Abstract
Circular RNA (circRNA) 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (circATIC; hsa_circ_0058058) was observed to be upregulated in multiple myeloma (MM) by former article. However, the function and exact mechanism of circATIC in MM development remain barely known. CircRNA-microRNA (miRNA)-messenger RNA (mRNA) axis was established through using bioinformatic databases (starbase, Circinteractome, and microT-CDS). Dual-luciferase reporter assay, RNA immunoprecipitation assay, and RNA-pull down assay were utilized to verify the target relationship between microRNA-324-5p (miR-324-5p) and circATIC or hepatocyte growth factor (HGF). CircATIC expression was upregulated in MM patients and cell lines. CircATIC interference notably hampered cell proliferation, migration, invasion, and glycolysis and induced cell apoptosis of MM cells. MiR-324-5p was a target of circATIC. CircATIC silencing-mediated effects in MM cells were largely overturned by the knockdown of miR-324-5p. HGF was a target of miR-324-5p, and circATIC upregulated the expression of HGF partly through sponging miR-324-5p in MM cells. MiR-324-5p suppressed the malignant behaviors of MM cells, which were largely counteracted by the overexpression of HGF in MM cells. CircATIC accelerated the proliferation, migration, invasion, and glycolysis and suppressed the apoptosis of MM cells through mediating miR-324-5p/HGF signaling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Multiple myeloma (MM) is a common hematologic disease with the over-activation of proliferation in plasma cells (Terpos et al. 2018). Despite diagnosis and treatment methods for MM have advanced, the prognosis remains unsatisfactory, especially in high-risk MM patients (Pawlyn and Morgan 2017). Therefore, discovering molecular mechanism behind MM progression is urgently needed to identify novel targets for MM diagnosis and prognosis, which also could be used as therapeutic targets.
Circular RNAs (circRNAs) engage in the initiation and progression of malignancies through sponging microRNAs (miRNAs) to regulate cellular malignant phenotypes (Yin et al. 2019; Zhao et al. 2019). Many circRNAs have been reported to regulate the progression of hematological disorders. For instance, circ_0009910 accelerated the progression of acute myeloid leukemia through sponging miR-20a-5p (Ping et al. 2019). Circ_0000142 elevated the malignant potential of MM cells through binding to miR-610 to induce AKT3 expression (Liu et al. 2020). CircRNA 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (circATIC; hsa_circ_0058058) was reported to be upregulated in MM (Zhou et al. 2020). We intended to investigate the biological significances behind the high expression of circATIC in MM progression.
MiRNAs exert regulatory roles in the progression of malignancies through degrading target messenger RNAs (mRNAs) (Guo et al. 2016). For instance, miR-342 and miR-363 restrained MM progression through decreasing Runx2 expression (Gowda et al. 2018). MiR-218 elevated the drug resistance of MM cells through downregulating LRRC28 (Chen et al. 2021). MiR-324-5p was predicted as a potential target of circATIC via bioinformatic database. MiR-324-5p exerted an anti-tumor role in many cancers, including MM (Jiang et al. 2018; Lin et al. 2018; Gu et al. 2019; Zhang et al. 2018). For instance, Zhang et al. claimed that miR-324-5p restrained the motility of MM cells (2018). In this study, we further explored the working mechanism of miR-324-5p in MM progression.
Based on bioinformatic analysis, hepatocyte growth factor (HGF) is a potential target of miR-324-5p. HGF is a multidomain glycoprotein, and it is a ligand of MET receptor (Ghiso and Giordano 2013). The oncogenic role of HGF has been reported in multiple malignancies, including blood tumors (Borset et al. 1996) and solid tumors (Graziano et al. 2011; Bu et al. 2020; Lennerz et al. 2011). As for MM, HGF was observed to be upregulated in the serum samples of MM patients, and it was a prognostic factor for MM patients (Seidel et al. 1998). Yang et al. found that HGF was upregulated in MM patients and cell lines, and HGF promoted the proliferation and restrained the apoptosis of MM cells (Yang and Chen 2019). The target relation between HGF and miR-324-5p was tested in our study, and their functional relevance in MM progression was investigated.
CircATIC was abnormally upregulated in MM patients and cell lines. CircATIC silencing restrained the proliferation, migration, invasion, and glycolysis and induced the apoptosis of MM cells. Subsequently, bioinformatic analysis was used to predict the downstream targets of circATIC, and the working mechanism of circATIC was confirmed through conducting rescue experiments.
Materials and Methods
Clinical Samples
A total of 37 MM patients and 8 healthy bone marrow (BM) donors were enrolled in ShangRao People’s Hospital. This study was conducted with the permission of the Medical Ethics Committee of ShangRao People’s Hospital and in accordance with the Declaration of Helsinki. Mononuclear cells were isolated from BM samples via gradient density centrifugation followed by the purification of plasma cells using CD138-coated magnetic beads (Miltenyi Biotech, Bergisch Gladbach, Germany). All subjects had signed written informed consents prior to sample collection.
