Abstract
Genetic instability is invoked in explaining the cell phenotype changes that take place during cancer progression. However, the coexistence of a vast diversity of distinct clones, most prominently visible in the form of non-clonal chromosomal aberrations, suggests that Darwinian selection of mutant cells is not operating at maximal efficacy. Conversely, non-genetic instability of cancer cells must also be considered. Such mutation-independent instability of cell states is most prosaically manifest in the phenotypic heterogeneity within clonal cell populations or in the reversible switching between immature “cancer stem cell-like” and more differentiated states. How are genetic and non-genetic instability related to each other? Here, we review basic theoretical foundations and offer a dynamical systems perspective in which cancer is the inevitable pathological manifestation of modes of malfunction that are immanent to the complex gene regulatory network of the genome. We explain in an accessible, qualitative, and permissively simplified manner the mathematical basis for the “epigenetic landscape” and how the latter relates to the better known “fitness landscape.” We show that these two classical metaphors have a formal basis. By combining these two landscape concepts, we unite development and somatic evolution as the drivers of the relentless increase in malignancy. Herein, the cancer cells are pushed toward cancer attractors in the evolutionarily unused regions of the epigenetic landscape that encode more and more “dedifferentiated” states as a consequence of both genetic (mutagenic) and non-genetic (regulatory) perturbations—including therapy. This would explain why for the cancer cell, the principle of “What does not kill me makes me stronger” is as much a driving force in tumor progression and development of drug resistance as the simple principle of “survival of the fittest.”
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
“Instability” of the genome in tumor cells has long been associated with cancer progression—both as a cause and a consequence [1]—and provides the basis for the idea that tumor progression is driven by the somatic evolution of cells that follows the Darwinian scheme of mutation and selection [2–5]. Genetic instability is now joined by non-genetic instability in view of the growing acceptance of the “plasticity” [6] of the cancer cell, as most lucidly epitomized by the rapid and reversible switching between the “cancer stem cell” (CSC) state and the more mature states of cancer cells within a tumor [7–12]. Such dynamical transitions between phenotypic states have more in common with developmental changes driven by regulatory pathways than with evolutionary changes driven by genetic mutations [13]. Neoplastic cells often fail to adhere to the cellular phenotype [14] to which cells are committed through the normal paths of differentiation and are prone to transdifferentiate [15]—supporting Virchows’s old idea that cancer is a disease of development [16].
Thus, both genetic and non-genetic instabilities obviously contribute to the phenotype instability and versatility of cancer cells. Yet, many cancer researchers fail to explicitly make this distinction—mostly because of a tacit denial of non-genetic cell state dynamics and the comfortable explanation for phenotypic changes afforded by the concept of genetic mutations. But when considering short timescales of hours or days in which the mutations barely spread in a cell population, it is clear that non-genetic instability dominates in accounting for the ubiquitous heterogeneity of cancer cell populations. Non-genetic heterogeneity is at full display in the rapid, spontaneous phenotypic diversification of a clonal (isogenic) cell population within a homogenous environment and has recently attracted much attention. If genetic instability is readily acknowledged because of the tangible nature of DNA mutations and chromosome aberrations, non-genetic instability is more evasive. What is the basis for non-genetic instability? How are these two connected? What is “instability” in the first place?
Often, we take for granted the meaning of a technical term, using it without considering that it evokes a different imagery and conceptualization than intended when it arrives in the mind of our conversation partner. “Instability” means different things to a psychiatrist than to an engineer. The meaning of a word depends on the context of personal experience, worldview, and mental occupation of the recipient [17]. Hence, the intended meaning is often “lost in translation” even in scientific communication within the same language (English) because in the increasingly multidisciplinary, fragmented life science research, we actually often speak different dialects that separate even a cell biologist from a geneticist. Terms, such as “instability,” “epigenetic,” or “network” [18], with which we are concerned here, are among those that have most suffered from semantic mistreatment and, hence, have caused miscommunication and misunderstanding. The lack of a common precise definition of concepts behind these terms has, unbeknownst to many cancer biologists, hampered cancer research. The problem is magnified as new discoveries are communicated using these terms.
The concept of “instability,” both of the “genetic” but, mostly, “non-genetic” type, must take center stage if we are to frame the problem of tumor progression in a formal way. Thus, the goals of this article are to help clarify and delineate the “true” meaning of “instability” and its relationship to gene regulatory networks and to epigenetics and to explain the underlying fundamental principles of dynamical systems as applied to integrated gene regulation and cell phenotype control. This will hopefully expose to all practitioners of cancer biology the importance of non-genetic instability in the evasiveness of tumors which accounts for therapy failure. We will end by offering a formal and molecular explanation for the plausible, widely suspected possibility that has been too outlandish to articulate: namely, that cancer therapy itself may in fact render a portion of cancer cells more malignant—at a rate that is much faster than can be explained by selection for the more resilient and malignant mutant cells in a Darwinian manner.
To achieve this, we will start with discussing the distinction between genetic and non-genetic instability, both of which are often, if explicitly differentiated at all, treated separately. But even before we can talk about instability, we need to first establish a technically precise notion of “stability.” We will thus offer a unifying conceptual framework that is rooted in the first principles of systems dynamics—applied to gene regulatory networks. The theory will be presented in a qualitative and accessible yet conceptually accurate manner. This will involve an excursion in quite general and abstract matters, but without use of equations, and lead to an integration of the metaphors of the ”fitness landscape,” used in evolutionary biology, and the “epigenetic landscape,” used in cell and developmental biology. In the second part, we will apply the general concepts to tumor progression which involves these two types of change: evolution and development. We hope that the cancer biologist who espouses specific, tangible, proximate explanations in terms of molecular mechanisms but eschews the abstract, generic, integrative concepts will be willing to endure the theory. In doing so, she will be rewarded with a novel type of reasoning that allows her to explain without hand waving, why it is only natural that therapeutic interventions in principle must make those cancer cells that escape the cytotoxic attack more malignant.
2 Phenomenology: genetic and non-genetic instability
Before we enter the more profound deliberations on (in)stability and its formal foundations, let us quickly review what is generally meant when cancer biologists talk about “genetic instability” and its less well-known complement, “non-genetic instability” of tumor cells.
2.1 Genetic instability of cancer cells
The term “genetic instability” (or, more concretely, genomic instability when emphasizing its material substrate) is used to refer to the simple observation that cancer cells are defective in the fidelity of replicating DNA and its proper partitioning to the daughter cells during cell division—be it at the level of single nucleotides (leading to point mutations) to entire chromosomes (leading to chromosome aberrations and karyotype anomalies) [1, 19, 20]. This is due to defects in DNA mismatch repair, in various check points of the cell cycle, in the proper function of mitotic spindle, etc., and the impaired ability to send cells with DNA and chromosomal damage into apoptosis due to defects in the p53 pathway [20]. As we will see, genome instability is both a consequence and a cause of tumor progression.
The immediate consequence of an instable genome is the “mutator” phenotype—a cell with dramatically increased mutation rate [21, 22]. Whether the mutator phenotype is necessary for tumorigenesis and progression or not has been the subject of an intense and interesting debate in the past two decades and will not be discussed here, for such discussion was framed by the narrow concept of Darwinian (somatic) evolution of cancer (selection of random mutant cells). We will later argue that this scheme of evolution plays a limited role [13].
The concept of genetic instability is particularly attractive because the consequences of a mutation can be understood in molecular terms, namely, in the following two ways: (1) a mutation can simply affect an effector gene, for instance, inactivate a cell cycle suppressor gene or an apoptosis gene (e.g., p16INK4 or Bcl2) or render the activity of a protein of a mitogenic pathway constitutive (e.g., point mutation in ras, amplification of EGFR); (2) the mutation may have less obvious effects when it affects components of the regulatory core, such as signaling or transcription factors, which often can possess opposite effects on cell growth, as is the case with myc, ras, etc., such that depending on the cellular context, they can promote apoptosis, differentiation, or senescence or enhance cell proliferation (see [23] for a review). The immediate effect in promoting cancer progression is often not readily explained in this second group. But with the formalism of gene regulatory networks (GRN) and attractors, we can make the general statement that mutations of this second type that affect regulators but not effectors essentially rewire the GRN. The rewiring of the GRN topology will be of central importance below when we introduce formal arguments on how a rewired network distorts the epigenetic landscape and thereby alter cell state dynamics.
2.2 Non-genetic instability
Instability of the phenotype that does not depend on mutations, that is, the drift of phenotype without alteration of the genotype, is what we refer to as “non-genetic.” We introduce this term here to describe the dramatic versatility of the tumor cell phenotype that is manifest in the ubiquitous heterogeneity of tumor cell populations in vivo as well as in the cell culture where it is inevitable even in clonal (isogenic) populations of cells [24, 25]. Hence, a large proportion of the apparent phenotypic instability is non-genetic in its origin. Non-genetic heterogeneity will be at the center of our discussion. The observation that cancer cells can dynamically switch their phenotype, as most prominently epitomized in the reversible transition between the cancer stem cell state and the more differentiated state [7, 8, 11, 12, 26] or during epithelial–mesenchymal transition (EMT) [27, 28] and its inverse (MET), has also led to the concept of “plasticity”—notably in conjunction with stem cells [29–33]. “Plasticity” deserves brief discussion because it is again an ad hoc label adopted more in a metaphorical manner without deeper reflection. Below, we will delineate the difference between plasticity and instability in terms of principles of dynamical systems.
In this phenomenological introduction, it suffices to raise the following point: if the tendency of cancer cells to drift away from some “ideal phenotype” defined by the lineage a cell has committed to [14] is non-genetic, i.e., not driven by mutations, this will by necessity require that one genotype (one stable invariant genome) does not map uniquely into one phenotype but can produce many distinct stable phenotypes. This one-to-many mapping is of course a fundamental property of metazoan since each of the diverse cell types in the body is produced by (essentially) the same genome. The ability of the regulatory control structures of a system to produce more than one stable system state is called multi-stability. This central concept will be discussed later. While consciously, that is, when explicitly confronted, no biologist would admit that she entertains the one-genotype-one phenotype view, in realty, the paradigm of such a 1:1 mapping subliminally still reigns in our minds and commands much of our thinking about cancer. In cancer research, this has prevented non-genetic instability from being placed on equal footing as genomic instability—whose material embodiment in DNA mutations makes it easy to comprehend.
Nowhere is the dominance of the mindset of a 1:1 mapping between genotype and phenotype as evident as in the unquestioned search for THE genetic mutation to explain every newly observed malignant phenotype in tumorigenesis: the acquisition of a single malignant trait by a neoplastic cell, such as its invasive capacity or autonomy of proliferation, is by default explained by a mutation [34, 35]. And nowhere is the myopia of such a view as evident as in the surprise evoked by the failure of cancer genome sequencing projects to identify universal, readily explained “driver mutations.”
To be able to maintain the 1:1 genotype–phenotype mapping principle when explaining tumor progression, one obviously also has to suppress the notion that the very same genome normally produces an unfathomable yet taken-for-granted diversity of stable, discrete cellular phenotypes: the variety of cell types in the metazoan body, as diverse as liver cells, neurons, muscle fibers, etc. Non-genetic instability is required to allow the genetic program in our genome to unfold and diversify and robustly commit the undifferentiated zygote to produce the thousands of distinct, stable, and self-reproducing cell types that behave akin to species in a balanced ecosystem that is our body. Who would still explain the transformation of an immature multipotent stem cell in higher mammals into one of the highly specialized post-mitotic cell with genetic mutations? Can such mutation-independent “plasticity” of cell phenotype be harnessed by the cancer cell? Are all cancer cell behaviors, such as migration, stimulation of angiogenesis, and other features that one likes to call “hallmarks” [36], really that abnormal and specific to cancer? Or are they rather déjà vu programs of development, reactivated in a new context? How does the non-genetic instability that drives normal development accelerate the acquisition of these hallmarks?
3 Dynamical systems formalism: stability vs. instability
3.1 Natural notion of stability
Before we enter the formal discourse on instability, it is obviously essential to first define “stability” (see Fig. 1a). A state of a system that is stable is more than just stationary or in a steady state. Simply not changing as time goes by (time-invariant) does not qualify as “stable” because stability implies more than stationarity: a stable system is resilient to perturbations in the sense that after suffering an external influence that changes its state, the system returns to its original state. Concretely, let us consider an air-conditioned room designed to offer “stable” temperature to its occupants, which is the set-point temperature T* that we dial into the thermostat. After an externally imposed change of the room temperature (“perturbation”), such as opening a window or warming of the weather, the temperature in the room will stay at the set point after a brief transient departure from it due to the perturbation (Fig. 1b, top). The faster the return back to the set point after the perturbation, the more “stable” is a system. In biology, the body temperature, or, more precisely, the state of a homeotherm organism with respect to its temperature, is stable: it also has a “set point”, namely, T* = 37 °C for humans, which is kept at constant level and resists external influence—to some extent. A cell type is also a system that can be assigned a stability—with respect to gene expression: in order to maintain its type identity, the expression level x i of each gene i in the genome, as can now be approximately determined by measuring the transcriptome, also has to be stable to perturbations, such as biochemical fluctuations of the cellular environment and molecular noise in gene expression machinery that alter expression of genes—the original cell type identity defining gene expression profile will be reestablished after perturbations. In brief, stable systems are not sensitive to changes in environments—up to some limits. Thus, the stability of a system in this formal definition is what biologists associate with resilience, homeostasis, tolerance, or memory of its original state.
3.2 Basic concepts from dynamical systems theory
To formalize, and thereby facilitate, our discussions on stability, we now introduce the following concepts: a “state,” a “state variable,” and the “state space.” Talking about stability in the above sense, the “thing” that can lose it and become unstable is the state S of a system. The state is the entity to which we attach the attributes “stable” or “unstable” (and not the system itself—which exhibits a different type of stability to be discussed below). A state S and its stability are defined with respect to a state variable: x, such as the temperature in our examples above, or the expression level of one or a set of genes in a cell. For instance S(x = 37 °C) is the set point and is a stable state S of the system body, whereas a state at T = 38 °C is unstable because homeostasis (body temperature control) will return it to 37 °C (see discussion of fever below).
