Abstract
Chromium is a non-essential and well-known toxic metal for microorganisms and plants. The widespread industrial use of this heavy metal has caused it to be considered as a serious environmental pollutant. Chromium exists in nature as two main species, the trivalent form, Cr(III), which is relatively innocuous, and the hexavalent form, Cr(VI), considered a more toxic species. At the intracellular level, however, Cr(III) seems to be responsible for most toxic effects of chromium. Cr(VI) is usually present as the oxyanion chromate. Inhibition of sulfate membrane transport and oxidative damage to biomolecules are associated with the toxic effects of chromate in bacteria. Several bacterial mechanisms of resistance to chromate have been reported. The best characterized mechanisms comprise efflux of chromate ions from the cell cytoplasm and reduction of Cr(VI) to Cr(III). Chromate efflux by the ChrA transporter has been established in Pseudomonas aeruginosa and Cupriavidus metallidurans (formerly Alcaligenes eutrophus) and consists of an energy-dependent process driven by the membrane potential. The CHR protein family, which includes putative ChrA orthologs, currently contains about 135 sequences from all three domains of life. Chromate reduction is carried out by chromate reductases from diverse bacterial species generating Cr(III) that may be detoxified by other mechanisms. Most characterized enzymes belong to the widespread NAD(P)H-dependent flavoprotein family of reductases. Several examples of bacterial systems protecting from the oxidative stress caused by chromate have been described. Other mechanisms of bacterial resistance to chromate involve the expression of components of the machinery for repair of DNA damage, and systems related to the homeostasis of iron and sulfur.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Occurrence of chromium
Chromium is the seventh most abundant element on earth and although occurs in oxidation states ranging from Cr(II) to Cr(VI), the trivalent Cr(III) and hexavalent Cr(VI) are the most stable and abundant forms (Cervantes et al. 2001). Cr(VI) is a strong oxidizing agent, commonly present in solution as the hydrochromate (HCrO −4 ), chromate (CrO 2−4 ) or dichromate (Cr2O 2−7 ) oxyanions, depending on the pH (US EPA 1998). Cr(VI) exists as water-soluble anions that may persist in water for long periods and is considered as a key contaminant at the US Department of Energy waste sites (Riley et al. 1992). Cr(III) derivatives are much less mobile and exist in the environment mostly forming stable complexes with both organic and inorganic ligands (Zayed and Terry 2003). In addition, Cr(III) is less toxic because is less soluble at physiological pH. At neutral pH, Cr(III) tends to precipitate as hydroxide [Cr(OH)3] or hydrated oxide (Cr2O.H2O) (Ehrlich 2002). The low solubility of Cr(III) [mostly as Cr2O3 and Cr(OH)3] is likely the major reason why Cr(III) makes up a small percentage of the total chromium concentration in polluted groundwater (US EPA 1998). Mobilisation of the Cr(OH)3 precipitate is slow, unless enhanced by dissolution in strongly acidic environments or by being complexed with organic compounds (Rai et al. 1987). The widespread use of Cr in diverse industrial processes has converted it into a serious contaminant of air, soil and water (Khasim et al. 1989).
Biological properties of chromium
Transmembrane transport of chromium
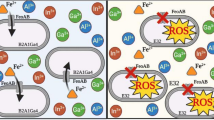
It has been demonstrated in a variety of bacterial species that chromate actively crosses biological membranes by means of the sulfate uptake pathway, which reflects the chemical analogy between these two oxyanions (Fig. 1) (Cervantes and Campos-García 2007). Cr(III) crosses cell membranes with a low efficiency because it forms insoluble compounds (Cary 1982). Inside the cell, Cr(VI) is readily reduced to Cr(III) by the action of various enzymatic or nonenzymatic activities; the Cr(III) generated may then exert diverse toxic effects in the cytoplasm (Fig. 1) (Cervantes et al. 2001).
Mechanisms of chromate transport, toxicity and resistance in bacterial cells. Mechanisms of damage and resistance are indicated by thin and heavy arrows, respectively. (A) Chromosome-encoded sulfate uptake pathway which is also used by chromate to enter the cell; when it is mutated (X) the transport of chromate diminishes. (B) Extracellular reduction of Cr(VI) to Cr(III) which does not cross the membrane. (C) Intracellular Cr(VI) to Cr(III) reduction may generate oxidative stress, as well as protein and DNA damage. (D) Detoxifying enzymes are involved in protection against oxidative stress, minimizing the toxic effects of chromate. (E) Plasmid-encoded transporters may efflux chromate from the cytoplasm. (F) DNA repair systems participate in the protection from the damage generated by Cr derivatives
Toxic effects of Cr(VI)
The biological effects of Cr strongly depend on its oxidation state and cellular localization. Cr(VI) is considered the most toxic form of Cr (for a review on the toxic effects of Cr see US EPA 1998) and is known to cause irritation of the skin and the respiratory tract, and lung carcinoma in humans. Occupational exposure to chromate is considered a serious toxicological problem, as it has been demonstrated that Cr(VI) is a human carcinogen (Riveros-Rosas et al. 1997; De Flora 2000). In bacteria, at the extracellular level, Cr(VI) is highly toxic because it rapidly enters to the cytoplasm where it may exert its toxic effects (Wong and Trevors 1988; Katz and Salem 1993). In the cytoplasm, Cr toxicity is mainly related to the process of reduction of Cr(VI) to lower oxidation states [i.e., Cr(III) and Cr(V)] in which free radicals may be formed (Fig. 1) (Shi and Dalal 1990; Kadiiska et al. 1994). Oxidative damage to DNA is probably responsible for the genotoxic effects caused by chromate (Kawanishi et al. 1986; Aiyar et al. 1991; Itoh et al. 1995; Luo et al. 1996).