Cell Lines
H929 MM cell line, OPM2 MM cell line, and normal plasma cell line nPCs were all acquired from BeNa Culture Collection (Beijing, China). All cell lines were maintained in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco, Carlsbad, CA, USA) plus 10% fetal bovine serum (FBS; Gibco), 2 mM l-glutamine (Gibco), 1% 100 U/mL penicillin (Invitrogen, Carlsbad, CA, USA), and 1% 100 μg/mL streptomycin (Invitrogen) in a 5% CO2 incubator at 37 °C.
Cyclization Validation of circATIC
RNase R (3 U/μg; Epicentre Technologies, Madison, WI, USA) was incubated with RNA sample (3 μg) for 30 min at room temperature, and the expression of circATIC and linear ATIC mRNA was measured by qRT-PCR.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Complementary DNA (cDNA) of circRNA and mRNA was synthesized using the Prime Script RT Master Mix (Takara, Dalian, China), while reverse transcription of miRNA was conducted using the miRNA First-Strand cDNA Synthesis Kit (TIANGEN, Beijing, China). PCR amplification of circRNA and mRNA was conducted using FastStart Universal SYBR Green Master (Roche, Mannheim, Germany), while the amplification of miRNA was performed with Taqman MicroRNA Assay Kit (Applied Biosystems, Foster City, CA, USA). U6 for miR-324-5p and β-actin for circATIC, ATIC, and HGF were used as house-keeping genes. Specific primers in qRT-PCR assay are listed in Table 1. The relative expression of these molecules was analyzed as the 2−ΔΔCt method.
Cell Transient Transfection
CircATIC-specific small interfering RNA (si-circATIC), scramble siRNA negative control (si-NC), circATIC overexpression plasmid (circATIC), negative control pLCDH-cir empty vector (NC), HGF overexpression plasmid (HGF), and pcDNA were obtained from Genepharma (Shanghai, China). MiR-324-5p mimics (miR-324-5p), miR-NC, miR-324-5p inhibitor (in-miR-324-5p), and in-miR-NC were chemically synthesized by Genechem Company (Shanghai, China). Transfection was conducted using Lipofectamine 3000 (Invitrogen), and MM cells were harvested for the following experiments at appropriate time points.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
After transfection for 0 h, 24 h, 48 h, or 72 h, MM cells in 96-well plates in sextuplicate were incubated with 20 μL 2.5 mg/mL MTT solution (Life Technologies, Waltham, MA, USA) for 4 h. 150 μL dimethyl sulfoxide (DMSO; Sangon Biotech, Shanghai, China) was then added to dissolve the formazan crystals. The absorbance at the wavelength of 570 nm was measured.
Flow Cytometry for Cell Cycle Analysis
MM cells were collected and then immobilized in 80% ice-cold ethanol. RNA content was removed through incubating with RNAase (2 mg/mL; Sigma, St. Louis, MO, USA) at 37 °C for 30 min. The DNA content was stained through incubating with propidium iodide (20 mg/mL; Sigma) for 20 min. The proportion of MM cells at G0/G1, S, and G2/M phases was analyzed by flow cytometry.
Transwell Migration and Invasion Assays
Transwell chambers were purchased from Costar (Corning, NY, USA). To determine cell migration ability, MM cells (3 × 104 cells) suspended in 100 μL serum-free medium were added into the upper chambers, and 600 μL culture medium added with 20% FBS (chemotactic factor) was used to fill the lower chambers. Migrated MM cells were counted after 24 h incubation. Magnification: 100 times.
To analyze cell invasion ability, MM cells (1 × 105 cells) suspended in 100 μL serum-free medium were added into Matrigel (Sigma)-coated upper chambers. Transwell invasion assay was carried out as similar as transwell migration assay. Magnification: 100 times.
Flow Cytometry for Cell Apoptosis Analysis
MM cells after transfection for 72 h were collected and resuspended in 100 μL binding buffer (Solarbio, Beijing, China). Annexin V-combined fluorescein isothiocyanate (Annexin V-FITC; 5 μL; Solarbio) and PI (5 μL; Solarbio) were added to incubate with MM cells for 15 min at room temperature in the dark. After staining, MM cells were immediately collected for apoptotic analysis on FACSCalibur (Becton Dickinson, Franklin Lakes, NJ, USA) using Cell Quest software (Becton Dickinson). The percentage of MM cells with FITC+/PI− and FITC+/PI+ was counted as apoptosis rate.
Glycolytic Analysis
The glycolysis of MM cells was analyzed through measuring the production of lactate and ATP and the utilization of glucose using fluorescence-based glucose or lactate assay kit (Biovision, Milpitas, CA, USA) and ATP Colorimetric Assay kit (Biovision).
Western Blot Assay
Cell lysates were harvested using cell lysis buffer (Abcam, Cambridge, MA, USA). Equal amount of protein samples (30 μg) in loading buffer were loaded onto sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to the polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). After incubating with 5% (w/v) skimmed milk for 1 h, the membrane was incubated with primary antibodies at 4 °C overnight, including anti-glucose transporter type 1 (anti-GLUT1; SAB4200519; Sigma), anti-hexokinase 2 (anti-HK2; HPA028587; Sigma), anti-lactate dehydrogenase A (anti-LDHA; SAB1100050; Sigma), anti-HGF (HPA040360; Sigma), and anti-β-actin (SAB5500001; Sigma). Appropriate horseradish peroxidase (HRP)-labeled secondary antibody (Sigma) was then incubated with the membrane. Protein bands were visualized using the enhanced chemiluminescent (ECL) Kit (Amersham Biosciences, Piscataway, NJ, USA). Quantity One software (Bio-Rad, Hercules, CA, USA) was utilized to analyze the densities of protein bands.