In contrast to this one-dimensional state variable x = temperature T, for the cell phenotype, a state S can be defined (crudely, for the sake of argumentation) by the expression levels of all the genes; hence, we have a high-dimensional state: S = [x 1, x 2,… x n ] that is defined by a set of variables, x i , which, in this simplified definition of a cell phenotypic state, is the cellular abundance of a protein i or the expression level of gene i (Fig. 2).
From gene regulatory network structure to epigenetic landscape. Overview of the elementary dynamical systems concepts applied on gene regulatory network (GRN) dynamics. The network of regulatory interactions between the N genes (a) determines the permissive gene expression profiles (b), which constitute a network state S and, in a simplified view, a cell phenotype. The state S maps into a point (green ball) in the N-dimensional state space (c)—spanned by the N axis (blue), each representing the expression level of a gene of the network. The N-dimensions are schematically depicted as a three-dimensional cube. Change of gene expression profile manifest as a movement (stroboscopic series of green balls) of S along the trajectory (green curve). Depicted are two trajectories that converge onto an attractor state. Hypothetical projection (dimension reduction) of the N-dimensions into two dimensions (d) or one dimension (e) permits the display of the axis that represent the quasi-potential U(S) for each state S as an “elevation.” A population of cells, each in slightly different “micro-states,” can be represented as a cloud of points in state space (not shown) or a “pile” of green balls in an attractor
All possible states of a system, that is, in the case of body temperature, the continuum of all the temperature values, constitute the “state space” of the system. It can be represented by a one-dimensional axis—from low (e.g., 0 °C) to high (e.g., 100 °C) temperature. A given state is a point in the state space (on this axis) and a change in temperature is a displacement of that state. In the case of the cell phenotype, we have a higher dimensional state space. If a cell contains only two genes, then their expression levels (activity of their encoded proteins), represented by x 1 and x 2, jointly define the cellular state S = [x 1, x 2] (Fig. 3). The state space will then be a two-dimensional plane in which a given cellular state is a point. It contains the entire spectrum, that is, all possible combinations of values of x 1 and x 2—all theoretically possible states that the two-gene cell can occupy. Change in a cell phenotype involves alterations in x 1 and x 2, thus, is a walk in this x 1–x 2 plane.
Concept of the quasi-potential for a two-gene system. Axes representing the expression levels of the two genes, x 1 and x 2, and spanning the planar state space. The third vertical dimension can be used to display the value of the quasi-potential U for each state S, giving rise to a landscape. Example state S 1 is shown, with its trajectory (green arrow) representing its future state, the spontaneous change of the state S toward an attractor state (local minimum on the landscape) driven by the stability-seeking behavior that originates in the gene regulatory network, here the interaction between x 1 and x 2
The concept of state space is of eminent importance for understanding the rest of this piece. It converts the abstract notion of a change of state (phenotype) into the more tangible image of a motion: a change of state is a shift in position in state space, a motion along a trajectory (Figs. 2c and 3). This is particularly useful for higher dimensional states, such as cell states that are determined by a gene expression profile of thousands of genes (for a more detailed introduction, see [37, 38]). In this state space description, stability then is manifest as the spontaneous movement of unstable states toward the stable state S*(x = 37 °C) in state space, e.g., the states S(x = 36.0 °C) and S(x = 38.5 °C) will, in healthy individuals, eventually move toward the stable set point state S*(x = 37°C): the stable set point “attracts” neighboring states.
3.3 Attractors
For our purposes, here, we can say that a state is stable (with respect to the variables x of interest) if (the relevant region of) the state space spanned by these variables x contains an attractor state S*. The attractor state is a stable steady state and its “position” in state space is a characteristic of a system (Figs. 1 and Fig. 2d, e): it is the “set point” to which the system returns after perturbation. An attractor state “attracts” systems in states that depart from it through “forces” that are inherent to the system: If the room is below the set-point temperature, the heater kicks in; if the room temperature is above the set point, the air conditioning is turned on—both ensure the movement of S in state space toward the attractor in response to external perturbations. This behavior is a property of the system and is encoded in its internal wiring: in the case of a thermostat system, the wiring connects the sensor and feeds room temperature information to a microprocessor that operates the cooling and heater engines according to the deviation from the attractor state (set point). The key characteristics that ensure stable temperature in this wiring structure are a negative feedback loop.
3.4 Cell types as attractors
The maintenance of a gene expression profile to ensure homeostatic stability of a cell phenotype is very similar to the example of maintaining system temperature, as discussed above, although it constitutes a higher dimensional system because the attractor state is defined not by one variable but a set of variables: the level of expression of all the genes that define a gene expression profile that establishes the characteristic of a particular phenotypic state (Fig. 2b). This self-stabilizing gene expression profile is encoded by an attractor state of the gene regulatory network, S* = [x 1, x 2,… x N ]. This particular set of expression levels x i obeys the intrinsic network constraints that dictate which genome-wide configuration of gene expression is allowed and stable. If for instance gene x 1 is a suppressor of x 2, then any state with the configuration in which the expression level x 1 is very high and x 2 is also very high would be “forbidden” by the network’s regulatory rules and would be very unlikely, i.e., “unstable”; states with such a configuration would not be able to hold their position in state space, but instead move away from it toward a stable attractor state. Thus, attractor states ensure homeostatic stability of the gene expression profiles.
Gene regulatory circuits of the genome-wide GRN (Fig. 2a) are replete with feedback loops that produce stable attractor states that the network (the cell) has to occupy. As we will see below, when we introduce instability, a network can produce multiple attractor states. Then, each distinct phenotypic state of cell, such as a cell type, which is determined by a specific gene expression configuration across the genome that must be stable, is naturally an attractor state of the GRN (for an overview, see [38, 39]). This central thesis is further discussed below in conjunction with “multi-stability” and will play an important role in understanding non-genetic (in)stability in cancer.
3.5 Regions in state space
The state space idea allows us to define regions in that space, such as the domain or basin of attraction. For instance, in living organisms, reducing the temperature will trigger thermogenic activity to bring the body temperature back to the set point. As long as such a response to the perturbation occurs, the system is in the basin of attraction of the attractor state S* = 37 °C. But at a critical threshold temperature, T crit, the system collapses: the organism enters in system failure and the body temperature will further decay until the organism dies and its temperature equilibrate with the external temperature, which represents the death of the organism. T crit is a critical point in the continuous state space; in this example case, T crit marks the border of the basin of attraction of the attractor state. Thus, the domain of attraction is limited by precisely defined boundaries around the attractor state.
Within these boundaries of the basin of attraction, all states will eventually “end up” in the attractor. “Gene expression noise,” the random fluctuations in the abundance level of proteins in the cell [40–42], translates into a wiggling movement, akin to a random walk, around the attractor state if the fluctuations are not large enough to cross the boundary of the basin of attraction. For a population of nominally identical cells (Fig. 2e), every cell will be in the same attractor, but a snapshot of the population will appear as a cluster of points (“cloud”) in state space, around the attractor state [43].
3.6 The quasi-potential landscape of a state space
The concept of a potential of states is central for understanding why landscape metaphors are more than just metaphors (Fig. 3). Thus, we now extend the state space representation by another dimension to even more intuitively and tangibly display the dynamical properties of a system with respect to stability and instability and the “drive” of a system toward the stable states and away from unstable states: let us assume for the sake of visual imagination, again a two-dimensional state space (Fig 3). We can now assign to each state S(x 1, x 2) a quasi-potential value, U(S) = U(x 1, x 2) that reflects the “relative instability” of that state S. Since the state S will be attracted to the most stable state when seeking stability, S moves in state space in the direction that decreases U most until it ends at the lowest value of U, which is found at the attractor state S*. Plotted as “elevation” for each point S in the x 1–x 2 plane, that is, as the third dimension in our two-dimensional state space, a landscape will emerge: a topography over the x 1–x 2 state space, in which the elevation represents the quasi-potential U(S) for each theoretically possible state S(x 1, x 2). This third dimension and the movements down the slopes of the landscape capture the intuition of spontaneous movement in state space toward the most stable state that is accessible from a given point.
This picture is of course reminiscent of a potential energy landscape in classical physics. But we need to insert a big caveat here: complex systems such as gene regulatory networks, or wired control systems, are “non-conservative systems” far away from the thermodynamic equilibrium, and hence, the potential landscape picture is to be taken with caution. In other words, the slopes of the landscape do not represent “true” gradients as in classical physical systems (hence the prefix “quasi”). For instance, the gene expression profile can actually “circle” in a flat region of the landscape. For more formal, still accessible explanation of the details of the computation of the quasi-potential landscape for non-equilibrium system and associated problems, see [18, 44].
With a landscape picture, the “urge” toward the attractor state can now be imagined as that of a ball that represents a state S through its position projected onto the state space—rolling down the valleys toward the lowest energy point—the attractor state.
4 Integrating stability and instability
With the above minimal formal tools, we can now discuss more interesting aspects of stability and instability that have deep roots in the theory of dynamical systems. Below, we define instability more formally, point to important distinctions to be made between related phenomena, and examine the relationships between stability and instability, but we will focus on the practical consequences.
4.1 What is instability?
We finally can define “instability” as the specific opposite stability as defined in the dynamical system formalism discussed above. A stationary or steady state can either be stable, as we noted above, or it can be instable (Fig. 1). Note that a system that is not in an attractor state but on the way to it is just “not stable”; it is a transient and not an unstable stationary state in the strict sense because instability is a property of a stationary state: in the absence of perturbations, an unstable steady state S* does not change; however, the tiniest perturbation will cause a system in such an unstable steady state to move away from the unstable stationary state.
Thus, in the quasi-landscape picture, whereas a stable steady state is represented by the bottom of a valley, the unstable state is at a hill top (Fig. 1, bottom). It is also locally flat and does not experience any intrinsic forces that would displace it. However, when an unstable state S* moves in any direction away from the point exactly on the summit, it will inevitably encounter a downhill slope; thus, that state will move away from the unstable steady state to the next accessible attractor state.
Back, briefly, to our temperature control system: the critical lower threshold temperature T crit for body heat generation represents an unstable state: if the system temperature has been lowered by a perturbation to a temperature still higher than T crit, the body will generate heat to push it back to the attractor temperature T* = 37 °C: homeostasis works. But if the system is slightly below T crit, the organism is too cold for the thermostat to work. The control system collapses and further cools down, as described above. At exactly T crit, the organism displays instability: slight fluctuations onto either side, T > T crit or T < T crit, can decide its fate. In dynamical systems terminology, the system state at T crit is unstable and sensitive to initial conditions (the point in state space at time zero of observation, here the temperature)—it is the “tipping point” or “hilltop” or “watershed” on the quasi-potential landscape at which the states space trajectories diverge.
Stability and instability, thus, are profoundly related in a way that is graphically captured by the state space formalism: both stable and instable states are stationary—they are not on slopes of the quasi-potential landscape but at the bottom of valleys or on hilltops. Stable attractor states (valleys) are separated by instable stats (hilltops). Attractors in multistable systems are disjoint but adjacent—separated by unstable states. This will become clear when we discuss the epigenetic landscape.
4.2 Beware of plasticity
In view of the versatile and dynamical phenotype of stem and tumor cells, the term “plasticity” is often used to describe their behavior [6, 29–32]. In the dynamical systems terminology, “plasticity,” however, if used at all, refers to a specific behavior, a weaker kind of stability than what is captured by the concept of an attractor state (“Lyapunov stability”): a system may, after a transient perturbation, not return to the original state (as in the case of an stable attractor), but also not escape from it (as in the case of instability), but instead it “tracks” the environmental change. There is neither restoring nor repelling force. The system stays where the perturbation leaves it, keeping the constellation that external factors impart on it—akin to the deformation of a plastic material where there is no recoil or elasticity. The system has no memory of the original state—only of the perturbed state. For instance, if the external temperature suddenly increases by 10 °C, the room would “adapt” and its temperature rises by 10 °C. In the picture of the quasi-potential landscape, plasticity thus is represented by regions of flat panes where the ball can be placed to any position in the state space and stays there because it experiences no intrinsic force (Fig. 1).
As we will see, “genome instability” is, strictly speaking, according to the above definitions, a misnomer—the mutating genome exhibits plasticity rather than instability (in the genotype space).
4.3 Multistability
We now discuss more explicitly the concept of multi-stability of dynamical systems theory that is central to its application to biology. Given that stability and instability map into valleys and hills, respectively, and that a quasi-potential landscape can contain multiple valleys, a system can thus contain multiple attractor states (Figs. 2 and 3). The coexistence of multiple alternative stable attractor states within a single system is called multi-stability and was first proposed as a principle that affords a single biochemical network or gene regulatory network the capacity to produce two distinct cellular states (bistability) by Max Delbruck in 1949 and Monod and Jacob in 1961 (for an overview and reference, see [38, 39]). This was the first formal concept to explain the phenomenon of cell differentiation. For details on the mathematical basis for how certain interactions produce bistability, see, e.g. [45, 46]. With multiple attractors, the question immediately arises as to which state a system—which of course can be only in one state at a time—will chose to be in.
The state space and its associated quasi-potential landscape U(S) represent the entire phenotypic repertoire of one system, determined by its internal structure—the wiring of sensors, processors, and actuators in the thermostat control and the “wiring” diagram of the gene–gene regulatory interactions in the genome. Again, since a system (e.g., a cell) can only be at one place on the landscape at a time, the ball that represents its state on the landscape can only be in one valley at a time. Where on the landscape (relative to the “watersheds”) it is placed initially (its initial state) will determine in which attractor state it will “end up.” The set of attractors thus represent alternative choices of system states for a given system and under given conditions. The “relative stability” of each attractor state S* is, crudely, manifest in the relative size and depth of an attractor basin (Fig. 3)—or valley of the quasi-potential landscape (other details also matter) [44]: it is intuitively plausible that the deeper and bigger the valley of a given attractor state S*, the larger perturbations (=displacement of the system state from the attractor state) can be tolerated without leaving the valley and the more likely is it for a system to end up in that valley after a random, massive displacement of the system state across attractors.
