Abstract
p53 is a tumor suppressor gene and plays an important role in the etiology of breast cancer. However, studies on the association between p53 polymorphisms and breast cancer risk have yielded conflicting results. We performed a meta-analysis to investigate the association between breast cancer and the p53 polymorphisms codon 72 (27,046 cases and 30,998 controls), IVS3 16 bp (3,332 cases and 3,700 controls) and IVS6+62A>G (8,787 cases and 9,869 controls) in different inheritance models. When all the eligible studies of codon 72 polymorphism were pooled into this meta-analysis, there was no evidence of significant association between breast cancer risk and p53 codon 72 polymorphism in any genetic model. However, in the stratified analysis for Indian population, significantly association was observed in additive model (OR = 0.62, 95% CI = 0.46–0.82, P value of heterogeneity test [P h] = 0.153) and recessive model (OR = 0.70, 95% CI = 0.50–0.92, P h = 0.463). IVS3 16 bp was significantly associated with breast cancer risk in a pooled 15 studies dataset (dominant model: OR = 1.14, 95% CI = 1.02–1.27, P h = 0.30; recessive model: OR = 1.61, 95% CI = 1.21–2.25, P h = 0.25; additive model: OR = 1.66, 95% CI = 1.24–2.21, P h = 0.28). No significant association was found between IVS6+62A>G polymorphism and breast cancer risk in a total of 14 studies. In summary, these results indicate that IVS3 16 bp is likely an important genetic marker contributing to susceptibility of breast cancer, and codon 72 homozygous mutants may be associated with decreased breast cancer risk in Indian population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer, a malignant proliferation of the epithelial cells that line the ducts or lobules of the breast, is the most common malignancy in women [1], accounting for approximately one-third of all cancers in women worldwide [2]. Although many risk factors for the development of breast cancer have been identified, such as the inherited genetic predisposition, the molecular mechanisms related to breast carcinogenesis remain under investigation [3, 4]. The disease seems to be the result of cumulative alterations of oncogenes and tumor suppressor genes that lead to clonal outgrowth of progressively malignant cells [5, 6].
Tumor suppressor gene p53 which located on 17p13 is one of the major markers of human tumor and one of the most commonly mutated genes in human cancer [7]. The p53 protein has a very important function in many physiological processes, such as cell cycle arrest, DNA repair, apoptosis, and gene transcription [8]. In addition to acquired mutations that alter its function in p53, there are many studies which have been identified in the p53 gene, the p53 codon 72 (rs1042522) polymorphism of exon 4 is a common single nucleotide polymorphism (SNP), where the variant encodes a proline (CCC) rather than an arginine (CGC) residue [9], it can affect p53 function. The two polymorphic variants have been indicated that their structure and biological properties were not the same [10].
Many studies have reported the role of p53 polymorphisms at codon 72 (rs1042522), IVS3 16 bp (rs17878362) and IVS6+62A>G (rs1625895) with breast cancer risk [15–68], but the results were inconclusive, some original studies thought that these polymorphisms were association with breast cancer risk, but others had different opinions. In addition, previous meta-analysis on p53 showed conflicting results. Hence, the correlation of this polymorphic gene remains unknown. In order to explore the association between p53 codon 72 (rs1042522), IVS3 16 bp (rs17878362) and IVS6+62A>G (rs1625895) polymorphisms with breast cancer risk, a Meta-analysis was conducted to summarize the data. Meta-analysis is a powerful tool for summarizing the different studies. It can not only overcome the problem of small size and inadequate statistical power of genetic studies of complex traits, but also provide more reliable results than a single case–control study.
Materials and methods
Search strategy and selection criteria
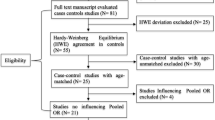
All the case–control studies were identified by a computerized literature search of the PubMed, EBSCO, and CGEMS database (prior to September 2010) using the following words and terms: “p53”, “TP53”, “polymorphism,” and “breast,” as well as their combinations. Only research articles were included and the language was not limited. The included studies have to meet the following criteria: (1) only the case–control studies and cohort studies were considered; (2) they were designed to evaluate the p53 codon 72, IVS3 16 bp (rs17878362) and IVS6+62A>G (rs1625895) polymorphisms and breast cancer risk, (3) the amount of published data was sufficient to allow estimation of an odds ratio (OR) with 95% confidence interval (CI); and (4) the distribution of genotypes among controls are in Hardy–Weinberg equilibrium (P < 0.01).
Data extraction
Information was carefully extracted from all eligible studies independently by two authors (He and Su) according to the inclusion criteria listed above. The following data were collected from each study: first author, year of publication, original country and ethnicity of the sample, source of controls, and genotype distribution. Disagreement was resolved by discussion between the two authors. If they could not reach a consensus, another author was consulted to resolve the dispute, and a final decision was made by two of this group of three authors. When a study reported results on different sub-populations according to ethnicity, we considered each sub-population as a separate study in the meta-analysis.