Bioinformatic Analysis
CircATIC–miRNA interactions were predicted by starbase software (http://starbase.sysu.edu.cn) and Circinteractome software (https://circinteractome.irp.nia.nih.gov), while the mRNA targets of miR-324-5p were sought by microT-CDS of DIANA TOOL (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS).
Dual-Luciferase Reporter Assay
The wild-type or mutant miR-324-5p-binding sequence in circATIC was synthesized and inserted into psiCHECK2 luciferase reporter vector (Promega, Madison, WI, USA) to generate circATIC WT or circATIC MUT. The wild-type or mutant miR-324-5p-binding sequence in the 3′ untranslated region (3′UTR) fragment of HGF was also amplified and cloned into psiCHECK2 luciferase reporter vector (Promega) to obtain HGF 3′UTR WT or HGF 3’UTR MUT. MM cells in 24-well plates were transfected with 10 nM of miR-NC or miR-324-5p and 40 ng luciferase reporter plasmids. Luciferase activities (Firefly intensity and Renilla intensity) were determined after transfection for 48 h using the dual-luciferase reporter assay system kit (Promega) on luminometer (Plate Chameleon V, Hidex, Finland). Renilla luciferase intensity was used as the control.
RNA Immunoprecipitation (RIP) Assay
MM cells were disrupted using RIP lysis buffer (Haoran Biotech, Shanghai, China) for 5 min. Cell extracts were incubated with magnetic beads (Bio-Rad) coated with argonaute 2 (AGO2) antibody (Abcam) or immunoglobulin G (IgG) antibody (Abcam) for 3 h. RNA enrichment of circATIC and miR-324-5p was assessed by qRT-PCR.
RNA-Pull Down Assay
A total of 2 μg cell lysates were incubated with 100 pmol biotinylated RNA (Bio-NC, Bio-circATIC WT and Bio-circATIC MUT). Streptavidin agarose beads were added to incubate for 1 h. After washing and boiling the beads, the enrichment of miR-324-5p was determined by qRT-PCR.
Statistical Analysis
Statistical results were shown as mean ± standard deviation (SD). Three experiments were performed with at least three replicates each. Student’s t test was utilized to evaluate the comparisons in two groups, and one-way analysis of variance (ANOVA) followed by Tukey’s test was used to assess the differences in multiple groups. Linear correlation among the expression of circATIC, miR-324-5p, and HGF was assessed using Spearman’s correlation coefficient. Differences were considered to be statistically significant with the P value of less than 0.05.
Results
CircATIC is Highly Expressed in MM
The expression characteristic of circATIC was analyzed in MM. CircATIC expression was higher in BM-derived plasma cells of MM patients (n = 37) than that in healthy donors (n = 8) (Fig. 1A). Compared with normal plasma cell line nPCs, circATIC expression was elevated in both MM cell lines (H929 and OPM2) (Fig. 1B). CircATIC expression was stable in RNase R+ group compared with Mock group (Fig. 1C, D), suggesting that circATIC was a circular transcript that was resistant to exonuclease digestion. The aberrant upregulation of circATIC might imply its important role in MM development.
CircATIC is highly expressed in MM. A qRT-PCR was implemented to measure the expression of circATIC in BM-derived plasma cells of MM patients (n = 37) and healthy bone marrow donors (n = 8). B The level of circATIC was determined in two MM cell lines (H929 and OPM2) and normal plasma cell line (nPCs) by qRT-PCR. C and D RNA samples isolated from H929 and OPM2 cells were equally divided into two parts, and these two parts were treated with RNase R or not, named Mock group and RNase R+ group. The relative expression of circATIC and linear ATIC mRNA was detected by qRT-PCR. *P < 0.05
CircATIC Silencing Restrains Cell Proliferation, Migration, Invasion, Glycolysis, and Triggers Cell Apoptosis of MM Cells
The specific siRNA targeting the junction sites of circATIC (si-circATIC) was utilized to conduct loss-of-function experiments to investigate the functions of circATIC in MM cells. CircATIC expression was notably reduced in si-circATIC-transfected MM cells compared with that in si-NC group (Fig. 2A, B). Cell proliferation ability was assessed by MTT assay and flow cytometry. Cell proliferation was significantly restrained with the interference of circATIC in MM cells (Fig. 2C, D). CircATIC silencing significantly reduced the percentage of MM cells in S phase but increased the percentage of MM cells in G0/G1 phase (Fig. 2E, F), suggesting that circATIC knockdown arrested cell cycle progression at G1/S transition. The numbers of migrated and invaded MM cells were notably reduced in circATIC-silenced group relative to si-NC group (Fig. 2G, H), suggesting that circATIC interference restrained the migration and invasion of MM cells. The apoptosis rate was significantly enhanced with the silencing of circATIC than that in si-NC group in MM cells (Fig. 2I). Furthermore, we assessed the glycolysis of MM cells through measuring the uptake of glucose and the production of lactate and ATP. CircATIC silencing markedly hampered the utilization of glucose and the production of lactate and ATP (Fig. 2J–L), suggesting that circATIC silencing restrained the glycolysis of MM cells. The expression of three glycolysis-associated key molecules (GLUT1, HK2, and LDHA) was measured via Western blot assay to further confirm the role of circATIC on the glycolysis of MM cells. As mentioned in Fig. 2M, N, circATIC silencing markedly downregulated the expression of GLUT1, HK2, and LDHA in MM cells, which further demonstrated that circATIC knockdown restrained the glycolysis of MM cells. Overall, circATIC interference blocked the malignant behaviors of MM cells.