4.4 Attractor transitions
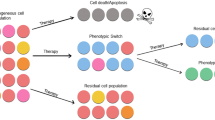
The existence of multiple alternative attractor states within one system now offers the possibility of choice between attractors. If attractors correspond to a cell phenotype, so is a state transition from one attractor to another a phenotype switch, for instance, between the stem cell state and the differentiated mature state or between the proliferative state, a quiescent phenotype, or an apoptotic state.
Regulatory signals, such as hormones or cytokines, that switch cells between such phenotypic states do so by altering the expression status of a set of genes. Since attractors represent the biologically realized system states, such transition between phenotypic states translates into the displacement of the system state S from one attractor to another. The need to cross boundaries of attractor basins, i.e., to jump from one valley to another, is manifest in the switch-like (quasi-discrete) nature of cell fate changes.
It is important to realize that because of the presence of attractors, even an “unspecific” perturbation will be able to trigger a switch-like transition into a specific attractor state. This is in line with our everyday experience: numerous nonspecific agents, such as DMSO, alcohols, sodium butyrate, pH, etc., can trigger the differentiation of cells in culture (references in [47]). The profound idea, which now can be comprehended in an intuitive way using our formal framework of state space, can be put as follows: cells always “have to go somewhere” when they are pushed around on the quasi-potential landscape. If they are forced to exit their current stable attractor state (for reasons explained below), they will find their way to the next accessible stable state—whatever gene expression profile it encodes. We will reencounter this phenomenon later when we discuss the response of cancer cell to “nonspecific” cytotoxic agents.
The multi-attractor quasi-potential landscape thus captures in an intuitively plausible yet formally accurate fashion how a cell’s gene regulatory network maps into its phenotypic behaviors in terms of phenotypic states, such as cell types, and their dynamical relationships. But most importantly, it epitomizes the inevitable fact, so fundamental as it is taken for granted, that one single genome, because of the principle of multi-stability in networks of interactions, produces a large set of discretely distinct, stable phenotypes. This is the reason for the necessary departure from a 1:1 mapping between genotypes and phenotypes discussed above.
4.5 Structural stability and the parameter space
We now introduce a more abstract type of change in dynamical systems such as the GRN (see Fig. 4). While a change of the cell phenotype is represented by a change of the system state S, i.e., a movement of S on the epigenetic landscape E 1 that is generated by genome G 1, we now consider a change of the landscape topography itself and thereby introduce an important concept. For this purpose, we need for a moment to separate two timescales. One timescale is the one in which the system state S moves in state space (or on the landscape) toward the attractors. This applies to cells reaching their destination attractor states as they differentiate into cell types, thus pertaining to the timescale of development. In contrast, the second timescale is much slower. It is the one in which the structure of the epigenetic landscape changes (Fig. 4a): a hill or valley may flatten and disappear, which will affect the course of the developmental trajectories. Such changes of the landscape topography per se takes place in the timescale of evolution because, since a genome G 1 maps uniquely into a landscape E 1 (Figs. 5 and 7), a change in landscape topography will require a mutation that changes the “wiring” diagram of the underlying genomic network G i which mathematically determines the landscape E i . For instance, the mutation that affects how a gene A and gene B interact changes a parameter that characterizes that interaction and is part of the network specification (Fig. 5c). This mutation alters the GRN/genome G i and may deepen or flatten a certain basin of attraction in its epigenetic landscape. In dynamical systems theory parlance, such changes are implemented by changes in the values of the control parameters.
Concept of parameter space, bifurcation, and structural stability. a Schematic representation of a system that exhibits a transition from a monostable to a bistable behavior: its epigenetic landscape (blue) changes from having one central valley (on the back) to two valleys (front) as a parameter k is gradually changed. Note the conversion of the central stable steady state S* into an instable steady state S′ and the birth of two new stable states. b Formal representation (“bifurcation diagram”) showing that the system undergoes a “pitch fork” bifurcation. The lines (“branches”) indicate the position of steady states in state space with respect to the state variable x (blue, vertical axis) for every parameter value k (red, horizontal axis) as well as their quality (stable or not stable). Regimes in which the monostable or the bistable behavior of the system is “structurally stable” are indicated by an asterisk and two asterisks, respectively. They are separated by the bifurcation point at a specific value of k
Relationship between the fitness landscape and the epigenetic landscape. The epigenetic landscape (blue) (a) represents the behavioral repertoire of one single genome or the gene regulatory network it encodes, in this case genome 1 (G1 in the text), which has epigenetic landscape 1 (E1). A genome or network in turn is one point in the fitness landscape (red) (b). In the epigenetic landscape, one point (green ball) is one network state = cell phenotype, and movement (change of state S) represents regulation. In the fitness landscape, one point (blue ball) is one genome = network. A movement (change of genome G) represents a mutation, here exemplified by a non-sense point mutation in gene A in genome 1 (c), which rewires the network (to genome 2). Note that in the epigenetic landscape, the states seek the lowest points available (valleys), whereas in the fitness landscape the genomes seek the highest point available (peaks), as indicated by the small arrows
Parameters can be used in models to represent the presence and the strength of interactions in the network. For each set of parameters, that is, for each given network structure, its associated dynamics can be simulated on a computer and the landscape recomputed. Analogous to the state space that contains all possible states S defined by the values of the state variables x i , here, the space that contains the entire range for the values of (a set of) parameter(s), and hence all network structures defined by the combinations of variations of these parameters, is the parameter space. Two distinct GRNs (which are distinctly wired), G a and G b, are thus two points in parameter space, much as two states S 1 and S 2 of the network/genome G a are two points in its state space or on its landscape E a. Thus, the parameter space, or the space of network structures/genomes, represents a hierarchically more encompassing category.
At a more abstract level (Fig. 4a), a movement along one axis in parameter space represents the change of the parameter value k, which, for instance, may specify the interaction strength for how gene A inhibits gene B (which is encoded in the genome). Such a shift in parameter space moves from one network/genome to another and changes the associated epigenetic landscape, which in turn may exhibit a shift in the position of an attractor state, e.g., the shifting of the actual set point to new values: in the case of fever, the set point of 37 °C is shifted to, e.g., 38 °C (Fig. 1b; see also more below).
In contrast to such continuous change of the position of attractors and their boundaries, one observes sudden, discontinuous events when traveling in parameter space: at some critical values for the parameter points, an attractor can disappear and be converted into a hilltop (Fig. 4). This is equivalent to flipping a stable steady state S* into an instable steady state, S′ (Fig. 4b). Such points in parameter space at which a sudden, qualitative change of the landscape occurs are called bifurcations. An attractor state (or any structure of the landscape of the state space) that persists in wide regions of the parameter space without encountering bifurcations is called structurally stable. (Do not confound bifurcation points in parameter space with the boundaries of attractors in state space (hilltops)—although both represent points of discontinuity).
Taken together, we need to make the following fundamental distinction in terms of “stability”: the return following (short-term) perturbations of a state S in a dynamical system G 1 (a specific GRN) to the attractor state (a cell type-defining gene expression profile) discussed so far is associated with dynamical stability (of that state) and is typically relevant at a timescale of the physiology and development of that system. In contrast, the existence of a particular attractor in a broad region of the parameter space is referred to as structural stability (of a system in comparison to a class of similar systems). Perturbations move a state in state space (the quasi-potential landscape) and test the dynamical stability of a state, whereas mutations move a network/genome in parameter space and thus test structural stability. The latter is lost if mutations change the control parameter such that the system crosses a bifurcation (where an attractor may disappear).
Let us apply this to a concrete example (Fig. 1b). A change in the system’s structure may shift the position of the attractor, which implies that the actual set point will change. For instance, in the case of body temperature control with the state variable “temperature T,” fever is not simply an imposed rise in temperature (perturbation), for were this the case, the body temperature homeostasis would immediately reverse the rise temperature by dissipating more or producing less heat. Instead, fever is caused by a shift in the set point (attractor), e.g., from S* = 37 °C to 38 °C. Suddenly, the state S = 37 °C is not an attractor state any more. It experiences a force of change that causes to shift to a state at higher temperature, the new set point S*2 = 38 °C, in order to minimize the energy on the quasi-potential landscape. The need to heat up is manifest in the fever chills in the early phase of a flu, which reflects the body’s attempt to generate heat through shivering in order to adjust the temperature to the new set point caused by the flu. In contrast, at resolution of the flue, the set point is set back to 37 °C and the body temperature must be cooled down, as manifest by sweating. Thus, fever is not a direct perturbation of the temperature state in state space but a transient shift in parameter space which displaces the attractor state for a while.
5 Two types of landscape—two types of instabilities
Equipped with the conceptual tools presented above, notably the awareness of the dualism of dynamical stability and structural stability, we now illustrate fundamental differences between these two and move on to contrast two widely used landscape metaphors that will be critical to understand the complementary roles of non-genetic and genetic instability in cancer progression: the epigenetic landscape and the fitness landscape (see Fig. 5).
5.1 Waddington’s epigenetic landscape—non-genetic instability
The multi-valley quasi-potential landscape U drawn over the (high-dimensional) space of all theoretical possible gene expression states of a genome and its network is the formal equivalent of Waddington’s famous epigenetic landscape [18] (Fig. 2d). This becomes clear upon analysis of the reason behind Waddington’s use of this elegant metaphor, which evolved over several decades: he introduced the landscape idea in the 1940s to explain the branching differentiation of cells into a variety of preexisting, stable, discretely distinct cell types. Figure 2d shows the best-known 1957 rendition [48]. Since this metaphor is a mathematical construction, it captures the ontological property that stable states (attractors) must be separated from instable states—simply because of the inevitable fact that neighboring valleys must be separated by hills.
The genomic regulatory network of metazoan is a complex network containing interconnected genes (Fig. 2a) in regulatory feedback control loops that produce a complex, rugged landscape in which each valley, each attracting region, corresponds to a cell type, as first proposed in the modern form by Stuart Kauffman in 1969 [38, 49]. Since each point in this N-dimensional state space, where N is ≈1,000–10,000s (roughly the number of regulatory “networked genes” in the genome, mostly transcription factors and signaling proteins and miRNAs), represents a network state, it also represents a gene expression profile across the genome, S = [x 1, x 2… x i ,… x N ] (Fig. 2b). The movement of the cell state S in this landscape directly manifests the constrained change of the gene expression profile: the landscape topography reflects the constraints imposed by the wiring diagram of the GRN and thus dictates in which direction a particular gene expression pattern can move spontaneously and how it responds to perturbations. The gene regulatory interactions in the wiring diagram of the GRN of course are encoded by the genome (since they are determined by the interaction domains of proteins and the sequence motifs of promoter elements, etc.). In brief, the genome encodes the shape of the epigenetic landscape, and the latter is invariant within a lifetime and is unique to an organism. Thus, remember (see Fig. 2): each genome G i has its unique GRN which uniquely maps into a landscape E i that contain a large multitude of attractor states S*—hence, one genome encodes for many distinct phenotypes.
Let us briefly recapitulate here the formal basis of the elevation U, the quasi-potential of the landscape (Figs. 2 and 3). To obtain the landscape, the high-dimensional state space of the network’s dynamics can be thought of as “compressed” (projected) onto a two-dimensional (x 1, x 2) plane for representation reasons (which is mathematically possible only to some extent and involves loss of some information). In this planar state space, every position (x 1, x 2) is a state S of the network G i (a gene expression profile of G i ) and the elevation in Z at a given position S(x 1, x 2) is the quasi-potential U of that state S that expresses its relative instability: the higher the altitude at a given position, the higher is the “potential” and the less stable is the gene expression profile S there, and, hence, the phenotype associated with it.
At this place, a brief note on the semantics may be necessary to avoid a common confusion [18, 50]. Waddington’s term “epigenetic” has little to do with its modern meaning used to describe DNA methylation and covalent chromatin modifications that modulate gene expression [51–53], although a causal link can be constructed (Pisco et al., in review). These molecular “epigenetic” modifications are not so much the cause of gene activation, but rather reflect the activation status of the gene locus as a consequence of the action of the gene regulatory network and of external inputs. In fact, the epigenetic status of a locus is reversible and often highly dynamic [54, 55]. Since the enzymes that control the epigenetic modification do not bind DNA, let alone recognize specific regulatory sequences, they need to be recruited to the gene locus by specific transcription factors. Thus, the unfolding of information of the genome is performed by the GRN—whose global dynamics can be formalized as the epigenetic landscape which reflects the action of a network. It is a logical necessity that a distinct gene expression profile across the genome can only arise if the activities at each locus are orchestrated. This cannot be achieved solely by local change of the epigenetic status, but require gene–gene coordination which can only be achieved by locus-specific actions of trans-acting factors which form the GRN [29, 56]. Epigenetic modifications are rather the local controllers of the specific gene activation kinetics [57], although such quantitative details can be essential.
5.2 The fitness landscape—genetic instability
If the epigenetic landscape represents an ensemble of all combinatorically possible system states S and is the property uniquely associated with one genome G 1 and the GRN it encodes, we now define a second type of landscape that appears at a much grander scheme, so to speak, the landscape of landscapes: the landscape, or space, that captures the ensemble of all possible genomes G i or, equivalently, of GRNs (for a given size of genome; Fig. 5b). The alert reader will note that this more encompassing space is essentially equivalent to the parameter space discussed above in conjunction with structural stability in that each position in it represents a distinct wiring diagram G i such that the landscape allows us to compare a quasi-continuum of networks or genomes.