Statistical analysis
The strength of association between p53 polymorphisms and breast cancer risk was assessed by Crude ORs with the corresponding 95% CIs. The pooled ORs were performed for an additive model (CC vs. YY), recessive model (CC vs. CY+YY) and a dominant model (CC+CY vs. YY). Heterogeneity among studies was checked by the Q test; the P value of more than 0.1 for the Q test indicates a lack of heterogeneity among studies, so the pooled OR was calculated by the fixed-effects model [69]. Otherwise, a random-effects model was used [70]. If heterogeneity was present we might use meta-regression analysis in exploring sources of heterogeneity [71]. In addition, subgroup analyses were conducted by ethnicity and resource of controls. Sensitivity analyses were performed to estimate the robustness of the summary estimate of alteration in breast cancer risk conferred by p53 codon 72 (rs1042522), IVS3 16 bp (rs17878362), and IVS6+62A>G (rs1625895) polymorphisms. Begg’s funnel plots [72] and Egger’s linear regression test [73] were used to assess publication bias. In the control group, Hardy–Weinberg equilibrium (HWE) was tested for using a goodness-of-fit chi-square test. All of the calculations were performed using STATA version 10.0 (STATA Corporation, College Station, TX).
Results
Study characteristics
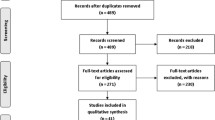
Table 1 listed the main characteristics and genotype distribution of codon 72 polymorphism (rs1042522), with a total of 56 eligible studies met the inclusion criteria, including first author, published year, ethnicity, original country, source of controls, and genotype distribution. However, the study of Pharoah et al. [50] and the study of Samson et al. [51] were excluded because subjects had been included by Baynes et al. [38] and Rajkumar et al. [44]. The distribution of genotypes in the controls was consistent with Hardy–Weinberg equilibrium in all studies except for two studies (P < 0.01) [21, 65], these studies were excluded in this meta-analysis. Hence, leaving 52 eligible studies (27,046 cases and 30,998 controls) that had assessed the association between the codon 72 polymorphism and breast cancer risk.
Table 2 listed the main characteristics and genotype distribution of IVS3 16 bp polymorphism (rs17878362), with a total of 15 eligible studies (3,332 cases and 3,700 controls) for investigating IVS3 16 bp polymorphism and breast cancer risk.
Table 3 listed the main characteristics and genotype distribution of IVS6+62A>G polymorphism (rs1625895), with a total of 14 eligible studies (8,787 cases and 9,869 controls) for investigating VS6+62A>G polymorphism and breast cancer risk.
Meta-analysis results
Codon 72 polymorphism
Table 4 listed the main results of the meta-analysis of codon 72 polymorphism and breast cancer risk. When all the eligible studies were pooled into this meta-analysis of codon 72, there was no evidence of significant association between breast cancer risk and p53 codon 72 polymorphism in any genetic model (dominant model: odds ratio [OR] = 0.97, 95% confidence interval [CI] = 0.90–1.05, P value of heterogeneity test [P h] < 0.001; recessive model: OR = 0.96, 95% CI = 0.88–1.06, P h = 0.009; additive model: OR = 0.95, 95% CI = 0.85–1.07, P h < 0.001). Significant between-study heterogeneity was detected in any genetic model. Hence, we performed the stratified analysis according to ethnicity, source of controls, and sample size. In the stratified analysis for India population, significantly decreased risk of breast cancer was observed in additive model (OR = 0.62, 95% CI = 0.46–0.82, P = 0.001, P h = 0.153, Fig. 1) and recessive model (OR = 0.70, 95% CI = 0.50–0.92, P h = 0.463, Fig. 2).
Previous codon 72 polymorphism
Three meta-analyses have been previously published for codon 72 polymorphism and breast cancer risk [11–14]. Table 4 listed the main results of meta-analysis of previous codon 72 polymorphism and breast cancer risk.
The study of Zhang et al. [11] had 39 studies, when all the eligible studies were pooled into the meta-analysis, significantly decreased risk of breast cancer was observed in dominant model (OR = 0.90, 95% CI: 0.82–0.99). In the stratified analysis by ethnicity, significantly decreased risk was also observed in European populations (dominant model: OR = 0.88, 95% CI: 0.80–0.98). In the stratified analysis by source of controls, they found that the variant genotypes were associated with a significantly decreased breast cancer risk in dominant model and additive model (dominant model: OR = 0.87. 95% CI: 0.78–0.97; homozygote comparison: OR = 0.88, 95% CI: 0.78–1.00).
The study of Hu et al. [12] had 37 studies, significantly decreased risk of breast cancer was found between Mediterranean and Northern European populations (recessive model: OR = 0.32, 95% CI: 0.24–0.44; additive model: OR = 0.35, 95% CI: 0.21–0.60). The data for this meta-analysis only included 375 cases and 389 controls from 6 studies between Mediterranean and Northern European populations.
The study of Ma et al. [13] had 21 studies, when all the eligible studies were pooled into the meta-analysis, significantly increased risk of breast cancer was observed in dominant model (OR = 1.179, 95% CI = 1.020–1.362). In the stratified analysis by source of controls, significantly increased risk was also observed by population-based study (dominant model: OR = 1.23, 95% CI = 1.05–1.43; recessive model: OR = 1.16, 95% CI = 1.01–1.33; additive model: OR = 1.28, 95% CI = 1.04–1.59).