CircATIC silencing restrains cell proliferation, migration, invasion, and glycolysis and triggers cell apoptosis of MM cells. A–N H929 and OPM2 cells were transfected with si-NC or si-circATIC. A and B The expression of circATIC was determined in transfected MM cells by qRT-PCR. C and D Cell proliferation curve was generated through measuring the OD value after transfection for 0 h, 24 h, 48 h, or 72 h via MTT assay. E and F Cell cycle progression was assessed by flow cytometry. G and H Transwell assays were performed to analyze cell migration ability and invasion ability. I Cell apoptosis rate was analyzed via flow cytometry. J–L Cell glycolysis was analyzed through measuring the production of lactate and ATP and the consumption of glucose using fluorescence-based glucose assay kit, fluorescence-based lactate assay kit, and ATP Colorimetric Assay kit. M and N Western blot assay was applied to analyze the expression of glycolysis-associated proteins (GLUT1, HK2, and LDHA) in transfected MM cells. *P < 0.05
CircATIC Interacts with miR-324-5p in MM Cells
CircRNAs could function as miRNA sponges (Panda 2018). Bioinformatic analysis using starbase and Circinteractome softwares was conducted to seek the candidate miRNA targets of circATIC. As shown in Fig. 3A, there were six miRNAs (miR-324-5p, miR-335-5p, miR-338-3p, miR-337-3p, miR-421, and miR-668-3p) that were predicted to be possible targets of circATIC by both starbase and Circinteractome softwares. The overexpression efficiency of circATIC plasmid was high in H929 and OPM2 cells (Fig. 3B, C). CircATIC overexpression reduced the expression of miR-324-5p and miR-338-3p (Fig. 3D, E), and we selected miR-324-5p for the following experiments due to its more obvious negative regulatory relationship with circATIC relative to miR-338-3p. The putative binding sequence between miR-324-5p and circATIC is shown in Fig. 3F. We mutated four bases “GGAU” in the putative binding sites of circATIC by “CCUA” to test if miR-324-5p bound to circATIC via the predicted sequence (Fig. 3F). MiR-324-5p transfection markedly reduced the luciferase activity in circATIC WT group when compared with miR-NC and circATIC WT group (Fig. 3G, H), suggesting that miR-324-5p was a target of circATIC in MM cells. Besides, luciferase activities remained unchanged in miR-NC+circATIC MUT group and miR-324-5p+circATIC MUT group (Fig. 3G, H), suggesting that circATIC bound to miR-324-5p via the predicted sequence. Subsequently, RIP assay and RNA-pull down assay were carried out to further confirm the target interaction between circATIC and miR-324-5p. MiR-324-5p and circATIC were both enriched in the precipitated complex when using AGO2 antibody rather than IgG antibody (Fig. 3I, J), suggesting the binding relation between circATIC and miR-324-5p. MiR-324-5p was markedly enriched when using Bio-circATIC WT compared with that in Bio-NC group and Bio-circATIC MUT group (Fig. 3K, L), which further validated the binding relation between circATIC and miR-324-5p. MiR-324-5p expression was decreased in BM-derived plasma cells of MM patients (n = 37) in comparison with that in healthy donors (n = 8) (Fig. 3M). MiR-324-5p expression was negatively correlated with the level of circATIC in MM patients (Fig. 3N). Also, there was a significant reduction in miR-324-5p expression in two MM cell lines compared with nPCs (Fig. 3O). MiR-324-5p level was elevated with the interference of circATIC, and the overexpression of circATIC reduced the expression of miR-324-5p in MM cells (Fig. 3P). Taken together, miR-324-5p was a target of circATIC, and it was negatively regulated by circATIC in MM cells.
