (p < 0.001; Fig. 4K). Western blot showed that sh-HCP5 obviously decreased PLAGL2 protein expression, and miR-128-3p inhibitor obviously increased PLAGL2 protein expression (Fig. 4L). These data indicated HCP5 to upregulate PLAGL2 expression by sponging miR-128-3p.
HCP5 activated Wnt/β-catenin signaling by regulating PLAGL2
HCP5 influence in β-catenin/cyclin D1 axis was studied through the TOP/FOP flash luciferase assay. The data revealed sh-HCP5 highly thwarted TOP/FOP flash reporter activities (p < 0.01), and HCP5 (overexpression of HCP5) promoted Wnt/β-catenin signaling in two cells (p < 0.001) (Fig. 5A). Meanwhile, western blot highlighted that sh-HCP5 obviously decreased the protein expression of PLAGL2, β-catenin, and cyclin D1, while HCP5 overexpression markedly elevated PLAGL2, β-catenin, and cyclin D1 expression within two cell lines (Fig. 5B). Moreover, co-transfection of sh-HCP5 and pcDNA3.1-PLAGL2 (PLAGL2, overexpression of PLAGL2) obviously reversed the effects of sh-HCP5 on TOP/FOP flash reporter functions (Fig. 5C), together with attenuating PLAGL2, β-catenin, and cyclin D1 inhibition within two cell lines (Fig. 5D). Such data indicates HCP5-triggered Wnt/β-catenin signaling through regulating PLAGL2.
HCP5 activated Wnt/β-catenin signaling by regulating PLAGL2. A, B NCI-H929 and MM.1S cells were transfected with sh-HCP5, HCP5 (pcDNA3.1-HCP5 overexpression plasmid), and corresponding negative controls. A Dual-luciferase assays for TOP/FOP activity. B The protein expression of PLAGL2, β-catenin, and cyclin D1 was detected by western blot. C, D NCI-H929 and MM.1S cells were co-transfected with sh-HCP5 and pcDNA3.1-PLAGL2 (overexpression of PLAGL2). C Dual-luciferase assays for TOP/FOP activity. D The protein expression of PLAGL2, β-catenin, and cyclin D1 was detected by western blot. **p < 0.01, ***p < 0.001. Each experiment was repeated three times
PLAGL2 overexpression reversed the inhibitory effects of sh-HCP5 in MM cell lines
Next, PLAGL2 knockdown or overexpression was conducted in NCI-H929 and MM.1S cells, and the transfection efficiency was validated by western blot (Supplementary Fig. 1A). By performing CCK-8, colony formation, EdU staining assays, and flow cytometry, we found that si-PLAGL2 significantly decreased the proliferative ability of MM.1S (p < 0.05), while PLAGL2 overexpression exacerbated NCI-H929 cell proliferation (p < 0.05) (Supplementary Fig. 1B–E). Furthermore, the rescue experiments were carried out. CCK-8 assay indicated that co-transfection of sh-HCP5 and PLAGL2 overexpression vector reversed that sh-HCP5 significantly inhibited the viability of the two cell lines (p < 0.01; Fig. 6A). Meanwhile, co-transfection of sh-HCP5 and PLAGL2 reversed sh-HCP5-directed inhibition of cellular proliferative rate (p < 0.01; Fig. 6B, C). In addition, co-transfection of sh-HCP5 and PLAGL2 significantly promoted cell cycle progression compared to sh-HCP5 (p < 0.05; Fig. 6D). These results indicated that HCP5 served as a competing endogenous RNA (ceRNA) for miR-128-3p by modulating PLAGL2 expression to exacerbate MM development.