The study of Francisco et al. [14] had 42 case–control studies reporting an association between the p53 codon 72 polymorphism and breast cancer. When all the eligible studies were pooled into the meta-analysis, no significant association of breast cancer risk was found in any genetic model. In the stratified analysis by source of country, significantly decreased risk was observed in Indian population (dominant model: OR = 0.75, 95% CI = 0.61–0.93; Arg/Arg vs. Pro/Pro: OR = 0.70, 95% CI = 0.53–0.91; recessive model: OR = 0.77, 95% CI = 0.61–0.97) in Indian population.
IVS3 16 bp polymorphism
Table 5 listed the main results of the meta-analysis of the IVS3 16 bp polymorphism and breast cancer risk. When all the eligible studies were pooled into the meta-analysis, significantly increased risks of breast cancer were observed in any genetic model (dominant model: OR = 1.14, 95% CI = 1.02–1.27, P = 0.017, P value of heterogeneity test [P h] = 0.30, Fig. 3; recessive model: OR = 1.61, 95% CI = 1.21–2.25, P = 0.001, P h = 0.25, Fig. 4; additive model: OR = 1.66, 95% CI = 1.24–2.21, P = 0.001, P h = 0.28, Fig. 5). Moreover, significant between-study heterogeneity was not detected in the meta-analysis of the IVS3 16 bp polymorphism and breast cancer under any genetic model.
IVS6+62A>G polymorphism
Table 6 listed the main results of the meta-analysis of the IVS6+62A>G polymorphism and breast cancer risk. When all the eligible studies were pooled into the meta-analysis, no significant association of breast cancer risk was found in any genetic model (dominant model: OR = 1.03, 95% CI = 0.91–1.18, P = 0.82, P value of heterogeneity test [P h] = 0.009; recessive model: OR = 0.93, 95% CI = 0.76–1.14, P = 0.49, P h = 0.85; additive model: OR = 0.93, 95% CI = 0.76–1.14, P = 0.51, P h = 0.80). Moreover, significant between-study heterogeneity was not detected in the meta-analysis of the IVS6+62A>G polymorphism and breast cancer under any genetic model, except for dominant model (P = 0.009 for heterogeneity).
Next, we performed stratified analysis by source of controls and ethnicity, in stratified subgroup meta-analysis, the IVS6+62A>G polymorphism was not found to be associated with breast cancer risk too.
Sensitive analysis
We tested the inclusion criteria of this meta-analysis by a sensitivity analysis. Sensitivity analysis were conducted to determine whether modification of the inclusion criteria of this meta-analysis affected the results, A single study involved in the meta–analysis was deleted each time to reflect the influence of individual data set to the pooled ORs, and the corresponding pooled ORs were not essentially altered (data not shown), indicating that our results were statistically robust.
Publication bias
Both Begg’s funnel plot and Egger’s test were performed to access the publication bias of this meta-analysis. Begg’s funnel plots did not reveal any evidence of obvious asymmetry in any genetic model in the overall meta-analysis (Figures not shown). The Egger’s test results suggested no evidence of publication bias in the meta-analysis of codon 72 (P = 0.259 for dominant model, P = 0.514 for recessive model, P = 0.328 for additive model); IVS3 16 bp (P = 0.869 for dominant model, P = 0.694 for recessive model, P = 0.744 for additive model) and IVS6+62A>G (P = 0.663 for dominant model, P = 0.566 for recessive model, P = 0.426 for additive model), indicating that our results were statistically robust.
Discussion
Many studies have reported the role of p53 polymorphisms at codon 72 (rs1042522), IVS3 16 bp (rs17878362) and IVS6+62A>G (rs1625895) with breast cancer risk [15–68], but the results were inconclusive, some original studies thought that these polymorphisms were associated with breast cancer risk, but other original studies thought no association with breast cancer risk. In addition, previous meta-analysis on codon 72 polymorphism showed conflicting results. Hence, a meta-analysis was conducted to explore the association between p53 codon 72, IVS3 16 bp and IVS6+62A>G polymorphisms and breast cancer risk.
Our present meta-analysis, which included 27,046 cases and 30,998 cases from 52 studies, explored the association between the p53 codon 72 polymorphism and breast cancer risk. The results indicated that codon 72 polymorphism may be not associated with breast cancer risk in Caucasian population. In the stratified analysis for Indian population, significantly decreased risk of breast cancer was observed in additive model (OR = 0.62, 95% CI = 0.46–0.82, P = 0.001, P h = 0.153) and recessive model (OR = 0.70, 95% CI = 0.50–0.92, P h = 0.463). The result indicated that codon 72 polymorphism may be associated with breast cancer risk, but there are only four studies in Indian population, to determine whether codon 72 polymorphism be applied to clinical genotyping for risk assessment still require large scale breast cancer case–controls researches in Indian population.
In this meta-analysis, significant association of the IVS3 16 bp polymorphism and breast cancer risk was found (dominant model: OR = 1.14, 95% CI = 1.02–1.27, P = 0.017, P value of heterogeneity test [P h] = 0.30; recessive model: OR = 1.61, 95% CI = 1.21–2.25, P = 0.001, P h = 0.25; additive model: OR = 1.66, 95% CI = 1.24–2.21, P = 0.001, P h = 0.28). The result indicated that IVS3 16 bp polymorphism may increase risk of developing breast cancer. To determine whether this marker should be applied to clinical genotyping for risk assessment still require large scale breast cancer case–control researches.