CircATIC interacts with miR-324-5p in MM cells. A The miRNA targets of circATIC were predicted through using starbase software and Circinteractome software. Six miRNAs that predicted to be candidate targets of circATIC by both bioinformatic softwares were listed. B and C The overexpression efficiency of circATIC ectopic expression plasmid (circATIC) was tested by qRT-PCR, and its negative control empty pLCDH-cir vector (NC) was regarded as the control. D and E The expression of six miRNAs (miR-324-5p, miR-335-5p, miR-338-3p, miR-337-3p, miR-421, and miR-668-3p) was detected in H929 and OPM2 cells transfected with NC or circATIC by qRT-PCR. F The putative binding sites between circATIC and miR-324-5p predicted by starbase software and Circinteractome software are shown. We mutated “GGAU” sequence by “CCUA” sequence in circATIC. G and H Dual-luciferase reporter assay was implemented to verify if miR-324-5p was a target of circATIC in MM cells. Luciferase activities were measured in two MM cell lines transfected with miR-NC+circATIC WT, miR-324-5p+circATIC WT, miR-NC+circATIC MUT, or miR-324-5p+circATIC MUT. I and J RIP assay was employed to analyze the target interaction between miR-324-5p and circATIC in MM cells. K and L RNA-pull down assay was conducted to confirm the binding relation between circATIC and miR-324-5p in MM cells. M MiR-324-5p expression was measured in BM-derived plasma cells of MM patients (n = 37) and healthy bone marrow donors (n = 8) by qRT-PCR. N Linear correlation between the levels of circATIC and miR-324-5p in MM patients was assessed by Spearman’s correlation coefficient. O qRT-PCR was applied to measure the expression of miR-324-5p in MM cell lines and nPCs. P The level of miR-324-5p was detected in H929 and OPM2 cells transfected with si-NC, si-circATIC, NC, or circATIC by qRT-PCR. *P < 0.05
MiR-324-5p Silencing Largely Attenuates circATIC Knockdown-Mediated Influences in MM Cells
CircATIC silencing-mediated upregulation in miR-324-5p expression was largely counteracted by the addition of in-miR-324-5p in MM cells (Fig. 4A). We conducted rescue experiments to test if circATIC functioned through sponging miR-324-5p. Through performing MTT assay and flow cytometry, we found that circATIC interference-mediated inhibitory effect in cell proliferation was alleviated by the addition of in-miR-324-5p in MM cells (Fig. 4B–E). MiR-324-5p silencing also counteracted the suppressive influences in cell migration and invasion abilities induced by circATIC silencing in MM cells (Fig. 4F, G). Cell apoptosis was triggered in circATIC-silenced group, and the introduction of in-miR-324-5p largely overturned circATIC silencing-triggered apoptosis of MM cells (Fig. 4H). CircATIC silencing suppressed the production of lactate and ATP and the utilization of glucose, and these suppressive effects were all attenuated by the addition of in-miR-324-5p in MM cells (Fig. 4I–K). The knockdown of miR-324-5p also largely rescued the expression of GLUT1, HK2, and LDHA in circATIC-silenced MM cells (Fig. 4L, M). Overall, circATIC silencing restrained the malignant phenotypes of MM cells partly through upregulating miR-324-5p.
MiR-324-5p silencing largely attenuates circATIC knockdown-mediated influences in MM cells. A–M MM cells were transfected with si-NC, si-circATIC, si-circATIC+in-miR-NC or si-circATIC+in-miR-324-5p. A qRT-PCR was applied to detect the expression of miR-324-5p in transfected MM cells. B and C MTT assay was conducted to analyze cell proliferation ability. D and E Flow cytometry was applied to assess cell cycle progression of transfected MM cells. F and G Cell migration and invasion abilities were measured by transwell assays. H Flow cytometry was conducted to analyze cell apoptosis rate in transfected MM cells. I–K Cell glycolysis was analyzed through assessing the uptake of glucose and the levels of lactate and ATP via fluorescence-based glucose or lactate assay kit and ATP Colorimetric Assay kit. L and M The protein levels of glycolysis-associated molecules (GLUT1, HK2, and LDHA) were examined by Western blot assay. *P < 0.05
HGF is a Target of miR-324-5p in MM Cells
microT-CDS database of DIANA TOOL was used to predict the possible mRNA targets of miR-324-5p, and HGF was predicted as one of the candidate targets of miR-324-5p (Fig. 5A). MiR-324-5p accumulation significantly decreased the luciferase activity in HGF 3’UTR WT group compared with miR-NC and HGF 3’UTR WT group (Fig. 5B, C), demonstrating the target interaction between miR-324-5p and HGF in MM cells. The mRNA and protein abundance of HGF were both increased in MM patients compared with healthy donors (Fig. 5D, E). Subsequently, we analyzed the linear correlation between the levels of HGF and miR-324-5p or circATIC. The results revealed a negative correlation between the expression of miR-324-5p and HGF (Fig. 5F), whereas there was a positive correlation between the expression of HGF and circATIC (Fig. 5G). HGF protein level was also found to be significantly elevated in MM cell lines compared with nPCs cell line (Fig. 5H). The transfection efficiency of miR-324-5p was high in both H929 and OPM2 cell lines (Fig. 5I). MiR-324-5p accumulation reduced the protein level of HGF in MM cells compared with that in miR-NC group (Fig. 5J). CircATIC overexpression enhanced the protein expression of HGF in MM cells, and this promoting effect was largely overturned by the addition of miR-324-5p (Fig. 5K). Taken together, miR-324-5p interacted with HGF and it negatively regulated the expression of HGF in MM cells.