Toxic effects of Cr(III)
Cr(III) is classified as an essential trace element for humans, since it seems to participate in the metabolism of glucose and lipids (Anderson 1997; Vincent 2004). However, Cr seems not to be required by microorganisms (Wong and Trevors 1988) or plants (Shanker et al. 2005). At the extracellular level, Cr(III) is relatively innocuous as a consequence of its insolubility and subsequent inability to cross cell membranes (Wong and Trevors 1988; Katz and Salem 1993).
Inside the cell, Cr(III) may generate toxic effects by its ability to bind to phosphates in DNA (Kortenkamp et al. 1991, Bridgewater et al. 1994, Plaper et al. 2002) (Fig. 1). The main forms of chromium-DNA adducts in mammalian cells are ternary complexes generated by cross-linking of cysteine and histidine to DNA via a phosphate-bound Cr(III) atom (Zhitkovich et al. 1996). Tyrosine and cysteine exhibited the highest activity in being complexed to DNA by Cr(III) in vitro (Salnikow et al. 1992). Cr(III) may exert additional toxic effects by its ability to bind to carboxyl and sulfhydryl groups in proteins (Levis and Bianchi 1982), and in human cells by competing with the transport of iron by transferrin (Moshtaghie et al. 1992). In Saccharomyces cerevisiae, oxidative damage to proteins has been established as a central mechanism of Cr toxicity (Sumner et al. 2005).
Bacterial mechanisms of chromate resistance
A variety of chromate-resistant bacterial isolates has been reported, and the mechanisms of resistance to this ion may be encoded either by plasmids or by chromosomal genes (Nies et al. 1998; Cervantes and Campos-García 2007). Usually, the genes located in plasmids encode membrane transporters, which directly mediate efflux of chromate ions from the cell’s cytoplasm (Fig. 1). On the other hand, resistance systems encoded within bacterial chromosomes are generally related to strategies such as specific or unspecific Cr(VI) reduction, free-radical detoxifying activities, repairing of DNA damage, and processes associated with sulfur or iron homeostasis (Fig. 1). Table 1 summarizes the bacterial strategies that have been related to chromate tolerance.
Transmembrane efflux of chromate
The efflux of chromate is a resistance mechanism conferred by the ChrA protein (Table 1). ChrA is encoded by plasmids pUM505 of Pseudomonas aeruginosa and pMOL28 from Cupriavidus metallidurans (previously Alcaligenes eutrophus and Ralstonia metallidurans) (Cervantes et al. 1990, Nies et al. 1990). ChrA from P. aeruginosa, of 416 amino acids (aa), displays a topology of 13 transmembrane segments (TMS) (Jiménez-Mejia et al. 2006). ChrA functions as a chemiosmotic pump that effluxes chromate from the cytoplasm using the proton motive force (Alvarez et al. 1999; Pimentel et al. 2002). In vitro and in vivo efflux of chromate showed saturation kinetics with similar Km values of 0.12 and 0.08 mM chromate, respectively (Alvarez et al. 1999; Pimentel et al. 2002). Efflux of chromate is inhibited by sulfate, suggesting that this analog oxyanion may also bind to the ChrA protein (Pimentel et al. 2002). In fact, it has been proposed that ChrA may function as a chromate/sulfate antiporter (Nies et al. 1998) nevertheless, sulfate transport by the ChrA proteins has not yet been determined.
Random mutagenesis of the P. aeruginosa chrA gene showed that most essential amino acid residues are located in the amino terminal end of ChrA (Aguilera et al. 2004). In agreement with this finding, phylogenetic analysis of ChrA homologs revealed that the amino terminal halves are more conserved than the carboxyl terminal halves (Díaz-Pérez et al. submitted). A similar situation was reported for transporters of the closely related major facilitator superfamily (MFS) (Pao et al. 1998). These data suggest that the two halves of ChrA carry out different roles in their transporting functions.