Meanwhile, no significant association of the IVS6+62A>G polymorphism and breast cancer risk was found. Hence, IVS6+62A>G may have no strong association with breast cancer risk, at least in our meta-analysis.
Previous meta-analysis on p53 codon 72 showed conflicting results. We have read with great interest the article “No significant association between the p53 codon 72 polymorphism and breast cancer risk: a meta-analysis of 21 studies involving 24,063 subjects” Published online on May 23, 2010 issue of “Breast Cancer Research and Treatment” [13]. The study of Ma [13] have 21 case–control studies, his conclusion indicate that it provided strong evidence that the P53 codon 72 polymorphism is not association with the risk of developing breast cancer. Ma et al. [13] concluded that no significant association was found between the P53 codon 72 polymorphism and breast cancer risk when all the eligible studies were pooled into the meta-analysis, but significant risk of breast cancer was observed in dominant model (OR = 1.179, 95% CI = 1.020–1.362). Similarly, Ma et al. [13] demonstrated that no significant association was observed for any of the genetic models in the stratified analysis by source of controls. But in the stratified analysis by source of controls, significant increased risks were observed by source of controls (dominant model: OR = 1.23, 95% CI = 1.05–1.43; recessive model: OR = 1.16, 95% CI = 1.01–1.33; additive model: OR = 1.28, 95% CI = 1.04–1.59). Hence, the ongoing uncertainty still existed and the conclusion by Ma et al. [13] was not entirely credible. In addition, several sizeable eligible studies have not been included in this meta-analysis, we thought that these studies satisfied the search criteria. Importantly, the data reported by Ma et al. [13] for the study by Schimit et al. [41] do not seem in line with the data provided by Schimit et al. [41] in their original publication. The numbers reported by Ma et al. [13] for Arg/Arg, Arg/Pro, Pro/Pro, in cases and controls, are 2797-2008-386 and 2024-1523-287, respectively. Interestingly enough, after carefully studying the data presented by Schimit et al. [41], the frequencies that we have retrieved on the 8,345 cases and 6,849 controls were 4499-3228-618 and 3661-2677-511, respectively. The data reported by Ma et al. [13] for the study by Sjalander et al. [16] do not seem in line with the data provided by Sjalander et al. [16] in their original publication too. The numbers reported by Ma et al. [13] for Arg/Arg, Arg/Pro, Pro/Pro, in cases and controls, are 24-93-95 and 61-253-375, respectively. Interestingly enough, after carefully studying the data presented by Sjalander et al. [16], the frequencies that we have retrieved on the 212 cases and 689 controls were 95-93-24 and 375-253-61, respectively. The data reported by Ma et al. [13] for the study by Sprague et al. [42] do not seem in line with the data provided by Sprague et al. [42] in their original publication too. The numbers reported by Ma et al. [13] for Arg/Arg, Arg/Pro, Pro/Pro, in cases and controls, are 823-570-89 and 705-490-83, respectively. Interestingly enough, after carefully studying the data presented by Sprague et al. [42], the frequencies that we have retrieved on the 1,653 cases and 1,854 controls were 909-644-100 and 1021-704-129, respectively. The data reported by Ma et al. [13] for the study by Weston et al. [17] do not seem in line with the data provided by Weston et al. [17] in their original publication too. The numbers reported by Ma et al. [13] for Arg/Arg, Arg/Pro, Pro/Pro, in cases and controls, are 6-27-32 and 3-42-72, respectively, in Caucasian. Interestingly enough, after carefully studying the data presented by Weston et al. [17], the frequencies that we have retrieved on the 65 cases and 117 controls were 32-27-6 and 72-42-3 in Caucasian, respectively.
Secondly, we have also read great interest the recent meta-analysis by Zhang et al. [11], the study of Zhang [11] have 39 case–control studies, the results suggested that p53 codon 72 polymorphism may contribute to susceptibility to breast cancer, especially in Europeans. Zhang et al. [11] concluded that significant association was found between the TP53 codon 72 polymorphism and breast cancer risk in the stratified analysis by ethnicity (Arg/pro vs. Arg/Arg: OR 0.89, 95% CI 0.80–0.99; dominant model: OR 0.88, 95% CI 0.80–0.98) and source of controls (Arg/pro vs. Arg/Arg: OR 0.88, 95% CI 0.78–0.98; dominant model: OR 0.87, 95% CI 0.78–0.97). But P value of Q test for heterogeneity test <0.001, when heterogeneity was very big, the results cannot be concluded that p53 codon 72 polymorphism may contribute to susceptibility to breast cancer, especially in Europeans. Hence, the ongoing uncertainty still existed and the conclusion by Zhang et al. [11] was not entirely credible. In addition, the study of by Baynes et al. [38] and the study by Pharoah et al. [50] essentially represent the same study, two studies by Buyru et al. [24, 74] have been included in this meta-analysis; however, careful inspection of both studies reveals that the same cases hace been included in them. Hence, incorporating one of the two studies by Buyru et al. might seem more appropriate. Importantly, several sizeable eligible studies have not been included in Zhang et al. [11], we thought that these studies satisfied the search criteria.