HGF is a target of miR-324-5p in MM cells. A microT-CDS of DIANA TOOL was used to predict the mRNA targets of miR-324-5p, and the putative binding sites between HGF and miR-324-5p are shown. B and C Dual-luciferase reporter assay was implemented to verify the target interaction between miR-324-5p and HGF in MM cells. D and E qRT-PCR and Western blot assay were carried out to measure the expression of HGF mRNA and protein in MM patients and healthy donors. F and G The linear correlation between the expression of HGF and miR-324-5p or circATIC was analyzed using Spearman’s correlation coefficient. H The protein level of HGF was detected in H929, OPM2 and nPCs by Western blot assay. I The expression of miR-324-5p was examined in H929 and OPM2 cells transfected with miR-NC or miR-324-5p by qRT-PCR. J Western blot assay was utilized to measure the protein level of HGF in H929 and OPM2 cells transfected with miR-NC or miR-324-5p. K The protein level of HGF was examined in H929 and OPM2 cells transfected with NC, circATIC, circATIC+miR-NC, or circATIC+miR-324-5p by Western blot assay. *P < 0.05
MiR-324-5p Suppresses Cell Proliferation, Migration, Invasion, and Glycolysis and Promotes Cell Apoptosis of MM Cells Largely Through Downregulating HGF
MiR-324-5p overexpression reduced the protein level of HGF, and the addition of HGF overexpression plasmid largely rescued its expression in MM cells (Fig. 6A). Therefore, rescue experiments were conducted using the same treatment to explore if miR-324-5p suppressed MM progression through reducing HGF expression. MiR-324-5p transfection suppressed cell proliferation, and cell proliferation ability was partly recovered in miR-324-5p and HGF co-transfected group (Fig. 6B–E), suggesting that miR-324-5p hampered the proliferation ability of MM cells partly through downregulating HGF expression. According to the results of transwell assays, miR-324-5p-mediated suppressive effects in cell migration and invasion were largely attenuated by the introduction of HGF (Fig. 6F, G). Cell apoptosis was triggered with the overexpression of miR-324-5p, and the co-transfection with HGF plasmid alleviated miR-324-5p-induced apoptosis in MM cells (Fig. 6H). MiR-324-5p-mediated inhibitory influences in the production of lactate and the utilization of glucose were both largely counteracted by the introduction of HGF plasmid (Fig. 6I, J). HGF overexpression also largely recovered the production of ATP in miR-324-5p-overexpressed MM cells (Fig. 6K). MiR-324-5p overexpression reduced the expression of GLUT1, HK2, and LDHA, which was largely overturned by the accumulation of HGF (Fig. 6L, M). Overall, these findings confirmed the anti-tumor role of miR-324-5p in MM, and miR-324-5p restrained the malignant behaviors of MM cells partly through reducing HGF level.
MiR-324-5p suppresses cell proliferation, migration, invasion and glycolysis and promotes cell apoptosis of MM cells largely through down-regulating HGF. A–M H929 and OPM2 cells were transfected with miR-NC, miR-324-5p, miR-324-5p+pcDNA or miR-324-5p+HGF. A The protein level of HGF was examined in transfected MM cells by Western blot assay. B and C Cell proliferation ability was evaluated by MTT assay. OD value was measured after transfection for 0 h, 24 h, 48 h or 72 h to generate cell proliferation curve. D and E Cell cycle progression was evaluated by flow cytometry. F and G The capacities of migration and invasion were measured by transwell migration and invasion assays. H Flow cytometry was used to analyze cell apoptosis rate in transfected MM cells. I–K The production of lactate and ATP and the utilization of glucose were analyzed through using fluorescence-based glucose or lactate assay kit and ATP Colorimetric Assay kit. L and M Western blot assay was performed to analyze the expression of GLUT1, HK2 and LDHA in transfected MM cells. *P < 0.05
Discussion
Recently, non-coding RNAs (especially circRNAs) attach much attention in cancer field due to their important regulatory roles. For instance, circ_100395 suppressed the development of lung cancer through targeting miR-1228/TCF21 signaling (Chen et al. 2018). CircZNF609 contributed to the migration ability of colorectal cancer cells through targeting miR-150/Gli1 axis (Wu et al. 2018). Nevertheless, only few articles focused on the roles of circRNAs in MM progression. Feng et al. demonstrated that circ_0000190 suppressed the progression of MM through sponging miR-767-5p (2019). Liu et al. found that circSMARCA5 hampered MM development through sponging miR-767-5p (2019). CircATIC expression was reported to be upregulated in MM (Zhou et al. 2020). Consistent with former article (Zhou et al. 2020), circATIC expression was found to be significantly enhanced in MM patients and cell lines compared with healthy donors and nPCs. After silencing circATIC using its specific siRNA (si-circATIC), the proliferation, migration, invasion, and glycolysis of MM cells were all blocked while cell apoptosis was induced, suggesting that circATIC knockdown suppressed MM progression in vitro.