The ChrA protein (401 aa) from Cupriavidus displays a different topology of 10 TMS and shows 29% of identical aa with respect to ChrA from Pseudomonas (Nies et al. 1998). The resistance mechanism, however, seems to be the efflux of chromate ions like in Pseudomonas (Nies et al. 1990). In addition, the C. metallidurans chromosome contains the chrA2 gene, encoding a protein 84% identical to the product of its plasmid-encoded ChrA homolog (chrA1). Expression of the ChrA2 protein also confers chromate resistance (Juhnke et al. 2002). Each chrA determinant increased chromate resistance just two-fold, whereas in the presence of both chrA determinants resistance increased four-fold in low-sulfate medium and five-fold in high-sulfate medium (Juhnke et al. 2002). This is an indication of the importance of both determinants in the chromate resistance by C. metallidurans.
The complete sequencing of plasmid pB4 (79 kilobases) from a Pseudomonas sp. strain revealed the presence of a chrA homologous gene that shares high sequence similarity with the chromate resistance determinant from plasmid pUM505 (93% aa identity) (Tauch et al. 2003). Sequence analysis showed that the chrA gene of pB4 forms part of the Tn5719 transposon. Additionally, the analysis revealed that the chr homologous region in pUM505 still contains remnants of transposon sequences, which suggests that Tn5719 is an ancestor of the chromate resistance determinant of pUM505 (Tauch et al. 2003); chromate resistance by pB4 has not been determined.
In summary, the efflux of chromate seems to be an efficient and widespread mechanism of resistance, which prevents the accumulation of this toxic ion inside the cell.
The CHR superfamily of transporters
The CHR superfamily of transporters, classified as TC # 2.A.51 (Saier 2003), is a group of proteins probably involved in chromate or sulfate transport (Nies et al. 1998). The databases of the CHR protein family currently contain 135 sequences of homologs, including proteins from eukaryotes (Cervantes and Campos-García 2007). With the exception of the P. aeruginosa and C. metallidurans ChrA proteins, the function of other CHR homologs has not yet been analyzed in detail. CHR homologs exist in two sizes (Nies et al. 1998, Díaz-Pérez et al. submitted):
-
(1)
Small proteins, or SCHR (about 200 aa), possess only one domain. Sequence analysis suggests that these proteins may form a paralog group inside the CHR superfamily (see below).
-
(2)
Large proteins, or LCHR (about 400 aa, except eukaryotic proteins of 500–600 aa), with two homologous domains.
Further sequence analysis suggested that LCHR proteins may have derived from a gene duplication event, as occurred with members of other families of transporters (Pao et al. 1998). The fact that several genomes contain two separated tandem copies of the genes for amino- and carboxyl-terminal parts of a large CHR also supports the hypothesis of a different function for each protein half (Nies et al. 1998).
The LCHR proteins are arranged in six subfamilies from bacteria (LCHR1 to LCHR6), and one subfamily from fungi (Díaz-Pérez et al. submitted). The LCHR1 subfamily contains all the Gram positive homologs. The ChrA proteins from C. metallidurans and P. aeruginosa, with a demonstrated function in chromate efflux, are located into the LCHR2 and LCHR5 subfamilies, respectively, which also include mainly proteins from proteobacteria. The LCHR3 subfamily, closer to LCHR2 and LCHR5, may also contain functional chromate transporters. The LCHR4 subfamily includes a protein from Desulfovibrio vulgaris and the only protein from an Archaea (Methanococcus jannaschii). The fungal CHR subfamily contains six proteins from fungal species that are significantly larger than their bacterial counterparts, due to a large interdomain sequence, and are most closely related to the LCHR1 subfamily. No proteins from these subfamilies have yet been studied.
When the large proteins were divided into their moieties amino- and carboxyl-terminal domains and aligned with the small proteins, a separate distribution of the SCHR and the LCHR groups was found (Díaz-Pérez et al. submitted). This suggested that the SCHR proteins form a paralog group inside the CHR family and that they probably carry out a function different to chromate transport.
Thus, the CHR superfamily is a widespread group of proteins, which includes chromate transporters that probably evolved recently as a result of chromate exposure by bacteria.
Chromate reduction
Bacterial reduction of metallic ions has been shown to occur for U(VI), Se(VI), Cr(VI), Mo(VI), Se(IV), Hg(II), Ag(I) and others (Lovley 1993; Bradley and Obraztsova 1998). A wide range of bacteria has been identified that are capable of carrying out a complete reduction of Cr(VI) to Cr(III) by oxidation–reduction reactions of biotic and abiotic nature. Microbial reduction of Cr(VI) to Cr(III) can be considered as an additional chromate resistance mechanism which is not usually a plasmid-associated trait (Cervantes et al. 2001). Cr(VI) reduction outside the cell generates Cr(III) which cannot cross cellular membranes.