Thirdly, we have also read with great interest the recent meta-analysis by Hu et al. [12], the study of Hu et al. [12] had 37 case–control studies, the results suggest that codon 72 had a potential role in association with breast cancer risk within certain populations or regions. Significantly decreased risk was observed by source of Ethnicity (dominant model: OR = 0.32, 95% CI = 0.24–0.44; Pro/Pro vs. Arg/Arg: OR = 0.35, 95% CI = 0.21–0.60) in the Mediterranean studies. In the Mediterranean was Caucasian, in addition, all eligible study was small sample in the Mediterranean. Hence, the ongoing uncertainty still existed and the conclusion by Hu et al. [12] was not entirely credible.
Last, we have also read with great interest the recent meta-analysis by Francisco et al. [14], the study of Francisco et al. [14] had 42 case–control studies reporting an association between the p53 codon 72 polymorphism and breast cancer. Significantly decreased risk was observed in Indian population (dominant model: OR = 0.75, 95% CI = 0.61–0.93; Arg/Arg vs. Pro/Pro: OR = 0.70, 95% CI = 0.53–0.91; recessive model: OR = 0.77, 95% CI = 0.61–0.97) in Indian population. The study of Francisco et al. [14] had only five case–control studies in Indian population, which include 715 cases and 1,668 controls. However, in our present meta-analysis, which including four case–control studies in Indian population, significantly decreased risk was only observed in additive model and recessive model. Sample size was not large in our present meta-analysis and Francisco et al. [14], hence, the results should be interpreted with caution. To determine whether codon 72 polymorphism be applied to clinical genotyping for risk assessment still require large scale breast cancer case–controls researches in Indian population.
However, there are several limitations in this meta-analysis. Our results should be interpreted with caution. First, the controls were not uniformly defined. Although all the controls were healthy populations, most of them were common populations, some controls were Population-based; other controls were hospital-based. Hence, non–differential misclassification bias is possible. Second, in the subgroup analysis may have had insufficient statistical power to check an association, Third, we were also unable to examine the interactions among gene–environment, lacking of the original data of the included studies limited our further evaluation of potential interactions, which may be an important component of the association between p53 codon 72 polymorphism and environment and breast cancer risk. Four, it was much difficult to get the all articles published in various language. We only included the studies published in English and Chinese. Last, our results were based on unadjusted published estimates. Because of data limitations, we were unable to adjust them such as age, smoking, alcohol consumption et al.
Overall, our results indicated that IVS3 16 bp polymorphism may increase risk of developing breast cancer; and codon 72 homozygous mutants may be associated with decreased breast cancer risk in India population.
References
Berns EM, van Staveren IL, Look MP, Smid M, Klijn JGM, Foekens JA (1998) Mutations in residues of TP53 that directly contact DNA predict poor outcome in human primary breast cancer. Br J Cancer 77:1130–1136
Parkin DM, Bray F, Ferlay J et al (2001) Estimating the world cancer burden: Globocan 2000. Int J Cancer 94:153–156
Veronesi U, Boyle P, Golsdhirsch A, Orecchia R, Viale G (2005) Breast cancer. Lancet 365:1727–1741
Yager JD, Davidson NE (2006) Mechanism of disease: estrogen carcinogenesis in breast cancer. N Engl J Med 354:270–282
Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100:57–70
Dumitrescu RG, Cotarla I (2005) Understanding breast cancer risk—where do we stand in 2005? J Cell Mol Med 9:208–221
Hollstein M, Sidransky D, Vogelstein B et al (1991) p53 mutations in human cancers. Science 253:49–53
Oren M (2003) Decision making by p53: life, death and cancer. Cell Death 10:431–442
Denehower LA (2005) p53 guardian and suppressor of longevity? Exp Gerontol 40:7–9
Dumont P, Leu JI, Della Pietra ACIII et al (2003) The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 33:357–365
Zhang Z, Wang M, Wu D, Wang M, Tong N, Tian Y, Zhang Z (2010) P53 codon 72 polymorphism contributes to breast cancer risk: a meta-analysis based on 39 case–control studies. Breast Cancer Res Treat 120(2):509–517