Thus, more generally, imagine a space of all gene network structures, an all-encompassing parameter space, where every point is a particular genome G i (since each network corresponds to one genome) and the neighboring points represent genomes that are just “one mutation away” from each other or, equivalently, GRNs that have similar wiring diagrams, differing from each other by the presence/absence or strength of a particular regulatory interaction (Fig. 5c). This space of all genomes of some (abstract) ensemble H of genomes is also referred to as the genotype space. Again, we can now think of all points in this space of all genomes as projected onto one plane, preserving neighborhood relationships as best as we can. Much as the quasi-potential landscape of the state space above where the elevation represents the relative (in)stability U of a phenotypic state S, we can now also utilize the elevation over the plane of genomes as an additional axis, F, to represent a quantity assigned to each genome G in this parameter space of all GRN architectures. But what is this quantity?
We are interested in comparing the phenotype repertoires, or epigenetic landscapes E associated with every individual genome G i in an ensemble H of genomes that are separated by mutations. Thus, we compare the relative fitness of the organisms i which has genome G i and whose cell phenotype repertoire is captured in its landscape E i with that of other genotypes G j . Such relative fitness in the tissue is relevant for cancer, as can be seen in this concrete example: in a population of cells that is genetically heterogeneous, as is always the case in tumors, individual tumor cells i have their own versions of GRN, G i . Cells with an epigenetic landscape E i in which in a given environment the attractor that encodes robust proliferation of a progenitor state S*p is large and deep, hence not easily perturbed, would be fitter than a landscape in which the corresponding attractor for proliferation is less stable and surrounded by steep descents leading down to the differentiated, non-proliferative state. In other words, we compare the fitness F(G i ) of the individual networks (genomes) G i and their associated epigenetic landscape E i with respect to the robustness of their proliferative state attractor under perturbations. We plot the fitness F i associated with each network (genome G i ) as the elevation over the space of networks (genomes; Fig. 5b). In the resulting landscape H, the elevation F(G i ) at location G i is the fitness landscape of the ensemble H of genomes G i. H may comprise all the organisms in an ecosystem or all the cells in a tumor. Obviously, the shape of the fitness landscape depends on the nature of the selection pressure.
Since elevation F reflects fitness, the cells will, under selection, tend to climb to the hilltops (=higher fitness). If selection is strong and omnipresent and the population large enough, mutating genomes always move uphill and cannot actually move downhill—they would be outcompeted and go extinct [58, 59]. Note that the trend to climb upward to the hilltops rather than fall to the lowest points in the valleys (as in the epigenetic landscape) is only a notational detail due to the definition of the vertical (Z) axes where the elevation corresponds to higher fitness but lower phenotypic state stability. Yet, there are profound differences between these two landscapes, as detailed below (Fig. 5b).
The concept of a fitness landscape mapped on top of the space of all genotypes is older than Waddington’s epigenetic landscape—it pertains to the realm of the older discipline of evolution and macroscopic biology. It was first proposed by Sewall Wright in 1931 [60], again as a metaphor, to illustrate the evolution of species in population genetics: mutations drive the movement in genotype space and selection imparts directionality toward the peak. This metaphor has been utilized to illustrate many elementary properties and problems in evolutionary theory, notably the problems of (i) finding the global peak as opposed to getting stuck in smaller local peaks and (ii) crossing valleys of low fitness since the only guidance for the direction of the walk on the landscape is to always walk uphill (in the steepest direction at each point). In the 1970s, Stuart Kauffman proposed the NK landscape which placed the metaphor onto formal footing by using a computational model ensemble of gene networks with discrete (binary) valued genes as organisms [61]. The fitness landscape has served many educational purposes as much as it has drawn criticism and controversy, but it remains a useful metaphor.
5.3 Differences between the epigenetic landscape E of each genome and the fitness landscape F of a population of genomes, H
The epigenetic landscape is described by the function U(S) and is a property of one genome G i , whereas the fitness landscape is described by the function F(G i ) and is the property of a genome ensemble H (Fig. 5). Obviously, these two landscapes pertain to two entirely different scales of view: the phenotypic repertoire (all states S) of an individual system encoded by a network/genome G vs. the fitness of an ensembles of all such possible systems. But there are more fundamental differences. The engine that drives the movement of a point in the epigenetic landscape from S 1 to S 2, i.e., the change of a gene expression profile, is in the intrinsic structure of the network which imposes the constraints that enforce coordinated change in gene activities. The directionality of movement on the epigenetic landscape is governed by the stability-seeking behavior and follows trajectories preordained by the genome. External perturbations (forceful shifting of S to any position with respect to the underlying X–Y plane of the landscape) are resisted by the internal dynamics manifest in the steepness of hills that need to be climbed.
In contrast, in the fitness landscape, there is no intrinsic stability, no intrinsic constraints and directionality: mutations are the motor of displacement. Movement toward the peaks (stable states) is imparted by the environment via selection. Without it, locomotion occurs (almost entirely) at random. Mutations are (in general) independent from each other. Thus, the driving force of locomotion on the landscape and its directionality (toward the peaks) are separable—unlike movement on the epigenetic landscape. The change of position on the underlying X–Y plane is primarily free in the absence of selection (which is equivalent to flattening the landscape). Thus, the dynamics of change of genotype is inherently plastic in the above sense. There is no intrinsic recoil force; no homeostatic force conveys “attraction” toward predefined positions of the space of genotypes, as in the case of attractors in the state space of GRNs. The directional movement in genotype space (toward the peaks) comes from outside: selection. Thus, stability of a distinct genotype G i , or, in other words, for a genotype to act akin to an attractor and reconstitute itself after random mutations, a high and invariant selection pressure and a high proliferation must be present.
One particular aspect for the walk on the landscape is the “velocity,” which, if we follow our formalism, reflects the mutation rate. The latter is not constant: the mutator phenotype of cancer cells increases the speed of the random walk in genotype space, but in the absence of selection, such apparent “genomic instability” is actually plasticity. Of note is that the mutation rate can be tuned by external factors such as stress on cells that via specific pathways interferes with the function of the DNA repair system [62]. Many stressors have been shown to increase mutation rates in single-cell organisms—the potential evolutionary advantage of which is obvious. This effect of externally imposed stress must of course be distinguished from selection that biases the direction of the walk because induced mutation is, from all we know, still random with respect to the desired phenotype (although restricted in the range of its molecular nature). In cancer cells, as discussed below, unfavorable conditions also increase genomic instability.
Since the hills and valleys of the fitness landscape encountered in a walk in genotype space are not intrinsic to the system but imparted by selection forces from the environment, the topography of the fitness landscape changes with any change of the tissue microenvironment and the associated nature of selection pressure experienced by the individual, cancer cells (genotypes). A network G i that is on a hilltop may lose its fitness advantage when the nature of selection pressure changes, thereby flattening that hill. In contrast, the topography of the epigenetic landscape E 1 of the given genome G 1 (at least in the way we parametrize a network) changes if that network undergoes a structural change, that is, moves on the fitness landscape from position G 1 to G 2 due to mutations.
In the fitness landscape, the flat regions (Fig. 5b, right) that allow a set genotypes (cancer cell clones) to “reach each other” without incurring selection pressure represent the so-called neutral space in which “mutations do not matter”: here, the genomes, G i and G j , that are separated by a mutation have networks that have the same fitness F(G i ) = F(G j ). In such regions, the movement of the genome in any direction is equally likely.
In contrast, in the epigenetic landscape (due to a counterintuitive property related to non-integrability in non-equilibrium systems) [44], movement in the flat regions between equally stable states still have to obey the constraints of the network that defines the landscape topography. Movement on the epigenetic landscape may require, even without changing elevation (ΔU = 0), “effort,” a property referred to as “path dependence.”
With respect to the formal notion of (in)stability, we can summarize the difference between the two landscape representations as follows: the epigenetic landscape visualizes dynamical (in)stability, whereas the fitness landscape indirectly manifests structural (in)stability that is exposed by the selecting environment. However, the change of genotype in the absence of selection pressure epitomizes plasticity rather than instability. Thus, the term “genome instability,” is, strictly speaking, a misnomer when used to describe a dynamical system behavior and without invoking a specific environment and associated selection.
5.4 Criticism of the fitness landscape
With these elementary properties of the fitness landscape, we can now address a central criticism of the idea of the fitness landscape: it tacitly assumes a one-to-one mapping between genotypes and phenotypes. It assigns to each position in genome space (each genotype) G i , a single phenotype that is characterized by a single value—its fitness F(G i ) (in a given environment). All evolutionary theories that use the fitness landscape hence implicitly subscribe to this paradigm of a 1:1 mapping. The common error committed when using the fitness landscape metaphor is the failure to appreciate the fundamental phenomenon of multi-stability: the ability of one genome to produce multiple stable phenotypic states (that are inherited over cell generations).
Thus, if we zoom in on the fitness landscape F to a single point which represents a network G 1 and magnify that single point, we will see an epigenetic landscape E 1 which displays the entire behavioral repertoire of that network encoded by genome G i . And if we walk on the fitness landscape, still looking through the magnifier, we will see how the epigenetic landscape “morphs” as we proceed from one position G i to another, from genome to genome. This nested relationship between the two landscapes is shown in Fig. 5. It is at the core of the integration of epigenetic and fitness landscape, hence of developmental and evolutionary instability in cancer. This scheme of thought directly refutes the notion of a 1:1 mapping between genotype and phenotype.
6 Cancer on the fitness landscape and the epigenetic landscape
What connects the two landscape concepts, the fitness landscape and the epigenetic landscape, to cancer and to cancer progression?
6.1 Cancer as somatic evolution
The fitness landscape is widely used in explaining cancer progression because of the unquestioned paradigm of the Darwinian somatic evolution of tumor cells [63]. Thus, random mutations, perhaps at accelerated rates due to the mutator phenotype [21, 64], afford the cancer cell the ability to explore the fitness landscape under selection pressure that emanates from the tissue microenvironment. The latter includes conditions such as hypoxia, antitumor defense of the immune system, low levels of growth factors, etc., and is thought to select cells that express the “hallmarks” of malignancy. As mentioned above, and extensively discussed elsewhere [13], this paradigm of the Darwinian evolution as the engine that promotes the increasing malignancy of the cancerous phenotype has many hidden as well as obvious inconsistencies. A well-articulated inconsistency pertains to the acquisition of metastatic capacity, which, according to the classical somatic evolution view [35, 65], is the result of a multistep process in which the needed mutations are successively acquired by selection.
The fact that some tumors, as best studied in breast cancer, often seed micro-metastasis early in the process of malignancy [66] is one example of a fundamental property that has exposed the first cracks in this paradigm of a cumulative process running on the mutation/selection scheme [67, 68]. In addition, much as such a scheme already meets serious challenges in the macroscopic evolution of species [69–74], in cancer development several phenomena also challenge the Darwinian orthodoxy. For instance, the principle of punctuated equilibria (“evolution by jerks”) [75–77] may be at play and is most prominently manifest in the coexistence of a vast diversity of non-clonal chromosomal aberrations [78] in a tumor, indicating the (temporal) absence of clonal selection [79]. We will return to this later.
Thus, the entire picture of cancer progression cannot be painted with the broad brush strokes of the elementary Darwinian scheme of adaption through selection, which is in need of refinement in view of the complexity phenotypes that defy explanation by a 1:1 genotype–phenotype mapping. Along the same line is the even more striking recent observation in leukemia that a single chromothripsis event in which a chromosome is chaotically rearranged can result in a sudden jerk of progression to higher malignancy [80].
6.2 Cancer states are unused attractors—the developmental aspect of tumorigenesis
Cancer, long viewed as a developmental disease by Virchow, may now more precisely be seen as a disruption of normal differentiation and misdifferentiation into abnormal cell types that resemble those encountered in the early stages of organismal development, or even of evolutionary stage [81]. Indeed, it has been proposed that cancer is the reactivation of an atavistic state used by evolutionary ancestors [82, 83]. If we recall the imagery of the epigenetic landscape, notably the rugged topography, such models become naturally plausible: in such a landscape concept, each cell state, ranging from immature stem cells to the mature post-mitotic cell type, is an attractor state of the genomic GRN—a valley in Waddington’s epigenetic landscape. However, since the latter is a mathematical consequence of a complex regulatory network, one can readily show by computer simulations that there are many more attractors (valleys) than actually used by the cells. In other words, there are many more (meta)stable, distinct gene expression configurations in the gene expression state space of a genome—each encoding a theoretical cell phenotype that would ensue from the combinatorial set of the expressed but is never realized. In 1971, Stuart Kauffman had the deep insight that such unused attractors may correspond to cancer cells [84]. Since then this concept has been refined, as summarized in [13, 81, 85].
Thus, if normal cell types are attractors, cancer cells are abnormal cell types and are abnormal attractors (see cartoon of Fig. 6). But what is the fundamental different between the “normal” and the unoccupied cancer attractors? How do cancer cells enter these unused attractors? The central idea is that mutational change of the genome, which is equivalent to a shift of position on the fitness landscape and to a distortion of the epigenetic landscape, allows the normally unoccupied attractors in side valleys to be accessed. These unused attractors are, to some extent, shielded from main developmental trajectories (Waddington’s “chreods”) on the epigenetic landscape and, hence, are not visited during normal development. But after the mutational distortion of the epigenetic landscape and possible lowering of protective hills that may have evolved to ensure normal development, they can suddenly become accessible to the developing cells, which may thus deviate from the normal path of development.