PLAGL2 overexpression reversed the inhibitory effect of sh-HCP5 in MM cell lines. NCI-H929 and MM.1S cells were transfected with sh-HCP5, or co-transfected with sh-HCP5 and PLAGL2 (overexpression of PLAGL2). A Cell viability was evaluated by CCK-8 assay. B, C Cell proliferation was evaluated by colony formation assay (B) and EdU staining assay (C). D Cell cycle was evaluated by flow cytometry. *p < 0.05, **p < 0.01. Each experiment was repeated three times
HCP5 knockdown inhibited the development of MM in vivo
To assess the effects of HCP5 on MM in vivo, sh-NC + empty vector, sh-HCP5 + empty vector, and sh-HCP5 + PLAGL2 (overexpression PLAGL2) were stably transfected into NCI-H929 cells and MM.1S cells and injected into nude mice. We found that sh-HCP5 obviously decreased tumor weight/dimensions, and co-treatment of sh-HCP5 + PLAGL2 significantly reverted the inhibiting influences of sh-HCP5 (Fig. 7A–C). Moreover, IHC assay showed that Ki-67 positive cells in tumor tissues of mice injected with sh-HCP5 were reduced, and co-treatment of sh-HCP5 + PLAGL2 obviously reversed the sh-HCP5-induced inhibitory effect (Fig. 7D). In addition, sh-HCP5 obviously downregulated PLAGL2, β-catenin, and cyclin D1 in tumor tissues, and co-treatment of sh-HCP5 + PLAGL2 reversed the inhibitory effect of sh-HCP5 (Fig. 7E). These results demonstrated that HCP5/miR-128-3p/PLAGL2 was closely implicated in MM progression in vivo.
HCP5 knockdown inhibited the development of MM in vivo. A The representative images of xenograft subcutaneous tumors. B Tumor volume was measured every week for 4 weeks. C Tumor weight. D Cell proliferation in tumor tissues was evaluated by H&E staining and IHC staining. E The expression of PLAGL2, β-catenin, and cyclin D1 in tumor tissues was detected by western blot. ***p < 0.001. Six mice in each group. Each experiment was repeated three times
Discussion
Due to the serious social and human health hazards, it is more urgent to develop effective anti-tumor agents for preventing MM development (Laubach et al. 2016; Juli and Oliverio 2019). Hence, it is necessary to explore the detailed pathogenic mechanism involved in MM. Here, we found that HCP5 knockdown significantly inhibited the progression of MM. Mechanistically, HCP5 activated Wnt/β‐catenin signaling by targeting miR-128-3p/PLAGL2 to elevate cyclin D1 expression. Our results identified that HCP5 played an oncogenic role in MM progression.
It is known that HCP5 plays essential roles in different types of human cancers by regulating cell proliferation, apoptosis, invasion, and migration processes. For instance, HCP5 is upregulated in colon tumor tissues, and HCP5 knockdown blocks proliferative/migrative capacities of colon tumor cells (Yun et al. 2019). HCP5 is identified as a ceRNA to regulate proliferation, invasion, migration, cell apoptosis, and autophagy of pancreatic cancer cells (Liu et al. 2019b). A high HCP5 level is shown to be positively correlated to the metastasis of prostate cancer. Moreover, HCP5 downregulation suppresses proliferative rates and colony formation capacity while enhances apoptosis of prostate cancer cells (Hu and Lu 2020). HCP5 has been demonstrated to contribute to the epithelial-mesenchymal transition (EMT) process in colorectal cancer by activating ZEB1 (Yang et al. 2019). Based on previous studies, we speculated that HCP5 might also play significant roles in MM development. As expected, we confirmed the upregulation of HCP5 in MM. Moreover, HCP5 knockdown obviously inhibited MM progression. Except for those abnormally expressed lncRNAs, we identified a new lncRNA HCP5 and confirmed its oncogenic role in MM progression.
To investigate the potential target miRNAs, we searched the microRNA.org, selected all 928 downregulated miRNAs in GSE125363 profile, and found that there were two miRNA candidates (miR-299-3p and miR-128-3p) simultaneously existing in the two datasets, suggesting that the two downregulated miRNAs might be targets of HCP5. MiR-299-3p was identified as a tumor-suppressing miRNA in thyroid cancer (Chen et al. 2019), hepatocellular carcinoma (Dang et al. 2018), and cervical cancer (Yu et al. 2019). MiR-128-3p also was shown to regulate the progression of human cancers, including colorectal cancer (Liu et al. 2019a), glioma (Fu et al. 2018), and breast cancer (Zhao et al. 2019). However, the functions of these two miRNAs in MM remain unclear. Meanwhile, we revealed that HCP5 was a miR-128-5p-sponge.