Hu Z, Li X, Yuan R, Ring BZ, Su L (2010) Three common TP53 polymorphisms in susceptibility to breast cancer, evidence from meta-analysis. Breast Cancer Res Treat 120(3):705–714
Ma Y, Yang J, Liu Z, Zhang P, Yang Z, Wang Y, Qin H (2011) No significant association between the TP53 codon 72 polymorphism and breast cancer risk: a meta-analysis of 21 studies involving 24,063 subjects. Breast Cancer Res Treat 125(1):201–205
Francisco G, Menezes PR, Eluf-Neto J, Chammas R (2010) Arg72Pro TP53 polymorphism and cancer susceptibility: a comprehensive meta-analysis of 302 case-control studies. Int J Cancer. doi:10.1002/ijc.25710
Kawajiri K, Nakachi K, Imai K, Watanabe J, Hayashi S (1993) Germ line polymorphisms of p53 and CYP1A1 genes involved in human lung cancer. Carcinogenesis 14:1085–1089
Sjalander A, Birgander R, Hallmans G, Cajander S, Lenner P, Athlin L, Beckman G, Beckman L (1996) p53 polymorphisms and haplotypes in breast cancer. Carcinogenesis 17:1313–1316
Weston A, Pan CF, Ksieski HB, Wallenstein S, Berkowitz GS, Tartter PI, Bleiweiss IJ, Brower ST, Senie RT, Wolff MS (1997) p53 haplotype determination in breast cancer. Cancer Epidemiol Biomarkers Prev 6:105–112
Helland A, Langerod A, Johnsen H, Olsen AO, Skovlund E, Borresen-Dale AL (1998) p53 polymorphism and risk of cervical cancer. Nature 396:530–531
Wang-Gohrke S, Rebbeck TR, Besenfelder W et al (1998) p53 germline polymorphisms are associated with an increased risk for breast cancer in German women. Anticancer Res 18:2095–2099
Khaliq S, Hameed A, Khaliq T et al (2000) P53 mutations, polymorphisms, and haplotypes in Pakistani ethnic groups and breast cancer patients. Genet Test 4:23–29
Papadakis EN, Dokianakis DN, Spandidos DA (2000) p53 codon 72 polymorphism as a risk factor in the development of breast cancer. Mol Cell Biol Res Commun 3:389–392
Li T, Lu ZM, Guo M, Wu QJ, Chen KN, Xing HP, Mei Q, Ke Y (2002) p53 codon 72 polymorphism (C/G) and the risk of human papillomavirus-associated carcinomas in China. Cancer 95:2571–2576
Wang-Gohrke S, Becher H, Kreienberg R, Runnebaum IB, Chang-Claude J (2003) Intron 3 16 bp duplication polymorphism of p53 is associated with an increased risk for breast cancer by the age of 50 years. Pharmacogenetics 12:269–272
Buyru N, Tigli H, Dalay N (2003) P53 codon 72 polymorphism in breast cancer. Oncol Rep 10:711–714
Mabrouk I, Baccouche S, El-Abed R, Mokdad-Gargouri R, Mosbah A, Saïd S, Daoud J, Frikha M, Jlidi R, Gargouri A (2003) No evidence of correlation between p53 codon 72 polymorphism and risk of bladder or breast carcinoma in Tunisian patients. Ann N Y Acad Sci 1010:764–770
Huang XE, Hamajima N, Katsuda N, Matsuo K, Hirose K, Mizutani M, Iwata H, Miura S, Xiang J, Tokudome S, Tajima K (2003) Association of p53 codon Arg72Pro and p73 G4C14-to-A4T14 at exon 2 genetic polymorphisms with the risk of Japanese breast cancer. Breast Cancer 10:307–311
Katiyar S, Thelma BK, Murthy NS, Hedau S, Jain N, Gopalkrishna V, Husain SA, Das BC (2003) Polymorphism of the p53 codon 72 Arg/Pro and the risk of HPV type 16/18-associated cervical and oral cancer in India. Mol Cell Biochem 252:117–124
Suspitsin EN, Buslov KG, Grigoriev MY, Ishutkina JG, Ulibina JM, Gorodinskaya VM, Pozharisski KM, Berstein LM, Hanson KP, Togo AV, Imyanitov EN (2003) Evidence against involvement of P53 polymorphism in breast cancer predisposition. Int J Cancer 103:431–433
Menzel HJ, Sarmanova J, Soucek P, Berberich R, Gru¨newald K, Haun M, Kraft HG (2004) Association of NQO1 polymorphism with spontaneous breast cancer in two independent populations. Br J Cancer 90:1989–1994
Noma C, Miyoshi Y, Taguchi T, Tamaki Y, Noguchi S (2004) Association of p53 genetic polymorphism (Arg72Pro) with estrogen receptor positive breast cancer risk in Japanese women. Cancer Lett 210:197–203
Mahasneh AA, Abdel-Hafiz SS (2004) Polymorphism of p53 gene in Jordanian population and possible associations with breast cancer and lung adenocarcinoma. Saudi Med J 25:1568–1573
Tommiska J, Eerola H, Heinonen M, Salonen L, Kaare M, Tallila J, Ristimäki A, von Smitten K, Aittomäki K, Heikkilä P, Blomqvist C, Nevanlinna H (2005) Breast cancer patients with p53 Pro72 homozygous genotype have a poorer survival. Clin Cancer Res 11:5098–5103
Kalemi TG, Lambropoulos AF, Gueorguiev M, Chrisafi S, Papazisis KT, Kotsis A (2005) The association of p53 mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T polymorphisms with breast cancer in Northern Greece. Cancer Lett 222:57–65