Imaginary and cartoonish representation of the entire epigenetic landscape of the genome of a higher metazoan with unused attractors = cancer attractors. The horizontal axis represents the high-dimensional state space—schematically as one dimension (as in Fig. 2e). The global gradient—from mountain top to the low valleys—represents the axis of development; hence, the higher the elevation, the more immature the states. Note the trajectories (“chreods”) of normal development (green broad arrows) which descend along the smooth (evolutionarily carved) valleys, whereas the uncharted terrains, shown here on the sides of the landscape (orange areas), are rugged because they were not sculpt by evolution (orange areas). In these uncharted regions, the attractors, typically not very stable, are the inevitable by-products of the wiring of the network that are unused. Mutations distort the landscape topography and suddenly these unused attractors can become accessible (red dashed arrow). Cells that are trapped in these unused attractors are, by necessity, less mature. They expose the associated gene expression programs to (somatic) evolution, which optimizes their self-serving fitness and promotes malignancy
Importantly, since the alternative cell states of the unused attractors have never been expressed, the associated cell phenotypes have not been exposed to organismal evolution, which would have tuned the gene expression profiles associated with these unused attractors to avert undue phenotypes that may interfere with organismal fitness. Thus, developing cells literally get stuck in such pathological, non-evolved attractors because their fate has never been harmonized with the needs of the tissue by fine-tuning the access to differentiation, senescence, or natural apoptosis [13, 85].
6.3 Large numbers of unused attractors are in state space regions that encode immature phenotypes
Computational simulations of gene network evolution that mimic the growth of the genome by gene duplication [86] and the accompanying increase of the number of attractors encoded by the genome reveal that during evolution, an immense (still not precisely quantified) number of attractors are produced that are never occupied but reside often in the vicinity of existing used attractors [87]. Their location with respect to the entire epigenetic landscape is of relevance: the landscape has a global gradient since the newly added attractors that turn out to be useful for the organism (new cell type) must be accessible—they must have a lower quasi-potential U than the existing ones. As a consequence, the landscape grew during evolution “downwards” as the attractors diversified, giving rise to hierarchical ordering: attractors of embryonic cells at the mountain top and ontogenetically more mature and phylogenetically younger cells at the bottom (Fig. 6) [81]. Now, the unused attractors can be added as side products of evolution at any time during this downward appositive growth of the epigenetic landscape, and hence, they are necessarily more likely to be numerous in older regions of the landscape, that is, near the mountain top in regions of the state space whose gene expression profile would encode for ontogenetically less mature and phylogenetically older phenotypes. If expressed, these unused attractors would produce phenotypes that by necessity would resemble those of immature cells. Specifically, they would exhibit a strong proliferation because the cell cycle is a primordial cellular program [88] dating back to unicellular life.
6.4 Unused attractors did not have a chance to evolve the encoding of social behavior and are genetically unstable
A fundamental property of unused attractors is that they do not encode for healthy cellular phenotypes because they have not been exposed to macroevolution. These attractors offer neither optimal stability for the cell itself nor a phenotypic behavior that benefits the host as a society of cells, such as controlled apoptosis [88, 89], access to terminally differentiated states, etc. Regarding the former, these “unevolved cells” have no optimized cell cycle with functioning checkpoints and a DNA repair system, which jointly assure fidelity of DNA replication to prevent passing mutated genomes to daughter cells. Therefore, given the intrinsic complexity of the machineries of DNA replication and cell division, they will inevitably suffer from genomic instability in which the mutated genome is passed on.
In fact, any experimental perturbation in the cell cycle machinery almost always increases the mutation rate [90, 91]. It may even be that the sophisticated machinery that prevents the spread of DNA sequence errors or chromosomal anomalies through cell division evolved alongside with the evolutionary growth and molding an increasingly complex epigenetic landscape and the ascendance of attractors for a variety of specialized cell types. Embryonic stem cells appear to tolerate polyploidy, including polyploid mitosis; that is, they lack a checkpoint-induced apoptosis [92]. The latter is activated upon differentiation in that the polyploidy cells then undergo programmed cell death [92]. This tolerance of chromosomal aberrations in the normal immature cell states is consistent with a large “neutral space”: flat regions in both the epigenetic landscape (the high-altitude regions representing immature phenotypes) as well as in the fitness landscape, and is in line with Heng’s concepts of non-clonal chromosome aberrations [78, 79].
Thus, ontogenetic immaturity may be inherently linked to tolerance of genome instability. This can be understood from the perspective of the evolutionary expansion of the epigenetic landscape to produce the deep attractors at the low-altitude bottom. These newest attractors encode mature, phenotypically and genetically stable cells which have lost tolerance to genome instability. Phylogenetically, tolerance of polyploidy has been implicated in the transition from unicellular to multicellular organisms [93]. If true correct, this would represent a most extreme form of avatism in cancer.
7 The ease of developing malignancy and the “true” role of mutations
Putting all together, we now propose a more encompassing picture of tumor progression. The argumentation leading to it can be traced back by the inquisitive reader in a logically consistent fashion to the first principles of dynamical systems epitomized by the two types of landscapes explained in this article.
7.1 Non-genetic cancer: in principle possible, but unlikely
Since unused attractors are latently present, just inaccessible in the normal tissue, tumorigenesis does not actually have to create de novo a new cancerous gene expression program. The developing cell can reach the unoccupied, potentially neoplastic attractor by jumping the hills on the epigenetic landscape. In molecular terms, a jump in state space across hills, or barriers of the quasi-potential U, can, in principle, be achieved by altering the expression of a distinct set of multiple genes. Once the cells have left the chreods of normal development, they are trapped in a side valley in the rugged terrains of the epigenetic landscape that has not been fine-sculpt by evolution to allow smooth flow down to the meaningful phenotypes [13, 81, 85]. Because evolution has shaped the epigenetic landscape so as to prevent such accidental deviation into the unevolved areas of the state space, this would require an extraordinarily large and unusual perturbation of GRN dynamics, were it to occur on a normal epigenetic landscape. Thus, in principle, “mutationless tumorigenesis” is possible, but very unlikely, requiring particularly large and repeated perturbations of the GRN. The tumor-promoting effect of chronic exposure to non-mutagenic compounds or tissue irritation may act via such perturbations.
Macroevolution of metazoan development has carved the epigenetic landscape (by stepwise mutational rewiring of the GRN) such that the chreods of normal development act like highways to robustly and efficiently guide cells to the attractors that encode the terminally differentiated, mature, evolutionally optimized functional cell states—those that we recognize as physiological cell types. This is achieved by erecting steep hills around the valleys that contain the normal developmental trajectories and the cell type attractors, thereby preventing the developing cells from veering away from the “righteous paths” and getting stuck in an unused attractor in the rugged regions of the epigenetic landscape that encode the immature, instable phenotypes.
7.2 Tumorigenic mutations reshape the epigenetic landscape and provide access to unused attractors
Enter mutations (see Fig. 7). As explained above (Fig. 5c), a mutation formally shifts the position of a given epigenetic landscape E i in the space of all genomes; that is, it displaces a network G from position G 1 to G 2 in the fitness landscape, thereby altering the topography of the epigenetic landscape from E 1 to E 1. This formally grounded picture represents the creation of a new network (G 2) that is similar to the previous one (G 1), but wired differently at some points. The new epigenetic landscape E 2 may lose its evolved optimal shape (E 1) such that the chreods in it have reduced depth and, thus, diminished capacity to “canalize” (to use Waddington’s metaphor) the developing cells to the correct destination attractors. Once the evolutionarily optimized epigenetic landscape is altered, deviation into the side valleys with the unused attractors becomes more likely (Fig. 7), requiring only slight (non-genetic) perturbations, or may even be inevitable. With this predisposition, then, an abnormal tissue environment (such as chronic inflammation) imparts additional non-genetic perturbations which can push some cells into these unused attractors during development as they descend toward the attractors encoding mature cell types. The deviant cells are literally stuck in these unused attractor states that represent immature or incompletely matured stages and that have no (evolved) access to the chreods or normal attractors. Given the geographic vicinity with the normal embryonic cells at the mountain top, these cells will exhibit gene expression profiles that are likely to resemble those used in the past—both in the ontogenetic and phylogenetic past.
Entry into the rugged region of the epigenetic landscape and the relationship between genetic and non-genetic factors in tumorigenesis. Schematic representation of the interplay between (1) mutation (=change of shape of epigenetic landscape, blue, due to mutation-driven displacement on the fitness landscape, red) and (2) non-genetic perturbation (displacement on the epigenetic landscape into the region of cancer attractors, orange contours). Thus, instability is “non-genetic” when it refers to displacement on the epigenetic landscape and is due to perturbation (orange arrow), whereas instability is “genetic” when it refers to displacement on the fitness landscape and is caused by mutations (red arrow)
Computer simulations in artificial complex GRNs of up to 100 genes [86] have shown that a class of molecular network architectures that appear to be prevalent in living systems [94, 95] are in general structurally robust. Mutations are not necessarily destructive, but often preserve the basic shape of the attractor landscape (partitioning of the epigenetic landscape into the attractors). It is much more likely that a random mutation (simulated as random rewiring of the GRN) distorts an existing attractor basin such that access to it is shut down rather than completely destroyed. This finding may explain the persistence of ancient attractors as hidden (not accessed) states when a phenotype is lost during evolution. This appears to have happened to the attractors for tooth and brown fat in birds which lack these two tissues [96, 97]. If access to such shutdown attractors is later regained, they are manifest as atavism. According to such thinking cancer can be viewed as a form of cellular atavism [82].
7.3 Unused attractors harbor potential for somatic evolution of the cancer cells
Once an unused attractor is occupied, cancer progression can take its own course with the help of the somatic form of Darwinian evolution which now promotes and is driven by the selfish interests of the cells. Since these new phenotypes have not been exposed to evolution (at least not since recently), the potential to evolve is immense. The sudden phenotypic realization of phenotypes with ancient and immature traits now provides all at once the key ingredients required for tumorigenesis and tumor progression—without the need to evolve them de novo: robust proliferation, which often is thought to be a primordial cellular program [88], migration, invasion, tolerance to chromosomal anomalies [92], and the increased mutation rate due to genome instability. Thus, the landscape formalism explains the inevitable nature of cancer [85]—accounting for the counterintuitive coexistence of a complex constructive process that creates robust survival and growth by random mutations and selection alone with the chaotic disruption of an evolved, complex system.
This new perspective on the role of mutations provides a possible rationale for the diversity of types of random mutations that occur at all scales—from single nucleotides to chromosomal anomalies [98]. There is nothing particular to aneuploidy that warrants its distinction as a specific class of genomic anomaly from DNA mutations [99]—both essentially rewire the network by altering gene regulatory relationships (Fig. 5c) and, hence, distort the epigenetic landscape. In doing so, they both, to varying degrees, may open access to the barred, uncharted regions in it.
7.4 Somatic evolution is also punctuated—due to bifurcation events
We proposed (see 7.1.) that unused, “unevolved” regions of the epigenetic landscape that contain multiple (small) attractors not tuned for development are rough and lack access to the normal chreods and attractors. Cells that have entered this uncharted terrain thus are unlikely to spontaneously return to the physiological regions of the epigenetic landscape (although this can happen and may constitute the rare cases of spontaneous differentiation and reversion of early cancer) [100]. They are trapped and roam rather freely among the many metastable states—a behavior that manifests as phenotypic plasticity and cell population heterogeneity. Since the alteration of networks by mutation (the displacement of a genome in genotype space) may not be accompanied by a drastic change of the epigenetic landscape due to structural stability, moderate distortions may lead to only a subtle change of cellular fitness relative to selection pressure: these mutations map into displacements that take place in the flat regions of the fitness landscape. The associated heterogeneity of karyotypes can then be interpreted as a manifestation of such neutral evolution [79]. Accordingly, a sudden expansion of individual clones should not be explained simply by the hand-waving assumption of selection of a particularly fit mutant clone, as in the classical Darwinian scheme, but rather can now be understood in a more formal way: the random walk in neutral genotype space, as we remember, is equivalent to a walk in the parameter space of the GRN (Fig. 2c) and, hence, may occasionally cross bifurcation points that represent discontinuous qualitative changes, such as through splitting of an attractor valley into two or disappearance of a separating hill. Such landscape-distorting events can suddenly grant entrance to a new unused attractor. The random walk in (neutral) genotype space and the equally random entry into domains of existence of cancer attractors that harbor the potential for rapid somatic evolution described above would naturally explain the punctuated equilibrium evolution dynamics of cancer cells proposed by Heng et al. [79].
The widespread and tacitly accepted notion of “phenotype instability” of cancer cells can now be formally explained in molecular and systems theory terms: the loss of normal stability of cell states conferred by deep valleys in the normal epigenetic landscape is accompanied by the loss of genotype stability associated with the immature and unevolved phenotype encoded by unused attractors. This genetic instability further promotes the exploration of new epigenetic landscapes with possibly more abnormal topography. Thus, we encounter a mutual reinforcement of non-genetic and genetic instability that speeds up the random walk on the fitness landscape.
8 The inevitability of increased malignancy after therapeutic perturbations
What happens when the cancer cell experiences an environmental perturbation? Formally, a (non-mutagenic) perturbation of a cell, such as blockade of a signaling pathway or any change in gene expression activities, is a kick on the ball in the epigenetic landscape. The cell moves in state space and, if it does not die, has to go somewhere, to some stable state. But where? With the perspective of the state space and landscape formalism, we can view tumor cell response to therapy in a new light.
8.1 Response to therapy is a regulatory event—a network perturbation that shifts the cell’s position in state space
Anticancer therapy does not necessarily inflict an overwhelming lethal blow to a cell (which would be so toxic as to be unsuited for therapy). Rather, chemotherapy or irradiation causes cell death by inducing subtle but complex regulatory responses and thereby trigger the cell’s own suicide program. A nonspecific cytotoxic intervention in the cell’s regulatory network that does not cause an overwhelming lethal blow thus stimulates a complex series of regulatory events which often amount to global cell stress. For instance, spindle apparatus-disrupting drugs, such as taxol, appear to cause cell stress beyond interfering with mitosis since microtubules are involved in many cellular processes [101]. Cisplatin causes particular DNA damage which triggers changes of gene expression and signaling [102]. Even target-selective drugs, such as specific kinase inhibitors, cause broad transcriptome responses [103]. Thus, drug treatment is a broad, multipoint perturbation of the network that, unless it instantly kills the cell by overwhelming necrosis, will cause major regulatory events in the cell, which results in genome-scale gene activity change that we now can formalize as a significant shift in the cell’s position in state space.