By combining upregulated miRNAs in GSE125361 dataset, we found that some potential target mRNAs might be targets of miR-128-3p, including BTG2, CSF1, ELOVL4, EPHB2, FBLN2, GREM1, PLAGL2, RELN, SYT1, TFEB, TMEM25, and WDTC1. Among them, several mRNAs, including BTG2, CSF1, EPHB2, GREM1, RELN, and TFEB, are reported to participate in the progression of MM (Leone et al. 2013; Baghdadi et al. 2019; Liu et al. 2014; Clark and Hewett 2020; An et al. 2018; Cea et al. 2012). Although the remaining miRNAs have not been studied in MM, their roles and functions in other diseases have been studied in detail. For example, FBLN2 is required for basement membrane integrity of mammary epithelium (Ibrahim et al. 2018). SYT1 has been demonstrated to be associated with the development of neurodevelopmental disorders (Baker et al. 2018). TMEM25 has been identified as a candidate biomarker methylated and downregulated in colorectal cancer (Hrašovec et al. 2013). WDTC1 is the ortholog of the drosophila adipose gene and is shown to be involved in human obesity (Lai et al. 2009). Thus, a series of functional experiments were performed. The results demonstrated that PLAGL2 is directly targeted by miR-128-3p and revealed the regulatory networking by miR-128-3p/PLAGL2 in MM.
Previous studies indicated Wnt/β‐catenin signaling was activated through positive regulation of cyclin D1 by PLAGL2 in colorectal cancer (Zhou et al. 2018). In addition, PLAGL2 overexpression transcriptionally triggered Wnt/β‐catenin signaling and exacerbated tumor development in colorectal cancer (Li et al. 2019). Here, we explored the effect of HCP5 in cyclin D1/Wnt/β‐catenin signaling in MM and highlighted that HCP5 knockdown severely inhibited the Wnt signaling using TOP/FOP flash reporter assay. Finally, PLAGL2 overexpression obviously reverted the inhibiting functions of sh-HCP5. These data further validated HCP5/miR-128-3p/PLAGL2 to promote MM proliferative rate by triggering the Wnt/β‐catenin signaling pathway.
Conclusion
In summary, this is the first study to reveal the function of HCP5 in MM. Our results demonstrated that HCP5 promoted cell proliferation and tumor formation of MM cells by activating the Wnt/β‐catenin signaling pathway via sponging miR-128-3p to increase PLAGL2 expression.
Data availability
Some of the data that support the findings of this study are not publicly available due to their containing information that could compromise the privacy of research participants, but are available on request from the corresponding author: Ruixiang Xia. Department of Hematology, the First Affiliated Hospital of Anhui Medical University, 218 Jixi Road, Shushan District, Hefei 230,022, Anhui, China. Email address: RuixiangXiaHospita@163.com.
References
Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Investig. 2017;127(3):761–71. https://doi.org/10.1172/jci84424.
Amodio N, D’Aquila P, Passarino G, Tassone P, Bellizzi D. Epigenetic modifications in multiple myeloma: recent advances on the role of DNA and histone methylation. Expert Opin Ther Targets. 2017;21(1):91–101. https://doi.org/10.1080/14728222.2016.1266339.
An QM, Liu YG, Lin L, Liu Y, Lu J, Ge Q. Expression of extracellular matrix protein reelin in patients with multiple myeloma and its relationship with prognosis. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2018;26(3):789–95. https://doi.org/10.7534/j.issn.1009-2137.2018.03.026.
Baghdadi M, Ishikawa K, Nakanishi S, Murata T, Umeyama Y, Kobayashi T, et al. A role for IL-34 in osteolytic disease of multiple myeloma. Blood Advances. 2019;3(4):541–51. https://doi.org/10.1182/bloodadvances.2018020008.
Baker K, Gordon SL, Melland H, Bumbak F, Scott DJ, Jiang TJ, et al. SYT1-associated neurodevelopmental disorder: a case series. Brain. 2018;141(9):2576–91. https://doi.org/10.1093/brain/awy209.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Cao DW, Liu MM, Duan R, Tao YF, Zhou JS, Fang WR, et al. The lncRNA Malat1 functions as a ceRNA to contribute to berberine-mediated inhibition of HMGB1 by sponging miR-181c-5p in poststroke inflammation. Acta Pharmacologica Sinica. 2020;41(1):22–33. https://doi.org/10.1038/s41401-019-0284-y.
Cea M, Cagnetta A, Fulciniti M, Tai YT, Hideshima T, Chauhan D, et al. Targeting NAD+ salvage pathway induces autophagy in multiple myeloma cells via mTORC1 and extracellular signal-regulated kinase (ERK1/2) inhibition. Blood. 2012;120(17):3519–29. https://doi.org/10.1182/blood-2012-03-416776.