Three Cr(VI) reduction mechanisms have been described (Cervantes and Campos-García 2007):
-
(i)
In aerobic conditions, chromate reduction has been commonly associated with soluble chromate reductases that use NADH or NADPH as cofactors.
-
(ii)
Under anaerobiosis, some bacteria, like Pseudomonas fluorescens LB300 (Bopp and Ehrlich 1988), can use Cr(VI) as an electron acceptor in the electron transport chain.
-
(iii)
Reduction of Cr(VI) may also be carried out by chemical reactions associated with compounds such as amino acids, nucleotides, sugars, vitamins, organic acids or glutathione. For instance, ascorbate is capable of reducing Cr(VI), and riboflavin derivatives FAD and FMN are essential coenzymes for chromate-reducing flavoenzymes (Masayasu 1991).
Enzymatic reduction of chromate
Chromate reduction is carried out by diverse bacterial species (Ohtake and Silver 1994; Cervantes et al. 2001) (Table 1). This reduction may be associated with the cell membrane or with the soluble fraction, and may occur either under aerobic or anaerobic conditions. The first enzyme described with the ability to transform Cr(VI) to Cr(III) was a Cr(VI) reductase from chromate-resistant Enterobacter cloacae HO1 (Ohtake et al. 1990). This is a membrane-associated enzyme that transfers electrons to Cr(VI) by NADH-dependent cytochromes (Wang et al. 1990).
Several bacterial Cr(VI) reductases, some conferring resistance to chromate, have been subsequently characterized. These enzymes commonly show a NADH:flavin oxidoreductase activity and can use Cr(VI) as electron acceptor (Gonzalez et al. 2005). Ishibashi et al. (1990) suggested that the ability to reduce chromate may be a secondary function for Cr(VI) reductases, which have a different primary role other than Cr(VI) reduction. The nitroreductases NfsA/NfsB from Vibrio harveyi possess a nitrofurazone nitroreductase as primary activity and a Cr(VI) reductase activity as a secondary function (Kwak et al. 2003). Similarly, ferric reductase FerB from Paracoccus denitrificans uses both Fe(III)-nitrilotriacetate and Cr(VI) as substrates (Mazoch et al. 2004). These secondary functions may be related to the bacterial enzymatic adaptation as a result of the relatively recent increase of Cr(VI) content in the environment due to anthropogenic activities (Silver and Phung 1996).
ChrR from Pseudomonas putida is the currently best studied Cr(VI) reductase. ChrR is a soluble flavin mononucleotide-binding protein (Park et al. 2000). This enzyme functions as a 50-kDa dimer and shows a NADH-dependent reductase activity. This multifunctional protein, besides its role as Cr(VI) reductase, also reduces ferricyanide (Ackerley et al. 2004). Studies with enzyme mutants showed that ChrR protects against chromate toxicity; this is possibly because it preempts chromate reduction by the cellular one-electron reducers, thereby minimizing reactive oxygen species (ROS) generation (Ackerley et al. 2004). During Cr(VI) reduction, ChrR shows a quinone reductase activity that generates a flavin semiquinone. By this reaction, the enzyme transfers >25% of the NADH electrons to superoxide anion and probably produces the Cr(V) species transiently (Fig. 2). Indeed, ChrR in one pathway reduces Cr(VI) to Cr(III), generating intermediary Cr(V) and superoxide anion, and by an additional mechanism reduces quinones, which provide shielding against ROS (Fig. 2). ChrR contains the sequence signature LFVTPEYNXXXXXXLKNAIDXXS as a member of the COG0431 (prokaryotic Cluster of Orthologous Groups of proteins), or KOG4530 (eukaryotic orthologous groups) (Tatusov et al. 2003), named also as NAD(P)H-dependent FMN reductase family (Pfam accession number: PF0358) (Finn et al. 2006). This protein family is a member of the flavoprotein clan that includes FMN- or FAD-binding redox proteins.
Chromate reduction and protection mechanism by Pseudomonas putida ChrR chromate reductase. Cr(VI) is reduced to Cr(V) by ChrR, previously reduced by NADH; Cr(V) is next converted to Cr(III) by diverse biomolecules generating reactive oxygen species (ROS). ROS may be eliminated by alternative mechanisms (i.e. catalases or peroxidases) or by the additional function of ChrR. ChrR in a reduced status may reduce quinones (such as vitamin K or coenzyme Q) which may then detoxify previously formed ROS. ChrROx, ChrRRed, oxidized or reduced forms of the ChrR chromate reductase, respectively; quinonesOx, quinonesRed, oxidized or reduced forms of quinones, respectively; O2·, superoxide radical; LO2·, lipoperoxide radical; H2O2, hydrogen peroxide; Vit. K, vitamin K; CoQ, coenzyme Q. Model based on data from Ackerley et al. 2004 and Gonzalez et al. 2005
The Escherichia coli YieF Cr(VI) reductase shares sequence homology with the P. putida ChrR enzyme (Ackerley et al. 2004). Both soluble enzymes are members of a widespread family of proteins and show similar kinetic and physicochemical properties. YieF has a broad substrate range and can reduce, in addition to Cr(VI), substrates like ferricyanide, vanadium (V), molybdenum (VI), several quinones, 2,6-dichloroindophenol (Ackerley et al. 2004), and even the prodrugs mitomycin C and 5-aziridinyl-2,4-dinitrobenzamide (Barak et al. 2006). The action of YieF involves an obligatory four-electron reduction of Cr(VI) by the protein dimer (50 kDa), in which the enzyme simultaneously transfers three electrons to Cr(VI) to produce Cr(III) and one electron to molecular oxygen generating ROS; no flavin semiquinone is generated during this process (Ackerley et al. 2004). YieF may thus provide to E. coli an effective protection mechanism against chromate toxicity by forming a lower amount of ROS.