Siddique MM, Balram C, Fiszer-Maliszewska L, Aggarwal A, Tan A, Tan P, Soo KC, Sabapathy K (2005) Evidence for selective expression of the p53 codon 72 polymorphs: implications in cancer development. Cancer Epidemiol Biomarkers Prev 14:2245–2252
Ohayon T, Gershoni-Baruch R, Papa MZ, Distelman Menachem T, Eisenberg Barzilai S, Friedman E (2005) The R72P P53 mutation is associated with familial breast cancer in Jewish women. Br J Cancer 92:1144–1148
Damin AP, Frazzon AP, Damin DC, Roehe A, Hermes V, Zettler C, Alexandre CO (2006) Evidence for an association of TP53 codon 72 polymorphism with breast cancer risk. Cancer Detect Prev 30:523–529
Ma H, Hu Z, Zhai X, Wang S, Wang X, Qin J, Chen W, Jin G, Liu J, Gao J, Wang X, Wei Q, Shen H (2006) Joint effects of single nucleotide polymorphisms in P53BP1 and p53 on breast cancer risk in a Chinese population. Carcinogenesis 27:766–771
Baynes C, Healey CS, Pooley KA, Scollen S, Luben RN, Thompson DJ, Pharoah PD, Easton DF, Ponder BA, Dunning AM (2007) SEARCH breast cancer study: common variants in the ATM, BRCA1, BRCA2, CHEK2 and TP53 cancer susceptibility genes are unlikely to increase breast cancer risk. Breast Cancer Res 9(2):R27
Gochhait S, Bukhari SI, Bairwa N, Vadhera S, Darvishi K, Raish M, Gupta P, Husain SA, Bamezai RN (2007) Implication of BRCA2–26G>A 5′ untranslated region polymorphism in susceptibility to sporadic breast cancer and its modulation by p53 codon 72 Arg>Pro polymorphism. Breast Cancer Res 9:R71
Khadang B, Fattahi MJ, Talei A, Dehaghani AS, Ghaderi A (2007) Polymorphism of TP53 codon 72 showed no association with breast cancer in Iranian women. Cancer Genet Cytogenet 173:38–42
Schmidt MK, Reincke S, Broeks A, Braaf LM, Hogervorst FB, Tollenaar RA, Johnson N, Fletcher O, Peto J, Tommiska J, Blomqvist C, Nevanlinna HA, Healey CS, Dunning AM, Pharoah PD, Easton DF, Dörk T, Van’t Veer LJ (2007) Breast cancer association consortium: do MDM2 SNP309 and TP53 R72P interact in breast cancer susceptibility? A large pooled series from the breast cancer association consortium. Cancer Res 67:9584–9590
Sprague BL, Trentham-Dietz A, Garcia-Closas M, Newcomb PA, Titus-Ernstoff L, Hampton JM, Chanock SJ, Haines JL, Egan KM (2007) Genetic variation in TP53 and risk of breast cancer in a population-based case control study. Carcinogenesis 28:1680–1686
Zhang W, Jin MJ, Chen K (2007) Association of p53 polymorphisms and its haplotypes with susceptibility of breast cancer. Zhejiang Da Xue Xue Bao Yi Xue Ban 36:561–566
Rajkumar T, Samson M, Rama R, Sridevi V, Mahji U, Swaminathan R, Nancy N (2008) TGFb1 (Leu10Pro), p53 (Arg72Pro) can predict for increased risk for breast cancer in south Indian women and TGFb1 Pro (Leu10Pro) allele predicts response to neo-adjuvant chemo-radiotherapy. Breast Cancer Res Treat 112:81–87
Cox DG, Deer D, Guo Q, Tworoger SS, Hankinson SE, Hunter DJ, De Vivo I (2007) The p53 Arg72Pro and MDM2-309 polymorphisms and risk of breast cancer in the nurses’ health studies. Cancer Causes Control 18:621–625
Gaudet MM, Gammon MD, Bensen JT, Sagiv SK, Shantakumar S, Teitelbaum SL, Eng SM, Neugut AI, Santella RM (2008) Genetic variation of TP53, polycyclic aromatic hydrocarbonrelated exposures, and breast cancer risk among women on Long Island, New York. Breast Cancer Res Treat 108:93–99
Garcia-Closas M, Kristensen V, Langerod A, Qi Y, Yeager M, Burdett L, Welch R, Lissowska J, Peplonska B, Brinton L, Gerhard DS, Gram IT, Perou CM, Borresen-Dale AL, Chanock S (2007) Common genetic variation in TP53 and its flanking genes, WDR79 and ATP1B2, and susceptibility to breast cancer. Int J Cancer 121:2532–2538
Johnson N, Fletcher O, Palles C, Rudd M, Webb E, Sellick G, dos Santos Silva I, McCormack V, Gibson L, Fraser A, Leonard A, Gilham C, Tavtigian SV, Ashworth A, Houlston R, Peto J (2007) Counting potentially functional variants in BRCA1, BRCA2 and ATM predicts breast cancer susceptibility. Hum Mol Genet 16:1051–1057
Franeková M, Zúbor P, Stanclová A, Dussan CA, Bohusová T, Galo S, Dobrota D, Kajo K, Péc M, Racay P (2007) Association of p53 polymorphisms with breast cancer: a case–control study in Slovak population. Neoplasma 54:155–161
Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA, SEARCH Investigators (2007) Association between common variation in 120 candidate genes and breast cancer risk. PloS Genet 3:e42