8.2 Spread of initial states (non-genetic heterogeneity) predisposes to heterogeneity of response
The rugged regions of the epigenetic landscape in which cancer attractors are located contain many small (meta)stable attractors (Fig. 6). This leads to population heterogeneity and plasticity of cancer cells because they occupy these distinct attractors and are thus dispersed in a continuum of “micro-states” that cover large regions of the state space (Fig. 7). Formally, this results in distinct initial conditions for each cell with regard to their response to the same perturbation. The diversity of the initial states at the time of perturbation is mirrored by the diversity of the responses: the perturbation hits cells differently such that each individual cell moves into different directions in state space. Such heterogeneity of cell dynamics in response to an external signal is well document by single-cell resolution measurements of gene expression, but has long been studied as an isolated phenomenon [104]. More recently, it was realized that, indeed, subpopulations of clonal progenitor cells which appear phenotypically to be homogenous indeed exhibit distinct transcriptomes and are susceptible to distinct differentiation signals [105]. More relevant for cancer therapy, such non-genetic heterogeneity has been amply documented for cancer cell death dynamics [106, 107]. Although the spreading of cancer cell populations into state space following therapy awaits precise quantitation by single-cell transcriptomics, the inherent heterogeneity of responsiveness is already manifest, albeit indirectly, in the almost ubiquitous but rarely articulated observation that, at intermediate doses of a cytotoxic agent, only a subset of cells responds, even in clonal populations [68].
8.3 Surviving cells move to other cancer attractors
The response diversification of cell populations suggests that the fraction of cells that is not pushed into apoptosis by the therapeutic perturbation will likely not stay in the same cancer attractor. Instead, these cells are expected to “end up” somewhere else on the epigenetic landscape, likely in other unused attractors in the uncharted, unevolved rugged regions of the state space. Based on geometric considerations of the epigenetic landscape, it is plausible that some or most of the perturbed but surviving cells land in adjacent or nearby unused attractors that encode equally or even more immature phenotypes.
In the formalisms of the parameter space and of structural stability, one can also envision this scenario of diversification of the response to perturbation as follows (see Fig. 8): the perturbation of the GRN may be so “deep” that one can model its action as an alteration of the parameters of regulatory interactions and, hence, will affect not dynamical but structural stability (remember Fig. 4). This obviously would be the case if the drugs induced (random) mutations (e.g., via stressed-induced genomic instability) [62] or if they alter “epigenetic” chromatin and DNA modifications: then, the model could be formulated (parametrized) such that the treatment could change the topography of the epigenetic landscape rather than just shifting the position of cells on the landscape. The cancer attractor state that existed before therapy could be destabilized and converted from a valley to a hilltop by the therapeutic perturbation, which now is, as we have learned, modeled as a shift in parameter space that pushes the cell state across a bifurcation event. As seen earlier, a broad class of bifurcation events consists of a branching that splits the attractor state into two new nearby attractors onto which the cells in the original one are partitioned (Fig. 4). One branch of the bifurcation may encode the (intended) apoptosis state and is accessed by a majority of cells (since the drug was screened to cause apoptosis), while the other inevitably present branch may encode a new cancer attractor that is even further away from normal chreods and is accessed by a sizable but often neglected fraction of the cell (Fig. 8b).
Model of effect of drug-induced perturbation based on cancer attractors. a A drastic perturbation can be modeled, like mutations, as change in the interaction parameters of the GRN and, hence, as a change of the epigenetic landscape topography. Drugs selected for abrogating the cancer cell state (e.g., due to induction of differentiation or apoptosis, blue arrows) are expected to destabilize the main cancer attractor through a bifurcation. This opens not only access to the apoptosis attractor (blue arrows) but possibly also to other attractors in the state space neighborhood in the uncharted region that are unused (magenta arrows). These newly accessed unused attractors are likely to also be cancerous and may represent even less mature and more malignant cells since they are located further away from the normal chreods, and hence encode cancer stem cell-like phenotypes which may confer resistance. b Representation of drug-induced perturbation as a pitchfork bifurcation (introduced in Fig. 4), here annotated with the biological significance. If a drug-induced change of the network causes a bifurcation, then almost by mathematical necessity, a portion of the cell population will experience a change that is “opposite” to that of the desired response. Thus, perhaps, induction of progression in a fraction of the population is linked to therapy
8.4 Cells that do not respond to cancer therapy actually respond by becoming more malignant
Since a therapeutic perturbation of a cancer cell population residing in a cancerous attractor state pushes the non-dying cells into a diversity of never-used nearby attractors near the currently occupied cancer cell attractor, it is now tempting to speculate—but reasonable to assume—that among these newly accessed, unused attractors, some encode alternative immature phenotypes of equally, if not more, malignant nature because the cells are pushed further and further away from the physiological regions of the landscape. This may explain the increasing “dedifferentiation” of recurrent tumors. Some of these distant unused attractors may encode drug resistance—a trait that is part of the ancient xenobiotic defense mechanism of cells.
With this picture, we arrive at the conclusion of our argumentation: it is inevitable that therapeutic intervention of the cancerous state causes—to varying extents—a fraction of cells to behave in a more malignant way, even if the majority of the cells are killed. Thus, progression toward a resistant tumor is not only due to the selection of cells that are genetic or epigenetic variants and “happen” to survive because they express a phenotype that is more drug-tolerant. Instead, the switch toward a therapy-resistant and more malignant state is induced by the therapy itself. While genetic mutations can be selected for and accumulate, one also needs to consider the non-genetic dynamics on the epigenetic landscape. Thus, instead of relying on Darwin’s “survival of the fittest” [108] alone, the motto that cancer cells follow may rather be Nietzsche’s “what does not kill me makes me stronger” [109]. Cells encounter bifurcations when stressed by the therapy and face the choice not between death and mere survival but between death and becoming stronger. Remarkably, such behavior is immanent to the network dynamics of a genome that has evolved to produce robust development of a multicellular organism.
8.5 Cell state dynamic and a new meaning of “cancer stem cell”
If seen in light of the formal concepts of cell phenotype dynamics explained in this article, the debate around the phenomenon of the very existence of CSC or “tumor-initiating cells” [11, 110–114] becomes moot. Much of the controversy stems from remnants of an old-fashioned view in which cell phenotypes are seen as fixed and immutable, unless there is a mutation. This is another example of the tacit and deeply rooted default notion of a one-to-one mapping between genotypes and phenotypes. In reality, a tumor cell population, even if genetically uniform, is phenotypically highly heterogeneous, consisting of cells of distinct degrees of immaturity, including some that qualify as bona fide cancer stem cells [115, 116] or possess traits of the immature mesenchymal state (in epithelial tumors) [33, 111, 117–119]. These phenotypes can transition into each other, reversibly or irreversibly (in the latter case typically from the less mature to the more mature state—preserving the directionality of normal development). It suffices to remind here that the stem cell-like state is associated with increased drug resistance, a property that can be viewed as part of the (phylogenetically) evolved xenobiotic response that is required to protect the long-living cells [10, 120, 121].
Such non-genetic dynamics with differentiation between subtypes has first been articulated by Stuart Kauffman, who proposed the possibility of differentiation therapy long before the dawn of the artificial notion of “cancer stem cells” [84].
8.6 Evidence for drug-induced increase in malignancy
A long list of observations indicates precisely the postulated therapy-induced stemness or drug resistance. For instance, irradiation induces stem cell-like behavior and drug tolerance in leukemic cells [122] and in hepatocarcinoma cells, including the expression of key pluripotency genes, such as Sox2 and Oct4 [123]. It was also shown that the chemotherapy agent cisplatin (which may cause a genome-wide response; see above) can also induce the same response. The stochasticity with which the stem cell state is induced [122] supports the notion that because of the heterogeneity of initial states, only a fraction of cells are poised to respond by transitioning into the more immature neighboring attractor state.
Irradiation has also been shown to predispose breast cancer cells to EMT, i.e., the transition into the more immature and malignant mesenchymal phenotype [124].
That multi-drug resistance (MDR) is not simply due to the selection of cells that “happen” to express resistant genes (e.g., due to a mutation in one of the MDR loci) but rather just one trait of an entire new phenotypic cell state is supported by the association of drug resistance with the mesenchymal phenotype and the finding that such cell states were mediated by specific pathways, including Notch, NF-kB, and the EMT-controlling genes Twist and Snail [125–128]. Consistently, chronic treatment with cisplatin of colorectal carcinoma cells leads to resistance that is associated with EMT. The apparent induction of multidrug resistance genes of the MDR family by chemotherapy is well documented [129, 130]. The long-known fact that MDR genes are also upregulated following nonspecific cell stress, such has heat shock [131, 132], can now be placed within the context of attractor switching induced by cell stress. Such behavior can be interpreted as an adaptive response that exploits remnants of an ancestral stress response state that is particularly accessible to cancer cells located in the immature regions of the state space. Then, the capacity to switch to a xenobiotic and drug resistance stress response state may be as immanent to the cancer cell phenotype as its instability and its immaturity.
Importantly, all the above observations were made in bulk tumor cell populations. Thus, it cannot be excluded that what appears as “induction” could in principle be that an extremely rapid selection of either preexisting non-genetic variants (cells that already were in an alternative resistance conveying attractor) or, less likely, of genetic mutants (which would constitute true Darwinian somatic evolution) has taken place. However, the short time frame of the macroscopic change makes a direct induction of a cellular response, that is, a switch of attractor states induced by the therapeutic perturbation, more probable.
8.7 Uniting mutation-less and mutation-driven progression by using both landscape concepts
That a somatic form of Darwinian evolution (selection of genetic mutants) takes place at a longer timescale of tumor progression is undeniable given the results of whole-genome sequencing of tumors [133, 134]. Perhaps, the non-Darwinian induction of the resistance phenotype by therapy provides a bridge to transit the valleys in the genetic fitness landscape—a conceptual problem that has plagued evolution biology. Thus, therapy-induced attractor transitions may produce in genome G i a temporary phenocopy of an attractor that in a different network G j would be dynamically more stable. In other words, following the perturbation, the cell first moves in epigenetic landscape (but stays at the same position on the fitness landscape), thereby affording some time for genetic mutations to accumulate and selection to operate [68], which then would nudge the cell toward higher points on the fitness landscape where the associated new epigenetic landscape has a basin of attraction for that same resistance phenotype that is larger and deeper. Future single-cell resolution gene expression analyses and tracking of cancer cell pedigrees will determine the relative contribution of true induction of individual cells as opposed to selection within a population to the emergence of more malignant cells.
9 Conclusion
We presented here a formal approach that allows us to think about phenomena in cancer biology that traditionally have been dealt with using ad hoc metaphors and qualitative argumentation. We focused on cell-autonomous progression (ignoring, for simplicity, intercellular processes such as tumor–stroma interactions). We showed that vague concepts such as non-genetic and genetic phenotype changes or instability can be explained using a set of formal tools epitomized by the epigenetic landscape and the fitness landscape. They capture the constraints that limit the potentials of realizing all configurations in an infinite space of possibilities: the gene expression state space is compartmentalized by the gene regulatory interactions which create valleys and hills that mold the epigenetic landscape; the genotype space is constrained by the differential fitness of individual genomes in the presence of selection which creates peaks and valleys that mold the fitness landscape. In showing that these two landscapes are linked in a particular way, we exposed an often made conceptual error behind the idea of the fitness landscape: that it mistakenly assumes that one genotype maps into one phenotype—when in reality a single genotype produces a complex set of alternative stable phenotypes represented by the valleys of the epigenetic landscape. Such non-genetic dynamics of phenotypes is well familiar to developmental biologists, yet has hardly been embraced by cancer biologists. But if they do so, they will see that many counterintuitive features of cancer progression, such as non-genetic clonal variability [135], can be readily explained in a natural and logically consistent fashion without taking refuge in new ad hoc concepts. Here, we focused on the apparently paradoxical but inevitable increase in malignancy in response to therapy as an example of a counterintuitive behavior.
This is not all gray theory. Of practical relevance is that recognition of the phenomenon of drug-induced escalation of malignancy opens a new opportunity for therapeutic approaches designed to stifle therapy resistance: if development of resistance is not a Darwinian evolutionary progress fueled by random mutations but a response shaped by network dynamics that involves attractor transitions, then it is mediated by the molecular signaling pathways that, in the healthy part of the landscape, facilitate state transitions during development but now misdirect transitions into the wrong attractors. These pathways could serve as therapeutic targets. Blocking such pathways may be more effective than the attempt, so far failed, to block the effector molecules of resistance, such as the MDR proteins [136], because resistance is mediated by an entire (high-dimensional) attractor.
While still grossly oversimplified (in part to better present the fundamental principles in which the metaphoric concepts are anchored, in part due to lack of specific data), the hope is that the generic, detail-free concepts discussed here will provide a new perspective and guidance to the practitioners of cancer biology in designing experiments and interpreting the data delivered by the flood of omics measurements.
References
Lengauer, C., Kinzler, K. W., & Vogelstein, B. (1998). Genetic instabilities in human cancers. Nature, 396(6712), 643–649.
Cahill, D. P., Kinzler, K. W., Vogelstein, B., & Lengauer, C. (1999). Genetic instability and Darwinian selection in tumours. Trends in Cell Biology, 9(12), M57–M60.
Attolini, C. S., & Michor, F. (2009). Evolutionary theory of cancer. Annals of the New York Academy of Sciences, 1168, 23–51. doi:10.1111/j.1749-6632.2009.04880.x (Review).
Nowak, M. A., Michor, F., & Iwasa, Y. (2006). Genetic instability and clonal expansion. Journal of Theoretical Biology, 241(1), 26–32. doi:10.1016/j.jtbi.2005.11.012.
Heng, H. H., Stevens, J. B., Bremer, S. W., Ye, K. J., Liu, G., & Ye, C. J. (2010). The evolutionary mechanism of cancer. Journal of Cellular Biochemistry, 109(6), 1072–1084. doi:10.1002/jcb.22497.