Chen M, Li L. SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/β-catenin pathway via CTNNB1. Cancer Sci. 2019;110(9):2794–805. https://doi.org/10.1111/cas.14140.
Chen X, Qi M, Yang Q, Li JY. MiR-299-3p functions as a tumor suppressor in thyroid cancer by regulating SHOC2. European Review for Medical and Pharmacological Sciences. 2019;23(1):232–40. https://doi.org/10.26355/eurrev_201901_16769.
Clark KC, Hewett DR. Targeted disruption of bone marrow stromal cell-derived Gremlin1 limits multiple myeloma disease progression in vivo. 2020;12(8). doi:https://doi.org/10.3390/cancers12082149.
Dang S, Zhou J, Wang Z, Wang K, Dai S, He S. MiR-299–3p functions as a tumor suppressor via targeting Sirtuin 5 in hepatocellular carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2018;106:966–75. doi:https://doi.org/10.1016/j.biopha.2018.06.042.
Detappe A, Bustoros M, Mouhieddine TH, Ghoroghchian PP. Advancements in nanomedicine for multiple myeloma. Trends Mol Med. 2018;24(6):560–74. https://doi.org/10.1016/j.molmed.2018.04.005.
Fu C, Li D, Zhang X, Liu N, Chi G, Jin X. LncRNA PVT1 Facilitates tumorigenesis and progression of glioma via regulation of MiR-128–3p/GREM1 axis and BMP signaling pathway. Neurotherapeutics. 2018;15(4):1139–57. https://doi.org/10.1007/s13311-018-0649-9.
Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nature Reviews Drug Discovery. 2010;9(10):775–89. https://doi.org/10.1038/nrd3179.
Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. 2017;8(60):101984–93. https://doi.org/10.18632/oncotarget.21957.
Gupta N, Kumar R, Seth T, Garg B, Sharma A. Targeting of stromal versican by miR-144/199 inhibits multiple myeloma by downregulating FAK/STAT3 signalling. RNA Biol. 2020;17(1):98–111. https://doi.org/10.1080/15476286.2019.1669405.
Hrašovec S, Hauptman N, Glavač D, Jelenc F, Ravnik-Glavač M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Disease Markers. 2013;34(2):93–104. https://doi.org/10.3233/dma-120948.
Hu R, Lu Z. Long non-coding RNA HCP5 promotes prostate cancer cell proliferation by acting as the sponge of miR-4656 to modulate CEMIP expression. Oncology Reports. 2020;43(1):328–36. https://doi.org/10.3892/or.2019.7404.
Ibrahim AM, Sabet S, El-Ghor AA, Kamel N. Fibulin-2 is required for basement membrane integrity of mammary epithelium. Sci rep. 2018;8(1):14139. https://doi.org/10.1038/s41598-018-32507-x.
Juli G, Oliverio M. Anti-tumor activity and epigenetic impact of the polyphenol oleacein in multiple myeloma. Cancer (Basel). 2019;11(7):990. https://doi.org/10.3390/cancers11070990.
Kong X, Duan Y, Sang Y, Li Y. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019;234(6):9105–17. https://doi.org/10.1002/jcp.27587.
Lai CQ, Parnell LD, Arnett DK, García-Bailo B, Tsai MY, Kabagambe EK, et al. WDTC1, the ortholog of Drosophila adipose gene, associates with human obesity, modulated by MUFA intake. Obesity (silver Spring, Md). 2009;17(3):593–600. https://doi.org/10.1038/oby.2008.561.
Laubach J, Garderet L, Mahindra A, Gahrton G, Caers J, Sezer O, et al. Management of relapsed multiple myeloma: recommendations of the International Myeloma Working Group. Leukemia. 2016;30(5):1005–17. https://doi.org/10.1038/leu.2015.356.
Leone E, Morelli E, Di Martino MT, Amodio N, Foresta U, Gullà A, et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res. 2013;19(8):2096–106. https://doi.org/10.1158/1078-0432.ccr-12-3325.
Li N, Li D, Du Y, Su C, Yang C, Lin C, et al. Overexpressed PLAGL2 transcriptionally activates Wnt6 and promotes cancer development in colorectal cancer. Oncology Reports. 2019;41(2):875–84. https://doi.org/10.3892/or.2018.6914.