Samson M, Swaminathan R, Rama R, Sridevi V, Nancy KN, Rajkumar T (2007) Role of GSTM1 (Null/Present), GSTP1 (Ile105Val) and P53 (Arg72Pro) genetic polymorphisms and the risk of breast cancer: a case control study from South India. Asian Pac J Cancer Prev 8:253–257
Akkiprik M, Sonmez O, Gulluoglu BM, Caglar HB, Kaya H, Demirkalem P, Abacioglu U, Sengoz M, Sav A, Ozer A (2009) Analysis of p53 gene polymorphisms and protein over-expression in patients with breast cancer. Pathol Oncol Res 15:359–368
Singh V, Rastogi N, Mathur N, Singh K, Singh MP (2008) Association of polymorphism in MDM-2 and p53 genes with breast cancer risk in Indian women. Ann Epidemiol 18:48–57
Lum SS, Chua HW, Li H, Li WF, Rao N, Wei J, Shao Z, Sabapathy K (2008) MDM2 SNP309 G allele increases risk but the Tallele is associated with earlier onset age of sporadic breast cancers in the Chinese population. Carcinogenesis 29:754–761
Cavallone L, Arcand SL, Maugard C, Ghadirian P, Mes-Masson AM, Provencher D, Tonin PN (2008) Haplotype analysis of TP53 polymorphisms, Arg72Pro and Ins16, in BRCA1 and BRCA2 mutation carriers of French Canadian descent. BMC Cancer 8:96
Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, Schmitt F (2008) Importance of TP53 codon 72 and intron 3 duplication 16 bp polymorphisms in prediction of susceptibility on breast cancer. BMC Cancer 8:32
De Vecchi G, Verderio P, Pizzamiglio S, Manoukian S, Bernard L, Pensotti V, Volorio S, Ravagnani F, Radice P, Peterlongo P (2008) The p53 Arg72Pro and Ins16 bp polymorphisms and their haplotypes are not associated with breast cancer risk in BRCA-mutation negative familial cases. Cancer Detect Prev 32:140–143
Nordgard SH, Alnaes GI, Hihn B et al (2008) Pathway based analysis of SNPs with relevance to 5-FU therapy: relation to intratumoral mRNA expression and survival. Int J Cancer 123:577–585
Henrırquez-Hernandez LA, Murias-Rosales A, Herna′ndez GA, Cabrera DLA, Dı′az-Chico BN, Mori DSM, Ferna′ndez PL (2009) Gene polymorphisms in TYMS, MTHFR, p53 and MDR1 as risk factors for breast cancer: a case–control study. Oncol Rep 22:1425–1433
Kazemi M, Salehi Z, Chakosari RJ (2009) TP53 codon 72 polymorphism and breast cancer in northern Iran. Oncol Res 18(1):25–30
Denisov EV, Cherdyntseva NV, Litvyakov NV, Slonimskaya EM (2009) TP53 mutations and Arg72Pro polymorphism in breast cancers. Cancer Genet Cytogenet 192:93–95
Aoki MN, da Silva AHAC, Amarante MK, do Val Carneiro JL, Fungaro MH, Watanabe MA (2009) CCR5 and p53 codon 72 gene polymorphisms: implications in breast cancer development. Int J Mol Med 23:429–435
Song F, Zheng H, Liu B, Wei S, Dai H, Zhang L, Calin GA, Hao X, Wei Q, Zhang W, Chen K (2009) An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res 15(19):6292–6300
Lång A, Palmebäck Wegman P, Wingren S (2009) The significance of MDM2 SNP309 and p53 Arg72Pro in young women with breast cancer. Oncol Rep 22:575–579
Hrstka R, Beranek M, Klocova K, Nenutil R, Vojtesek B (2009) Intronic polymorphisms in TP53 indicate lymph node metastasis in breast cancer. Oncol Rep 22:1205–1211
Kara N, Karakus N, Ulusoy AN, Ozaslan C, Gungor B, Bagci H (2010) P53 codon 72 and HER2 codon 655 polymorphisms in Turkish breast cancer patients. DNA Cell Biol 29:7
Ebner F, Schremmer-Danninger E, Rehbock J (2010) The role of TP53 and p21 gene polymorphisms in breast cancer biology in a well specified and characterized German cohort. J Cancer Res Clin Oncol 136:1369–1375
Bisof V, Salihović MP, Narancić NS, Skarić-Jurić T, Jakić-Razumović J, Janićijević B, Turek S, Rudan P (2010) TP53 gene polymorphisms and breast cancer in Croatian women: a pilot study. Eur J Gynaecol Oncol 31(5):539–544
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Thompson SG, Higgins JPT (2002) How meta-regression analyses be undertaken and ihterpreted? Statist Med 21:1559–1573
Begg CB, Mazumdar M (1994) Operating characteristics of a rank correlation test for publication bias. Biometrics 50(4):1088–1101
Egger M, Smith DG, Schneider M (1997) Minder C Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Buyru N, Altinisik J, Demokan S, Dalay N (2007) p53 genotypes 153 and haplotypes associated with risk of breast cancer. Cancer Detect Prev 31:207–213
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
He, XF., Su, J., Zhang, Y. et al. Association between the p53 polymorphisms and breast cancer risk: meta-analysis based on case–control study. Breast Cancer Res Treat 130, 517–529 (2011). https://doi.org/10.1007/s10549-011-1583-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1583-2