Another E. coli enzyme, the Fre flavin reductase, reduces Cr(VI) by a different strategy that involves complexation of Cr(III) with the NAD+ cofactor (Puzon et al. 2002). This interaction may be related to the notorious ability of Cr(III) to form adducts with DNA.
In conclusion, Cr(VI) reduction seems to be an efficient system of resistance to chromate in bacteria; however, the use of alternative substrates in addition to Cr(VI) by chromate reductases suggests that this reduction activity has been an adaptive mechanism promoted by recent chromate exposure.
The NAD(P)H-dependent FMN reductase family
The NAD(P)H-dependent FMN reductase (FMN_red) protein family, which includes putative ChrR orthologs, currently comprises 243 homologous dimeric or tetrameric proteins that bind the FMN cofactor. Members of the group are widespread, suggesting an early evolutionary origin of this protein family. The utilization of NAD(P)H and the absence of a flavin semiquinone radical distinguish this protein family from flavodoxins, which adopt the same structural fold, i.e. a five-stranded β sheet sandwiched by five α helices (Deller et al. 2006). The FMN_red protein family may be divided into ten main clusters, where each one probably corresponds to different protein subfamilies. Only three of the above mentioned protein clusters include characterized proteins and can be defined as subfamilies, using criteria outlined previously for other protein groups (Riveros-Rosas et al. 2003).
Subfamily I is the most numerous group (73 homologous sequences), and is present mainly in proteobacteria (Fig. 3). The ChrR enzyme of P. putida (accession number AAK56852) is the only well-characterized protein included inside this subfamily (Fig. 3).
Phylogenetic analysis of the NAD(P)H-dependent FMN reductase protein family. Shown is the consensus tree constructed with the minimum evolution (ME) method using available protein sequences that belong to the three protein subfamilies containing at least one characterized protein member (highlighted in yellow). Trees were calculated using MEGA 3.1 (Kumar et al. 2004). Similar topologies were obtained using the neighbor-joining (NJ), maximum parsimony (MP) and unweighted pair-group method using arithmetic averages (UPGMA) methods. Entries include sequence access number, abbreviated species name, and amino acid length. Subfamily boundaries are indicated with yellow capped pins; subfamilies I, II and III are indicated with blue, red and brown lines, respectively. Dots bars indicate nodes supported in >70% (open), >80% (gray), or >90% (filled) of 1,000 random bootstrap replicates of all UPGMA, NJ, ME and MP trees. Scale bar represents 0.1 amino acid substitutions per site
Subfamily II, with 32 homologous proteins, is present in archaea, bacteria, mainly proteobacteria, and plants (Fig. 3). The YieF protein from E. coli (Ackerley et al. 2004), the FMN reductase from P. aeruginosa PAO1 (Agarwal et al. 2006) and the NAD(P)H:quinone reductase (NQR) of Arabidopsis thaliana (Sparla et al. 1999) are included in this subfamily (Fig. 3).
Subfamily III comprises nine homologous proteins reported in firmicutes, fungi, and mycetozoa (Dictyostelium discoideum) (Fig. 3). Two characterized proteins belong to this subfamily: the azoreductase from Bacillus sp. OY1-2 (Suzuki et al. 2001) and the dimeric S. cerevisiae YLR011wp protein (Liger et al. 2004) (Fig. 3). The Bacillus protein transforms azo dyes into colourless compounds, a reaction mediated by a reductase activity for the azo group in the presence of NADPH (Suzuki et al. 2001). YLR011wp from S. cerevisiae also shows a weak but specific reductive activity over azo dyes and nitrocompounds, in addition to a strong ferricyanide reductase activity (Liger et al. 2004).
In summary, the multifunctional abilities of the FMN_red family members make it unlikely that the primary role of this protein family is chromate reduction; this is even more possible if, as mentioned above, chromium in nature is present primarily as Cr(III), and the introduction of the Cr(VI) species in the environment is a relatively recent event.