Liang L, Xu J, Wang M, Xu G, Zhang N, Wang G, et al. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death & Disease. 2018;9(3):372. https://doi.org/10.1038/s41419-018-0382-7.
Liu H, Devraj K, Möller K, Liebner S, Hecker M, Korff T. EphrinB-mediated reverse signalling controls junctional integrity and pro-inflammatory differentiation of endothelial cells. Thrombosis and Haemostasis. 2014;112(1):151–63. https://doi.org/10.1160/th13-12-1034.
Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-transmitted miR-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Molecular Cancer. 2019a;18(1):43. https://doi.org/10.1186/s12943-019-0981-7.
Liu Y, Wang J, Dong L, Xia L, Zhu H, Li Z, et al. Long noncoding RNA HCP5 regulates pancreatic cancer gemcitabine (GEM) resistance by sponging Hsa-miR-214-3p to target HDGF. OncoTargets and Therapy. 2019b;12:8207–16. https://doi.org/10.2147/ott.s222703.
Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–7. https://doi.org/10.1016/j.jaci.2017.08.034.
Parks RN, Kim CJ, Al-Safi ZA, Armstrong AA, Zore T, Moatamed NA. Multiple myeloma 1 transcription factor is superior to CD138 as a marker of plasma cells in endometrium. Int J Surg Pathol. 2019;27(4):372–9. https://doi.org/10.1177/1066896918814307.
Pawlyn C, Bright MD, Buros AF, Stein CK, Walters Z, Aronson LI, et al. Overexpression of EZH2 in multiple myeloma is associated with poor prognosis and dysregulation of cell cycle control. Blood Cancer. 2017;7(3):e549. https://doi.org/10.1038/bcj.2017.27.
She X, Yu Z, Cui Y, Lei Q, Wang Z, Xu G, et al. miR-128 and miR-149 enhance the chemosensitivity of temozolomide by Rap1B-mediated cytoskeletal remodeling in glioblastoma. Oncology Reports. 2014;32(3):957–64. https://doi.org/10.3892/or.2014.3318.
Spaan I, Raymakers RA, van de Stolpe A, Peperzak V. Wnt Signaling in Multiple Myeloma: a Central Player in Disease with Therapeutic Potential. 2018;11(1):67. https://doi.org/10.1186/s13045-018-0615-3.
Wang JH, Zheng WW, Cheng ST, Liu BX, Liu FR, Song JQ. Correlation between microRNA-21 and sprouty homolog 2 gene expression in multiple myeloma. Molecular Medicine Reports. 2015;11(6):4220–4. https://doi.org/10.3892/mmr.2015.3280.
Wang L, Luan T, Zhou S, Lin J, Yang Y, Liu W, et al. LncRNA HCP5 promotes triple negative breast cancer progression as a ceRNA to regulate BIRC3 by sponging miR-219a-5p. Cancer Medicine. 2019;8(9):4389–403. https://doi.org/10.1002/cam4.2335.
Weiss GJ, Bemis LT, Nakajima E, Sugita M, Birks DK, Robinson WA, et al. EGFR regulation by microRNA in lung cancer: correlation with clinical response and survival to gefitinib and EGFR expression in cell lines. Ann Oncol. 2008;19(6):1053–9. https://doi.org/10.1093/annonc/mdn006.
Wu H, Liu C, Yang Q, Xin C, Du J, Sun F, et al. MIR145-3p promotes autophagy and enhances bortezomib sensitivity in multiple myeloma by targeting HDAC4. Autophagy. 2020;16(4):683–97. https://doi.org/10.1080/15548627.2019.1635380.
Xu B, Zhang X, Wang S, Shi B. MiR-449a suppresses cell migration and invasion by targeting PLAGL2 in breast cancer. Pathology, Research and Practice. 2018;214(5):790–5. https://doi.org/10.1016/j.prp.2017.12.012.
Yang C, Sun J, Liu W, Yang Y, Chu Z, Yang T, et al. Long noncoding RNA HCP5 contributes to epithelial-mesenchymal transition in colorectal cancer through ZEB1 activation and interacting with miR-139-5p. Am J Transl Res. 2019;11(2):953–63.
Yang J, Li C, Mudd A, Gu X. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Bioscience, Biotechnology, and Biochemistry. 2017;81(12):2301–6. https://doi.org/10.1080/09168451.2017.1387048.