CircRNAs could act as miRNA sponges in cancers (Panda 2018). For instance, circ_100269 expression was reduced in gastric cancer (GC), and it restrained GC tumor growth through sponging miR-630 (Zhang et al. 2017). CircHIPK3 contributed to the progression of colorectal cancer through sponging miR-7 (Zeng et al. 2018). We found that miR-324-5p was a target of circATIC in MM cells. MiR-324-5p functioned as an anti-tumor molecule in many cancers. For instance, miR-324-5p acted as the target of long non-coding RNA TPT1-AS1 to restrain cervical cancer development (Jiang et al. 2018). MiR-324-5p suppressed cell viability and triggered cell apoptosis of GC cells through targeting TSPAN8 (Lin et al. 2018). Gu et al. demonstrated that miR-324-5p hampered the proliferation and invasion of colorectal cancer cells through targeting ELAVL1 (2019). As for MM, Zhang et al. found that miR-324-5p restrained the migration and invasion abilities of MM cells through targeting the SCFβ−TrCP E3 ligase (Xu et al. 2018). MiR-324-5p level was reduced in MM patients and cell lines. Through performing rescue experiments, we found that circATIC silencing restrained the malignant potential of MM cells partly through upregulating miR-324-5p.
Subsequently, bioinformatic software (microT-CDS of DIANA TOOL) was utilized to determine the downstream targets of miR-324-5p. HGF was confirmed as a target of miR-324-5p on the basis of the results of dual-luciferase reporter assay. HGF was initially identified as a mitogenic protein in rat hepatocytes (Russell et al. 1984). HGF exhibited an important role in modulating the growth and regeneration of liver (Michalopoulos 1993). HGF also played an oncogenic function in multiple malignancies. For instance, Chen et al. demonstrated that HGF promoted the epithelial–mesenchymal transition and angiogenesis of non-small-cell lung cancer cells, and these effects were alleviated by miR-206 (2016). Bu et al. found that LINC00240 contributed to the malignant progression of hepatocellular carcinoma through upregulating HGF via acting as miR-4465 sponge (2020). As for MM, Yang et al. demonstrated that UCA1 accelerated MM progression by binding to miR-1271-5p to induce HGF expression (Yang and Chen 2019), suggesting the oncogenic role of HGF in MM progression. HGF enrichment was enhanced in MM patients and cell lines compared with their matching counterparts. HGF was negatively regulated by miR-324-5p in MM cells. Additionally, we found that circATIC enhanced HGF expression through sponging miR-324-5p in MM cells. The results of rescue experiments revealed that miR-324-5p suppressed cell proliferation, migration, invasion, and glycolysis and promoted cell apoptosis partly through downregulating HGF in MM cells.
Conclusion
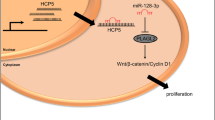
In conclusion, circATIC/miR-324-5p/HGF signaling was identified in our study for the first time. CircATIC accelerated cell proliferation, migration, invasion, and glycolysis and restrained cell apoptosis of MM cells through acting as miR-324-5p sponge to elevate HGF expression (Fig. 7). These findings suggested that circATIC/miR-324-5p/HGF signaling might be potential molecular target for MM treatment.
Data Availability
The analyzed datasets generated during the present study are available from the corresponding author on reasonable request.
References
Borset M, Lien E, Espevik T, Helseth E, Waage A, Sundan A (1996) Concomitant expression of hepatocyte growth factor/scatter factor and the receptor c-MET in human myeloma cell lines. J Biol Chem 271:24655–24661
Bu WJ, Fang Z, Li WL, Wang X, Dong MJ, Tao QY et al (2020) LINC00240 sponges miR-4465 to promote proliferation, migration, and invasion of hepatocellular carcinoma cells via HGF/c-MET signaling pathway. Eur Rev Med Pharmacol Sci 24:10452–10461
Chen QY, Jiao DM, Wu YQ, Chen J, Wang J, Tang XL et al (2016) MiR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met /PI3k/Akt/mTOR pathway. Oncotarget 7:18247–18261
Chen D, Ma W, Ke Z, Xie F (2018) CircRNA hsa_circ_100395 regulates miR-1228/TCF21 pathway to inhibit lung cancer progression. Cell Cycle 17:2080–2090
Chen H, Cao W, Chen J, Liu D, Zhou L, Du F et al (2021) miR-218 contributes to drug resistance in multiple myeloma via targeting LRRC28. J Cell Biochem 122:305–314
Feng Y, Zhang L, Wu J, Khadka B, Fang Z, Gu J et al (2019) CircRNA circ_0000190 inhibits the progression of multiple myeloma through modulating miR-767-5p/MAPK4 pathway. J Exp Clin Cancer Res 38:54
Ghiso E, Giordano S (2013) Targeting MET: why, where and how? Curr Opin Pharmacol 13:511–518
Gowda PS, Wildman BJ, Trotter TN, Xu X, Hao X, Hassan MQ et al (2018) Runx2 suppression by miR-342 and miR-363 inhibits multiple myeloma progression. Mol Cancer Res 16:1138–1148
Graziano F, Galluccio N, Lorenzini P, Ruzzo A, Canestrari E, D’Emidio S et al (2011) Genetic activation of the MET pathway and prognosis of patients with high-risk, radically resected gastric cancer. J Clin Oncol 29:4789–4795