Protection against oxidative stress
Since the generation of ROS occurs during Cr(VI) reduction to Cr(III) (Fig. 1), the participation of bacterial proteins in the defense against oxidative stress induced by chromate represents an additional mechanism of chromate resistance (Table 1).
E. coli displays several chromate protective systems, including the activation of enzymes such as superoxide dismutase (SOD) and catalase (Ackerley et al. 2006). Additionally, chromate exposure in E. coli led to the depletion of the pools of glutathione and other thiols, suggesting that these compounds have an important detoxifying role against Cr(VI) (Ackerley et al. 2006).
A microarray analysis of Cr(VI)-exposed cultures of Caulobacter crescentus, a bacterium known for their distinctive ability to live in polluted habitats, showed the up-regulation of genes involved in the response to heavy-metal toxicity such as those encoding SOD, glutathione S-transferase, thioredoxin, and glutaredoxin. This indicates that C. crescentus employs different processes to counteract oxidative stress upon exposure to chromate (Hu et al. 2005).
Shewanella oneidensisMR-1, a metal-reducing bacterium, when exposed to chromate for 90 min, up-regulated genes involved in cellular detoxification (Brown et al. 2006). In addition, a 24-h (chronic) Cr(VI) exposure of S. oneidensis, as revealed by transcriptome and proteome analysis, induced genes encoding thioredoxins and glutaredoxins. The induction of detoxification and stress response genes seems to play an important role in the adaptation of S. oneidensis under anoxic metal-reducing conditions (Chourey et al. 2006).
Besides chromosomal genes, plasmids may also encode systems devoted to protect bacterial cells from the oxidative stress caused by chromate. Plasmid pMOL28 from C. metallidurans, which encodes the ChrA chromate efflux pump, in addition encodes the ChrC and ChrE proteins that seem to be also involved in chromate resistance (Juhnke et al. 2002). ChrC (197 aa) shows homology to iron-containing SOD enzymes able to detoxify superoxide radicals; however, the low activity of this probable SOD enzyme precluded the authors to assign ChrC a clear function (Juhnke et al. 2002). ChrE (113 aa) may participate in the cleavage of chromium-glutathione complexes on the basis of its homology to members of the rhodanese superfamily (Juhnke et al. 2002). The involvement of the rhodanese RdhA in the acquisition of sulfur compounds was already reported in Synechococcus sp. (Laudenbach et al. 1991). Some proteins from the rhodanese superfamily are involved in detoxification of compounds causing oxidative stress, suggesting that ChrE may play a similar role in C. metallidurans.
In conclusion, enzymes, that participate in detoxification of ROS generated after Cr(VI) exposure and reduction may be involved in the protection against the deleterious effects of chromate.
DNA repair
Another defensive shield against Cr toxicity is the protection of bacterial cells from DNA damage caused by chromium compounds (Fig. 1). Cr(VI) has long been known to induce the E. coli SOS repair system that protects DNA from oxidative damage (Llagostera et al. 1986). Components of the recombinational DNA repair system, like DNA helicases RecG and RuvB, were also shown to participate in the response to DNA damage caused by chromate in P. aeruginosa (Miranda et al. 2005). Similarly, after 24 h of Cr(VI) exposure of S. oneidensis, the SO0368, uvrD, and hrpA genes, which encode helicases, were induced (Chourey et al. 2006). C. crescentus also showed the up-regulation of genes related to repair of DNA damage (endonucleases, RecA protein) in response to Cr(VI) treatment (Hu et al. 2005).
Other mechanisms of resistance to chromate
Additional protective systems of the Cr toxic effects are probably associated with a reduced uptake of Cr(VI) by the sulfate uptake pathway (Fig. 1) and with sulfur or iron homeostasis (Table 1). C. crescentus seems not to have a chromate efflux system, but Cr stress down-regulates a sulfate transport system probably reducing chromate uptake (Hu et al. 2005).
SrpC, encoded by plasmid pANL from Synechococcus sp., is a sulfur-regulated protein of 393 aa that shows 62% of amino acid identity with the P. aeruginosa ChrA protein (Nicholson and Laudenbach 1995). SrpC is located into the LCHR2 subgroup of the CHR superfamily but may be involved in sulfate uptake instead of extruding chromate ions. Interestingly, plasmid pANL also encodes the SrpA protein, with sequence similarity to catalases. SrpA was proposed to participate in the detoxification of hydrogen peroxide that may help diminishing Cr oxidative damages (Nicholson and Laudenbach 1995).
Exposure to Cr(VI) in S. oneidensis caused the up-regulation of genes involved in sulfate transport; this suggested the possibility of chromate-induced sulfur limitation, perhaps through the competitive inhibition of sulfate uptake by chromate (Brown et al. 2006). S. oneidensis also showed the enhanced expression of genes encoding proteins involved in sulfur metabolism (adenylyl sulfate kinase, sulfite reductase) and in iron binding (ferritin) and transport (siderophore biosynthesis, heme transport). It has been suggested that uptake of iron prevents the generation of highly reactive hydroxyl radicals via Fenton reactions thus lowering the toxic effects of chromate (Brown et al. 2006).