Yu Y, Shen HM, Fang DM, Meng QJ, Xin YH. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22(15):4812–9. https://doi.org/10.26355/eurrev_201808_15616.
Yu Y, Zhao JD, Yang H. MiR-299-3p inhibits proliferation and invasion of cervical cancer cell via targeting TCF4. Eur Rev Med Pharmacol Sci. 2019;23(13):5621–7. https://doi.org/10.26355/eurrev_201907_18296.
Yun WK, Hu YM, Zhao CB, Yu DY, Tang JB. HCP5 promotes colon cancer development by activating AP1G1 via PI3K/AKT pathway. European Review for Medical and Pharmacological Sciences. 2019;23(7):2786–93. https://doi.org/10.26355/eurrev_201904_17553.
Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl). 2009;87(1):43–51. https://doi.org/10.1007/s00109-008-0403-6.
Zhao J, Li D, Fang L. MiR-128–3p suppresses breast cancer cellular progression via targeting LIMK1. Biomed Pharmacother. 2019;115:108947. doi:https://doi.org/10.1016/j.biopha.2019.108947.
Zhou J, Liu H, Zhang L, Liu X, Zhang C, Wang Y, et al. DJ-1 promotes colorectal cancer progression through activating PLAGL2/Wnt/BMP4 axis. Cell Death & Disease. 2018;9(9):865. https://doi.org/10.1038/s41419-018-0883-4.
Zhou Z, Wu L, Liu Z, Zhang X, Han S, Zhao N, et al. MicroRNA-214-3p targets the PLAGL2-MYH9 axis to suppress tumor proliferation and metastasis in human colorectal cancer. Aging. 2020;12(10):9633–57. https://doi.org/10.18632/aging.103233.
Acknowledgements
We thank the major projects of Natural Science Research Projects of Universities in Anhui Province for the support.
Funding
This work was supported by the Anhui Medical University 2020 School Research Fund (2020xky163).
Author information
Authors and Affiliations
Contributions
Qinhua Liu, Ruixiang Xia: study concepts, literature research, clinical studies, data analysis, experimental studies, manuscript writing and review; Ruonan Ran: study design, literature research, experimental studies, and manuscript editing; Mingyue Song: definition of intellectual content, clinical studies, data acquisition, and statistical analysis; Xiaodan Li1: data acquisition, manuscript preparation, and data analysis; Zhengsheng Wu, Guanrong Dai: data acquisition and statistical analysis. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Informed consent was obtained from all individual participants included in the study. All producers were approved by the Human and Animal Ethics Committee of the First Affiliated Hospital of Anhui Medical University. Procedures operated in this research were completed in keeping with the standards set out in the Announcement of Helsinki and Laboratory Guidelines of Research in China.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
•HCP5 was significantly upregulated in MM tissues and cancer cell lines.
•HCP5 knockdown effectively inhibited proliferation and cell cycle of MM cell lines in vitro and suppressed tumor growth in vivo.
•HCP5 could function as competitive endogenous RNA (ceRNA) via directly sponging of miR-128-3p, which further regulates PLAGL2 expression.
Supplementary Information
Below is the link to the electronic supplementary material.

10565_2021_9628_MOESM1_ESM.jpg
Supplementary file1 PLAGL2 overexpression promoted proliferation of MM cell lines. NCI-H929 cells were transfected with PLAGL2 (pcDNA3.1-PLAGL2 overexpression plasmid) and corresponding negative control (the empty vector), and MM.1S cells were transfected with si-PLAGL2 and negative control si-NC. (A) Transfection efficiency was evaluated by Western blot. (B) Cell viability was evaluated by CCK-8 assay. (C and D) Cell proliferation was evaluated by colony formation assay (C) and EdU staining assay (D). (E) Cell cycle was evaluated by flow cytometry. * P < 0.05, ** P < 0.01, *** P < 0.001. Each experiment was repeated three times. (JPG 1329 KB)
Rights and permissions
About this article
Cite this article
Liu, Q., Ran, R., Song, M. et al. LncRNA HCP5 acts as a miR-128-3p sponge to promote the progression of multiple myeloma through activating Wnt/β‐catenin/cyclin D1 signaling via PLAGL2. Cell Biol Toxicol 38, 979–993 (2022). https://doi.org/10.1007/s10565-021-09628-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-021-09628-7