Gu C, Zhang M, Sun W, Dong C (2019) Upregulation of miR-324-5p inhibits proliferation and invasion of colorectal cancer cells by targeting ELAVL1. Oncol Res 27:515–524
Guo J, McKenna SL, O’Dwyer ME, Cahill MR, O’Driscoll CM (2016) RNA interference for multiple myeloma therapy: targeting signal transduction pathways. Expert Opin Ther Targets 20:107–121
Jiang H, Huang G, Zhao N, Zhang T, Jiang M, He Y et al (2018) Long non-coding RNA TPT1-AS1 promotes cell growth and metastasis in cervical cancer via acting AS a sponge for miR-324-5p. J Exp Clin Cancer Res 37:169
Lennerz JK, Kwak EL, Ackerman A, Michael M, Fox SB, Bergethon K et al (2011) MET amplification identifies a small and aggressive subgroup of esophagogastric adenocarcinoma with evidence of responsiveness to crizotinib. J Clin Oncol 29:4803–4810
Lin H, Zhou AJ, Zhang JY, Liu SF, Gu JX (2018) MiR-324-5p reduces viability and induces apoptosis in gastric cancer cells through modulating TSPAN8. J Pharm Pharmacol 70:1513–1520
Liu H, Wu Y, Wang S, Jiang J, Zhang C, Jiang Y et al (2019) Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer 19:937
Liu F, Wang YL, Wei JM, Huang ZD (2020) Upregulation of circ_0000142 promotes multiple myeloma progression by adsorbing miR-610 and upregulating AKT3 expression. J Biochem 169:327–336
Michalopoulos G (1993) HGF and liver regeneration. Gastroenterol Jpn 28(Suppl 4):36–39 (Discussion 53-6)
Panda AC (2018) Circular RNAs act as miRNA sponges. Adv Exp Med Biol 1087:67–79
Pawlyn C, Morgan GJ (2017) Evolutionary biology of high-risk multiple myeloma. Nat Rev Cancer 17:543–556
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z (2019) Silencing of circ_0009910 inhibits acute myeloid leukemia cell growth through increasing miR-20a-5p. Blood Cells Mol Dis 75:41–47
Russell WE, McGowan JA, Bucher NL (1984) Partial characterization of a hepatocyte growth factor from rat platelets. J Cell Physiol 119:183–192
Seidel C, Børset M, Turesson I, Abildgaard N, Sundan A, Waage A (1998) Elevated serum concentrations of hepatocyte growth factor in patients with multiple myeloma. The nordic myeloma study group. Blood 91:806–812
Terpos E, Ntanasis-Stathopoulos I, Gavriatopoulou M, Dimopoulos MA (2018) Pathogenesis of bone disease in multiple myeloma: from bench to bedside. Blood Cancer J 8:7
Wu L, Xia J, Yang J, Shi Y, Xia H, Xiang X et al (2018) Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J Buon 23:1343–1349
Xu H, Zhang Y, Qi L, Ding L, Jiang H, Yu H (2018) NFIX circular RNA promotes glioma progression by regulating miR-34a-5p via notch signaling pathway. Front Mol Neurosci 11:225
Yang Y, Chen L (2019) Downregulation of lncRNA UCA1 facilitates apoptosis and reduces proliferation in multiple myeloma via regulation of the miR-1271-5p/HGF axis. J Chin Med Assoc 82:699–709
Yin Y, Long J, He Q, Li Y, Liao Y, He P et al (2019) Emerging roles of circRNA in formation and progression of cancer. J Cancer 10:5015–5021
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T et al (2018) CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 9:417
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z et al (2017) CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (albany NY) 9:1585–1594
Zhang L, Liu D, Tang B, Xu A, Huang H, Su Y et al (2018) MicroRNA-324-5p suppresses the migration and invasion of MM cells by inhibiting the SCF(β-TrCP) E3 ligase. Oncol Lett 16:5331–5338
Zhao W, Dong M, Pan J, Wang Y, Zhou J, Ma J et al (2019) Circular RNAs: a novel target among non-coding RNAs with potential roles in malignant tumors (Review). Mol Med Rep 20:3463–3474
Zhou F, Wang D, Wei W, Chen H, Shi H, Zhou N et al (2020) Comprehensive profiling of circular RNA expressions reveals potential diagnostic and prognostic biomarkers in multiple myeloma. BMC Cancer 20:40
Acknowledgements
Not applicable.
Funding
No funding was received.
Author information
Authors and Affiliations
Contributions
Conceptualization and methodology were performed by FW and YW, formal analysis and data curation were performed by YW, XD, and WW, validation and investigation were performed by BW and FW, writing—original draft preparation and writing—review and editing were performed by BW, FW, and YW, and approval of final manuscript was performed by all authors.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical Approval
The present study was approved by the ethical review committee of ShangRao People’s Hospital.
Consent to Participate
Written informed consent was obtained from all enrolled patients.
Consent for Publication
Patients agree to participate in this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wu, B., Wang, F., Wang, Y. et al. CircATIC Contributes to Multiple Myeloma Progression via miR-324-5p-Dependent Regulation of HGF. Biochem Genet 60, 2515–2532 (2022). https://doi.org/10.1007/s10528-022-10228-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-022-10228-1