McCullough, K. D., Coleman, W. B., Ricketts, S. L., Wilson, J. W., Smith, G. J., & Grisham, J. W. (1998). Plasticity of the neoplastic phenotype in vivo is regulated by epigenetic factors. Proceedings of the National Academy of Sciences of the United States of America, 95(26), 15333–15338.
Gupta, P. B., Fillmore, C. M., Jiang, G., Shapira, S. D., Tao, K., Kuperwasser, C., et al. (2011). Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell, 146(4), 633–644. doi:10.1016/j.cell.2011.07.026 (Research support, non-U.S. government).
Chaffer, C. L., Brueckmann, I., Scheel, C., Kaestli, A. J., Wiggins, P. A., Rodrigues, L. O., et al. (2011). Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proceedings of the National Academy of Sciences of the United States of America, 108(19), 7950–7955. doi:10.1073/pnas.1102454108.
Roesch, A., Fukunaga-Kalabis, M., Schmidt, E. C., Zabierowski, S. E., Brafford, P. A., Vultur, A., et al. (2010). A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell, 141(4), 583–594. doi:10.1016/j.cell.2010.04.020.
Dean, M., Fojo, T., & Bates, S. (2005). Tumour stem cells and drug resistance. Nature Reviews. Cancer, 5(4), 275–284.
Hoek, K. S., & Goding, C. R. (2010). Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell & Melanoma Research, 23(6), 746–759. doi:10.1111/j.1755-148X.2010.00757.x (Comparative study review).
Sharma, S. V., Lee, D. Y., Li, B., Quinlan, M. P., Takahashi, F., Maheswaran, S., et al. (2010). A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell, 141(1), 69–80. doi:10.1016/j.cell.2010.02.027.
Huang, S. (2012). Tumor progression: chance and necessity in Darwinian and Lamarckian somatic (mutationless) evolution. Progress in Biophysics and Molecular Biology, 110(1), 69–86.
Holmberg, J., & Perlmann, T. (2012). Maintaining differentiated cellular identity. Nature Reviews. Genetics, 13(6), 429–439. doi:10.1038/nrg3209 (Research support, non-U.S. government. Review).
Sawyers, C. L., Denny, C. T., & Witte, O. N. (1991). Leukemia and the disruption of normal hematopoiesis. Cell, 64(2), 337–350 (Research support, non-U.S. government. Review).
Virchow, R. L. K. (1978). Cellular pathology (1859th ed., pp. 204–207). London: John Churchill.
Deutscher, G. (2010). Through the language glass: why the world looks different in other languages (1st ed.). New York: Macmillan.
Huang, S. (2012). The molecular and mathematical basis of Waddington’s epigenetic landscape: a framework for post-Darwinian biology. BioEssays, 34(2), 149–155.
Sieber, O., Heinimann, K., & Tomlinson, I. (2005). Genomic stability and tumorigenesis. Seminars in Cancer Biology, 15(1), 61–66. doi:10.1016/j.semcancer.2004.09.005.
Negrini, S., Gorgoulis, V. G., & Halazonetis, T. D. (2010). Genomic instability—an evolving hallmark of cancer. Nature Reviews. Molecular Cell Biology, 11(3), 220–228. doi:10.1038/nrm2858.
Beckman, R. A. (2010). Efficiency of carcinogenesis: is the mutator phenotype inevitable? Seminars in Cancer Biology, 20(5), 340–352. doi:10.1016/j.semcancer.2010.10.004 (Review).
Bielas, J. H., & Loeb, L. A. (2005). Mutator phenotype in cancer: timing and perspectives. Environmental and Molecular Mutagenesis, 45(2–3), 206–213.
Huang, S., & Ingber, D. E. (2000). Shape-dependent control of cell growth, differentiation, and apoptosis: switching between attractors in cell regulatory networks. Experimental Cell Research, 261(1), 91–103.
Huang, S. (2009). Non-genetic heterogeneity of cells in development: more than just noise. Development, 136(23), 3853–3862. doi:10.1242/dev.035139.
Altschuler, S. J., & Wu, L. F. (2010). Cellular heterogeneity: do differences make a difference? Cell, 141(4), 559–563. doi:10.1016/j.cell.2010.04.033.
Stewart, J. M., Shaw, P. A., Gedye, C., Bernardini, M. Q., Neel, B. G., & Ailles, L. E. (2011). Phenotypic heterogeneity and instability of human ovarian tumor-initiating cells. Proceedings of the National Academy of Sciences of the United States of America, 108(16), 6468–6473. doi:10.1073/pnas.1005529108.
Baum, B., Settleman, J., & Quinlan, M. P. (2008). Transitions between epithelial and mesenchymal states in development and disease. Semin Cell Dev Biol, 19(3), 294–308. doi:10.1016/j.semcdb.2008.02.001.
Singh, A., & Settleman, J. (2010). EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene, 29(34), 4741–4751. doi:10.1038/onc.2010.215.
Bonifer, C. (2005). Epigenetic plasticity of hematopoietic cells. Cell Cycle, 4(2), 211–214.
Frisen, J. (2002). Stem cell plasticity? Neuron, 35(3), 415–418.
Joshi, C. V., & Enver, T. (2002). Plasticity revisited. Current Opinion in Cell Biology, 14(6), 749–755.
Raff, M. (2003). Adult stem cell plasticity: fact or artifact? Annual Review of Cell and Developmental Biology, 19, 1–22.
Gotzmann, J., Mikula, M., Eger, A., Schulte-Hermann, R., Foisner, R., Beug, H., et al. (2004). Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutation Research, 566(1), 9–20.
Spencer, S. L., Gerety, R. A., Pienta, K. J., & Forrest, S. (2006). Modeling somatic evolution in tumorigenesis. PLoS Computational Biology, 2(8), e108.
Vogelstein, B., & Kinzler, K. W. (1993). The multistep nature of cancer. Trends in Genetics, 9(4), 138–141.
Hanahan, D., & Weinberg, R. A. (2011). Hallmarks of cancer: the next generation. Cell, 144(5), 646–674. doi:10.1016/j.cell.2011.02.013 (Research support, N.I.H. Extramural review).
Huang, S. (2011). Systems biology of stem cells: three useful perspectives to help overcome the paradigm of linear pathways. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 366(1575), 2247–2259. doi:10.1098/rstb.2011.0008.
Huang, S., & Kauffman, S. (2009). Complex gene regulatory networks—from structure to biological observables: cell fate determination. In R. A. Meyers (Ed.), Encyclopedia of complexity and systems science. Heidelberg: Springer, 1180–1213.
Huang, S. (2009). Reprogramming cell fates: reconciling rarity with robustness. BioEssays, 31(5), 546–560. doi:10.1002/bies.200800189.
Eldar, A., & Elowitz, M. B. (2010). Functional roles for noise in genetic circuits. Nature, 467(7312), 167–173. doi:10.1038/nature09326.
Kaern, M., Elston, T. C., Blake, W. J., & Collins, J. J. (2005). Stochasticity in gene expression: from theories to phenotypes. Nature Reviews. Genetics, 6(6), 451–464.
Raj, A., & van Oudenaarden, A. (2008). Nature, nurture, or chance: stochastic gene expression and its consequences. Cell, 135(2), 216–226. doi:10.1016/j.cell.2008.09.050.
Huang, S. (2010). Cell lineage determination in state space: a systems view brings flexibility to dogmatic canonical rules. PLoS Biology, 8(5), e1000380. doi:10.1371/journal.pbio.1000380.
Zhou, J. X., Aliyu, M. D., Aurell, E., & Huang, S. (2012). Quasi-potential landscape in complex multi-stable systems. Journal of the Royal Society, Interface, 9(77), 3539–3553. doi:10.1098/rsif.2012.0434.
Ferrell, J. E., & Xiong, W. (2001). Bistability in cell signaling: how to make continuous processes discontinuous, and reversible processes irreversible. Chaos, 11(1), 227–236. doi:10.1063/1.1349894.
Tyson, J. J., Chen, K. C., & Novak, B. (2003). Sniffers, buzzers, toggles and blinkers: dynamics of regulatory and signaling pathways in the cell. Current Opinion in Cell Biology, 15(2), 221–231.
Huang, S. (2001). Genomics, complexity and drug discovery: insights from Boolean network models of cellular regulation. Pharmacogenomics, 2(3), 203–222.
Waddington, C. H. (1957). The strategy of the genes. London: Allen and Unwin.
Kauffman, S. (1969). Homeostasis and differentiation in random genetic control networks. Nature, 224(215), 177–178.
Ptashne, M. (2007). On the use of the word ‘epigenetic’. Current Biology, 17(7), R233–R236.
Bird, A. (2007). Perceptions of epigenetics. Nature, 447(7143), 396–398.
Holliday, R. (2005). DNA methylation and epigenotypes. Biochemistry (Mosc), 70(5), 500–504.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693–705.
Kubicek, S., & Jenuwein, T. (2004). A crack in histone lysine methylation. Cell, 119(7), 903–906.
Trojer, P., & Reinberg, D. (2006). Histone lysine demethylases and their impact on epigenetics. Cell, 125(2), 213–217.
Bonifer, C., Hoogenkamp, M., Krysinska, H., & Tagoh, H. (2008). How transcription factors program chromatin—lessons from studies of the regulation of myeloid-specific genes. Seminars in Immunology, 20(4), 257–263. doi:10.1016/j.smim.2008.05.001.
Suter, D. M., Molina, N., Naef, F., & Schibler, U. (2011). Origins and consequences of transcriptional discontinuity. Current Opinion in Cell Biology, 23(6), 657–662. doi:10.1016/j.ceb.2011.09.004.
Lynch, M. (2007). Colloquium papers: the frailty of adaptive hypotheses for the origins of organismal complexity. Proceedings of the National Academy of Sciences of the United States of America, 104(Suppl 1), 8597–8604.
Nowak, M. A. (2006). Evolutionary dynamics: exploring the equations of life (1st ed.). Cambridge, MA: Belknap.
Wright, S. (1945). The differential equation of the distribution of gene frequencies. Proceedings of the National Academy of Sciences of the United States of America, 31(12), 382–389.
Kauffman, S. A. (1993). The origins of order. New York: Oxford University Press.
Galhardo, R. S., Hastings, P. J., & Rosenberg, S. M. (2007). Mutation as a stress response and the regulation of evolvability. Critical Reviews in Biochemistry and Molecular Biology, 42(5), 399–435. doi:10.1080/10409230701648502 (Research support, N.I.H. Extramural review).
Brutovsky, B., & Horvath, D. (2010). Optimization aspects of carcinogenesis. Medical Hypotheses, 74(5), 922–927. doi:10.1016/j.mehy.2009.10.019 (Research support, non-U.S. government).
Bielas, J. H., Loeb, K. R., Rubin, B. P., True, L. D., & Loeb, L. A. (2006). Human cancers express a mutator phenotype. Proceedings of the National Academy of Sciences of the United States of America, 103(48), 18238–18242.
Cairns, J. (1975). Mutation selection and the natural history of cancer. Nature, 255(5505), 197–200.
Klein, C. A. (2009). Parallel progression of primary tumours and metastases. Nature Reviews. Cancer, 9(4), 302–312. doi:10.1038/nrc2627.
Bernards, R., & Weinberg, R. A. (2002). A progression puzzle. Nature, 418(6900), 823.
Brock, A., Chang, H., & Huang, S. (2009). Non-genetic heterogeneity—a mutation-independent driving force for the somatic evolution of tumours. Nature Reviews. Genetics, 10(5), 336–342. doi:10.1038/nrg2556.
Gould, S. J. (1997). Darwinian fundamentalism. New York Review of Books (June 12), pp. 34–37.
Gould, S. J., & Lewontin, R. C. (1979). The spandrels of San Marco and the Panglossian paradigm: a critique of the adaptationist programme. Proceedings of the Royal Society of London. Series B: Biological Sciences, 205(1161), 581–598.
Lewontin, R. C. (2000). The triple helix: gene, organism, and environment (2001st ed.). Cambridge, MA: Harvard University Press.
Rose, S., Kamin, L. J., & Lewontin, R. C. (1985). Not in our genes: biology, ideology, and human nature. New York: Pantheon.
Pigliucci, M. (2009). An extended synthesis for evolutionary biology. Annals of the New York Academy of Sciences, 1168, 218–228. doi:10.1111/j.1749-6632.2009.04578.x.
Noble, D. (2011). Neo-Darwinism, the modern synthesis and selfish genes: are they of use in physiology? The Journal of Physiology, 589(Pt 5), 1007–1015. doi:10.1113/jphysiol.2010.201384 (Research support, non-U.S. government. Review).
Eldredge, N., & Gould, S. J. (1997). On punctuated equilibria. Science, 276(5311), 338–341 (Comment letter).
Caporale, L. H. (2009). Putting together the pieces: evolutionary mechanisms at work within genomes: can we suggest molecular underpinnings of punctuated equilibria and the Cambrian explosion? BioEssays, 31(7), 700–702. doi:10.1002/bies.200900067.
Gould, S. J. (1994). Tempo and mode in the macroevolutionary reconstruction of Darwinism. Proceedings of the National Academy of Sciences of the United States of America, 91(15), 6764–6771 (Review).
Heng, H. H., Bremer, S. W., Stevens, J., Ye, K. J., Miller, F., Liu, G., et al. (2006). Cancer progression by non-clonal chromosome aberrations. Journal of Cellular Biochemistry, 98(6), 1424–1435. doi:10.1002/jcb.20964 (Comparative study. Research support, non-U.S. government. Review).
Heng, H. H., Stevens, J. B., Bremer, S. W., Liu, G., Abdallah, B. Y., & Ye, C. J. (2011). Evolutionary mechanisms and diversity in cancer. Advances in Cancer Research, 112, 217–253. doi:10.1016/B978-0-12-387688-1.00008-9 (Research support, non-U.S. government, non-P.H.S. Review).