Conclusions
Microorganisms have evolved diverse resistance mechanisms to cope with chromate toxicity. These systems include direct strategies that involve the efflux of toxic chromate ions from the cytoplasm or the transformation of Cr(VI) to innocuous Cr(III) outside the cell. Several probable Cr(VI) membrane transporters have been identified and they have been grouped into a large superfamily, although only two bacterial homologous able to extrude chromate are well characterized. Many bacterial species are reported to reduce Cr(VI) to Cr(III), but the biochemical properties of only a few Cr(VI) reductases have been elucidated. The diverse characteristics of these ancient enzymes and their wide distribution support the hypothesis that reduction of chromate is a secondary role for Cr reductases.
Diverse bacterial species seem to display indirect systems of tolerance to Cr. After chromate exposure, these bacteria show a varied regulatory network that involves the expression of genes for several different metabolic processes as a Cr stress defensive strategy. These include genes for sulfur or iron homeostasis and ROS detoxification. These indirect systems of tolerance to Cr include mechanisms focused to maintain the integrity of the cells by protecting them from oxidative stress or to repair the damages caused by Cr derivatives.
References
Ackerley DF, Gonzalez CF, Park CH, Blake R, Keyhan M, Matin A (2004) Chromate-reducing properties of soluble flavoproteins from Pseudomonas putida and Escherichia coli. Appl Environ Microbiol 70:873–882
Ackerley DF, Barak Y, Lynch SV, Curtin J, Matin A (2006) Effect of chromate stress on Escherichia coli K-12. J Bacteriol 188:3371–3381
Agarwal R, Bonnano JB, Burley SK, Swaminathan S (2006) Structure determination of an FMN-reductase from Pseudomonas aeruginosa PAO1 using sulfur anomalous signal. Acta Crystallogr D 62:383–391
Aguilera S, Aguilar ME, Chávez MP, López-Meza JE, Pedraza-Reyes M, Campos-García J, Cervantes C (2004) Essential residues in the chromate transporter ChrA of Pseudomonas aeruginosa. FEMS Microbiol Lett 232:107–112
Aiyar J, Berkovits HJ, Floyd RA, Wetterhahn KE (1991) Reaction of chromium(VI) with glutathione or with hydrogen peroxide: identification of reactive intermediates and their role in chromium(VI)-induced DNA damage. Environ Health Perspect 92:53–62
Alvarez AH, Moreno-Sánchez R, Cervantes C (1999) Chromate efflux by means of the ChrA chromate resistance protein from Pseudomonas aeruginosa. J Bacteriol 181:7398–7400
Anderson RA (1997) Chromium as an essential nutrient for humans. Regul Toxicol Pharmacol 26:535–541
Barak Y, Thorne SH, Ackerley DF, Lynch SV, Contag CH, Matin A (2006) New enzyme for reductive cancer chemotherapy, YieF, and its improvement by directed evolution. Mol Cancer Ther 5:97–103
Bopp LH, Erlich HL (1988) Chromate resistance and reduction in Pseudomonas fluorescens strain LB300. Arch Microbiol 150:426–431
Bradley MT, Obraztsova AY (1998) Sulfate-reducing bacterium grows with Cr(VI), U(VI), Mn(IV) and Fe(III) as electron acceptor. FEMS Microbiol Lett 162:193–198
Bridgewater LC, Manning FC, Woo ES, Patierno SR (1994) DNA polymerase arrest by adducted trivalent chromium. Mol Carcinog 9:122–133
Brown SD, Thompson MR, VerBerkmoes NC, Chourey K, Shah M, Zhou J, Hettich RL, Thompson DK (2006) Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics 5:1054–1071
Cary EE (1982) Chromium in air, soil and natural waters. In: Langard S (ed) Biological and environmental aspects of chromium. Elsevier, Amsterdam, pp 48–64
Cervantes C, Ohtake H, Chu L, Misra T, Silver S (1990) Cloning, nucleotide sequence, and expression of the chromate resistance determinant of Pseudomonas aeruginosa plasmid pUM505. J Bacteriol 172:287–291
Cervantes C, Campos-García J, Devars S, Gutiérrez-Corona F, Loza-Tavera H, Torres-Guzmán JC, Moreno-Sánchez R (2001) Interactions of chromium with microorganisms and plants. FEMS Microbiol Rev 25:335–347
Cervantes C, Campos-Garcia J. (2007) Reduction and efflux of chromate by bacteria. In: Nies DH, Silver S (eds) Molecular Microbiology of Heavy Metals. Springer-Verlag, Berlin, pp 407–420