Stephens, P. J., Greenman, C. D., Fu, B., Yang, F., Bignell, G. R., Mudie, L. J., et al. (2011). Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell, 144(1), 27–40. doi:10.1016/j.cell.2010.11.055.
Huang, S., Ernberg, I., & Kauffman, S. (2009). Cancer attractors: a systems view of tumors from a gene network dynamics and developmental perspective. Seminars in Cell & Developmental Biology, 20(7), 869–876. doi:10.1016/j.semcdb.2009.07.003.
Davies, P. C., & Lineweaver, C. H. (2011). Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Physical Biology, 8((1), 015001. doi:10.1088/1478-3975/8/1/015001 (Research support, N.I.H., Extramural).
Ben-Jacob, E., Coffey, D. S., & Levine, H. (2012). Bacterial survival strategies suggest rethinking cancer cooperativity. Trends in Microbiology, 20(9), 403–410. doi:10.1016/j.tim.2012.06.001.
Kauffman, S. (1971). Differentiation of malignant to benign cells. Journal of Theoretical Biology, 31(3), 429–451.
Huang, S. (2011). On the intrinsic inevitability of cancer: from foetal to fatal attraction. Seminars in Cancer Biology, 21(3), 183–199. doi:10.1016/j.semcancer.2011.05.003.
Aldana, M., Balleza, E., Kauffman, S., & Resendiz, O. (2007). Robustness and evolvability in genetic regulatory networks. Journal of Theoretical Biology, 245(3), 433–448.
Torres-Sosa, C., Huang, S., & Aldana, M. (2012). Criticality is an emergent property of genetic networks that exhibit evolvability. PLoS Computational Biology, 8(9), e1002669. doi:10.1371/journal.pcbi.1002669 (Research support, non-U.S. government).
Sonnenschein, C., & Soto, A. M. (1998). The society of cells: cancer and control of cell proliferation. Philadelphia, PA: Garland Science.
Raff, M. C. (1992). Social controls on cell survival and cell death. Nature, 356(6368), 397–400.
Paulovich, A. G., Toczyski, D. P., & Hartwell, L. H. (1997). When checkpoints fail. Cell, 88(3), 315–321.
Hernando, E., Nahle, Z., Juan, G., Diaz-Rodriguez, E., Alaminos, M., Hemann, M., et al. (2004). Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature, 430(7001), 797–802.
Mantel, C., Guo, Y., Lee, M. R., Kim, M. K., Han, M. K., Shibayama, H., et al. (2007). Checkpoint–apoptosis uncoupling in human and mouse embryonic stem cells: a source of karyotpic instability. Blood, 109(10), 4518–4527. doi:10.1182/blood-2006-10-054247 (Research support, N.I.H., non-U.S. government. Extramural).
Erenpreisa, J., & Cragg, M. S. (2007). Cancer: a matter of life cycle? Cell Biology International, 31(12), 1507–1510. doi:10.1016/j.cellbi.2007.08.013 (Review).
Balleza, E., Alvarez-Buylla, E. R., Chaos, A., Kauffman, A., Shmulevich, I., & Aldana, M. (2008). Critical dynamics in genetic regulatory networks: examples from four kingdoms. PLoS One, 3, e2456.
Shmulevich, I., Kauffman, S. A., & Aldana, M. (2005). Eukaryotic cells are dynamically ordered or critical but not chaotic. Proceedings of the National Academy of Sciences of the United States of America, 102(38), 13439–13444.
Mitsiadis, T. A., Caton, J., & Cobourne, M. (2006). Waking-up the sleeping beauty: recovery of the ancestral bird odontogenic program. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution, 306(3), 227–233. doi:10.1002/jez.b.21094 (Research support, non-U.S. government. Comparative study).
Newman, S. A. (2011). Thermogenesis, muscle hyperplasia, and the origin of birds. BioEssays, 33(9), 653–656. doi:10.1002/bies.201100061 (Research support, U.S. government, non-P.H.S.).
Kitada, K., Taima, A., Ogasawara, K., Metsugi, S., & Aikawa, S. (2011). Chromosome-specific segmentation revealed by structural analysis of individually isolated chromosomes. Genes, Chromosomes & Cancer, 50(4), 217–227. doi:10.1002/gcc.20847.
Li, R., Yerganian, G., Duesberg, P., Kraemer, A., Willer, A., Rausch, C., et al. (1997). Aneuploidy correlated 100 % with chemical transformation of Chinese hamster cells. Proceedings of the National Academy of Sciences of the United States of America, 94(26), 14506–14511.
Clark, W. H. (1991). Tumour progression and the nature of cancer. British Journal of Cancer, 64(4), 631–644.
Mitchison, T. J. (2012). The proliferation rate paradox in antimitotic chemotherapy. Molecular Biology of the Cell, 23(1), 1–6. doi:10.1091/mbc.E10-04-0335 (Research support, N.I.H. Extramural).
Persons, D. L., Yazlovitskaya, E. M., Cui, W., & Pelling, J. C. (1999). Cisplatin-induced activation of mitogen-activated protein kinases in ovarian carcinoma cells: inhibition of extracellular signal-regulated kinase activity increases sensitivity to cisplatin. Clinical Cancer Research, 5(5), 1007–1014 (Research support, U.S. government, P.H.S. Comparative study).
Sims, A. H., Zweemer, A. J., Nagumo, Y., Faratian, D., Muir, M., Dodds, M., et al. (2012). Defining the molecular response to trastuzumab, pertuzumab and combination therapy in ovarian cancer. British Journal of Cancer, 106(11), 1779–1789. doi:10.1038/bjc.2012.176 (Research support, non-U.S. government).
Levsky, J. M., & Singer, R. H. (2003). Gene expression and the myth of the average cell. Trends in Cell Biology, 13(1), 4–6.
Chang, H. H., Hemberg, M., Barahona, M., Ingber, D. E., & Huang, S. (2008). Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature, 453(7194), 544–547.
Cohen, A. A., Geva-Zatorsky, N., Eden, E., Frenkel-Morgenstern, M., Issaeva, I., Sigal, A., et al. (2008). Dynamic proteomics of individual cancer cells in response to a drug. Science, 322, 1511–1516.
Spencer, S. L., Gaudet, S., Albeck, J. G., Burke, J. M., & Sorger, P. K. (2009). Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature, 459(7245), 428–432. doi:10.1038/nature08012.
Darwin, C. (1869). On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life (5th ed., pp. 91–92). London: John Murray.
Nietzsche, F. (1998). Twilight of the idols (D. Large, trans.). Oxford: Oxford University Press.
Dirks, P. (2010). Cancer stem cells: invitation to a second round. Nature, 466(7302), 40–41. doi:10.1038/466040a.
Gupta, P. B., Chaffer, C. L., & Weinberg, R. A. (2009). Cancer stem cells: mirage or reality? Nature Medicine, 15(9), 1010–1012. doi:10.1038/nm0909-1010.
Hill, R. P., & Perris, R. (2007). “Destemming” cancer stem cells. Journal of the National Cancer Institute, 99(19), 1435–1440.
Jordan, C. T. (2009). Cancer stem cells: controversial or just misunderstood? Cell Stem Cell, 4(3), 203–205. doi:10.1016/j.stem.2009.02.003.
Kelly, P. N., Dakic, A., Adams, J. M., Nutt, S. L., & Strasser, A. (2007). Tumor growth need not be driven by rare cancer stem cells. Science, 317(5836), 337. doi:10.1126/science.1142596.
Lobo, N. A., Shimono, Y., Qian, D., & Clarke, M. F. (2007). The biology of cancer stem cells. Annual Review of Cell and Developmental Biology, 23, 675–699. doi:10.1146/annurev.cellbio.22.010305.104154.
Reya, T., Morrison, S. J., Clarke, M. F., & Weissman, I. L. (2001). Stem cells, cancer, and cancer stem cells. Nature, 414(6859), 105–111.
Blick, T., Widodo, E., Hugo, H., Waltham, M., Lenburg, M. E., Neve, R. M., et al. (2008). Epithelial mesenchymal transition traits in human breast cancer cell lines. Clinical & Experimental Metastasis, 25(6), 629–642.
Deka, J., Wiedemann, N., Anderle, P., Murphy-Seiler, F., Bultinck, J., Eyckerman, S., et al. (2010). Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Research, 70(16), 6619–6628. doi:10.1158/0008-5472.CAN-10-0148.
Mani, S. A., Guo, W., Liao, M. J., Eaton, E. N., Ayyanan, A., Zhou, A. Y., et al. (2008). The epithelial–mesenchymal transition generates cells with properties of stem cells. Cell, 133(4), 704–715.
Donnenberg, V. S., & Donnenberg, A. D. (2005). Multiple drug resistance in cancer revisited: the cancer stem cell hypothesis. The Journal of Clinical Pharmacology, 45(8), 872–877.
Elliot, A., Adams, J., & Al-Hajj, M. (2010). The ABCs of cancer stem cell drug resistance. IDrugs, 13(9), 632–635.
Lee, G. Y., Shim, J. S., Cho, B., Jung, J. Y., Lee, D. S., & Oh, I. H. (2011). Stochastic acquisition of a stem cell-like state and drug tolerance in leukemia cells stressed by radiation. International Journal of Hematology, 93(1), 27–35. doi:10.1007/s12185-010-0734-2 (Research support, non-U.S. government).
Ghisolfi, L., Keates, A. C., Hu, X., Lee, D. K., & Li, C. J. (2012). Ionizing radiation induces stemness in cancer cells. PLoS One, 7(8), e43628. doi:10.1371/journal.pone.0043628 (Research support, non-U.S. government).
Andarawewa, K. L., Erickson, A. C., Chou, W. S., Costes, S. V., Gascard, P., Mott, J. D., et al. (2007). Ionizing radiation predisposes nonmalignant human mammary epithelial cells to undergo transforming growth factor beta induced epithelial to mesenchymal transition. Cancer Research, 67(18), 8662–8670. doi:10.1158/0008-5472.CAN-07-1294 (Research support, U.S. government, non-P.H.S.).
Arumugam, T., Ramachandran, V., Fournier, K. F., Wang, H., Marquis, L., Abbruzzese, J. L., et al. (2009). Epithelial to mesenchymal transition contributes to drug resistance in pancreatic cancer. Cancer Research, 69(14), 5820–5828. doi:10.1158/0008-5472.CAN-08-2819.
Li, Q. Q., Xu, J. D., Wang, W. J., Cao, X. X., Chen, Q., Tang, F., et al. (2009). Twist1-mediated adriamycin-induced epithelial–mesenchymal transition relates to multidrug resistance and invasive potential in breast cancer cells. Clinical Cancer Research, 15(8), 2657–2665. doi:10.1158/1078-0432.CCR-08-2372 (Research support, non-U.S. government).
Li, Q. Q., Chen, Z. Q., Cao, X. X., Xu, J. D., Xu, J. W., Chen, Y. Y., et al. (2011). Involvement of NF-kappaB/miR-448 regulatory feedback loop in chemotherapy-induced epithelial–mesenchymal transition of breast cancer cells. Cell Death and Differentiation, 18(1), 16–25. doi:10.1038/cdd.2010.103 (Research support, non-U.S. government).
Wang, Z., Li, Y., Kong, D., Banerjee, S., Ahmad, A., Azmi, A. S., et al. (2009). Acquisition of epithelial–mesenchymal transition phenotype of gemcitabine-resistant pancreatic cancer cells is linked with activation of the notch signaling pathway. Cancer Research, 69(6), 2400–2407. doi:10.1158/0008-5472.CAN-08-4312 (Research support, N.I.H., non-U.S. government. Extramural).
Chaudhary, P. M., & Roninson, I. B. (1993). Induction of multidrug resistance in human cells by transient exposure to different chemotherapeutic drugs. Journal of the National Cancer Institute, 85(8), 632–639.
Abolhoda, A., Wilson, A. E., Ross, H., Danenberg, P. V., Burt, M., & Scotto, K. W. (1999). Rapid activation of MDR1 gene expression in human metastatic sarcoma after in vivo exposure to doxorubicin. Clinical Cancer Research, 5(11), 3352–3356.
Stein, U., Jurchott, K., Walther, W., Bergmann, S., Schlag, P. M., & Royer, H. D. (2001). Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. The Journal of Biological Chemistry, 276(30), 28562–28569. doi:10.1074/jbc.M100311200 (Research support, non-U.S. government).
Chin, K. V., Tanaka, S., Darlington, G., Pastan, I., & Gottesman, M. M. (1990). Heat shock and arsenite increase expression of the multidrug resistance (MDR1) gene in human renal carcinoma cells. The Journal of Biological Chemistry, 265(1), 221–226.
Pleasance, E. D., Cheetham, R. K., Stephens, P. J., McBride, D. J., Humphray, S. J., Greenman, C. D., et al. (2010). A comprehensive catalogue of somatic mutations from a human cancer genome. Nature, 463(7278), 191–196. doi:10.1038/nature08658.
Yates, L. R., & Campbell, P. J. (2012). Evolution of the cancer genome. Nature Reviews. Genetics, 13(11), 795–806. doi:10.1038/nrg3317.
Kreso, A., O'Brien, C. A., van Galen, P., Gan, O. I., Notta, F., Brown, A. M., et al. (2013). Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science, 339(6119), 543–548. doi:10.1126/science.1227670.
Nobili, S., Landini, I., Mazzei, T., & Mini, E. (2012). Overcoming tumor multidrug resistance using drugs able to evade P-glycoprotein or to exploit its expression. Medicinal Research Reviews, 32, 1220–1262. doi:10.1002/med.20239.
Acknowledgments
The author would like to thank the Institute for Systems Biology and Alberta Innovates for supporting this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Huang, S. Genetic and non-genetic instability in tumor progression: link between the fitness landscape and the epigenetic landscape of cancer cells. Cancer Metastasis Rev 32, 423–448 (2013). https://doi.org/10.1007/s10555-013-9435-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-013-9435-7