Chourey K, Thompson MR, Morrell-Falvey J, VerBerkmoes NC, Brown SD, Shah M, Zhou J, Doktycz M, Hettich RL, Thompson DK (2006) Global molecular and morphological effects of 24-hour chromium(VI) exposure on Shewanella oneidensis MR-1. Appl Environ Microbiol 72:6331–6344
De Flora S (2000) Treshold mechanisms and site specificity in chromium (VI) carcinogenesis. Carcinogenesis 21:533–541
Deller S, Sollener S, Trenker-El-Toukhy R, Jelesarov I, Gubitz GM, Macheroux P (2006) Characterization of a thermostable NADPH:FMN oxidoreductase from the mesophilic bacterium Bacillus subtilis. Biochemistry 45:7083–7091
Ehrlich HL (2002) How microbes mobilize metals in ores: A view of current understandings and proposals for further research. Miner Metall Process 19:220–224
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Assmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251
Gonzalez CF, Ackerley DF, Lynch SV, Matin A (2005) ChrR, a soluble quinone reductase of Pseudomonas putida that defends against H2O2. J Biol Chem 280:22590–22595
Hu P, Brodie EL, Suzuki Y, McAdams HH, Andersen GL (2005) Whole-genome transcriptional analysis of heavy metal stresses in Caulobacter crescentus. J Bacteriol 187:8437–8449
Ishibashi Y, Cervantes C, Silver S (1990) Chromium reduction in Pseudomonas putida. Appl Environ Microbiol 56:2268–2270
Itoh M, Nakamura M, Suzuki T, Kawai K, Horitsu H, Takamizawa K (1995) Mechanism of chromium(VI) toxicity in Escherichia coli: is hydrogen peroxide essential in Cr(VI) toxicity? J Biochem 117:780–786
Jiménez-Mejía R, Campos-García J, Cervantes C (2006) Membrane topology of the chromate transporter ChrA of Pseudomonas aeruginosa. FEMS Microbiol Lett 262:178–184
Juhnke S, Peitzsch N, Hubener N, Groβe C, Nies DH (2002) New genes involved in chromate resistance in Ralstonia metallidurans strain CH34. Arch Microbiol 179:15–25
Kadiiska MB, Xiang QH, Mason RP (1994) In vivo free radical generation by chromium (VI): An electron resonance spin-trapping investigation. Chem Res Toxicol 7:800–805
Katz SA, Salem H (1993) The toxicology of chromium with respect to its chemical speciation: a review. J Appl Toxicol 13:217–224
Kawanishi S, Inoue S, Sano S (1986) Mechanism of DNA cleavage induced by sodium chromate (VI) in the presence of hydrogen peroxide. J Biol Chem 261:5952–5958
Khasim DI, Kumar NV, Hussain RC (1989) Environmental contamination of chromium in agricultural and animal products near a chromate industry. Bull Environ Contam Toxicol 43:742–746
Kortenkamp A, O’Brien P, Beyersmann D (1991) The reduction of chromate is a prerequisite of chromium binding to cell nuclei. Carcinogenesis 12:1143–1144
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kwak YH, Lee DS, Kim HB (2003) Vibrio harveyi nitroreductase is also a chromate reductase. Appl Environ Microbiol 69:4390–4395
Laudenbach DE, Ehrhardt D, Green L, Grossman A (1991) Isolation and characterization of a sulfur-regulated gene encoding a periplasmically localized protein with sequence similarity to rhodanese. J Bacteriol 173:2751–2760
Levis AG, Bianchi V (1982) Mutagenic and cytogenetic effects of chromium compounds. In: Langard S (ed) Biological and environmental aspects of chromium. Elsevier, Amsterdam, pp 171–208
Liger D, Graille M, Zhou CZ, Leulliot N, Quevillon-Cheruel S, Blondeau K, Janin J, Van Tilbeurgh H (2004) Crystal structure and functional characterization of yeast YLR011wp, an enzyme with NAD(P)H-FMN and ferric iron reductase activities. J Biol Chem 279:34890–34897
Llagostera M, Garrido S, Guerrero R, Barbé J (1986) Induction of SOS genes of Escherichia coli by chromium compounds. Environ Mutagen 8:571–577
Lovley DR (1993) Dissimilatory metal reduction. Annu Rev Microbiol 47:263–290
Luo H, Lu Y, Shi X, Mao Y, Dalal NS (1996) Chromium (IV)-mediated Fenton-like reaction causes DNA damage: implication to genotoxicity of chromate. Ann Clin Lab Sci 26:185–191
Masayasu S (1991) Effects of vitamins on chromium(VI)-induced damage. Environ Health Perspect 92:63–70
Mazoch J, Tesarik R, Sedlacek V, Kucera I, Turanek J (2004) Isolation and biochemical characterization of two soluble iron (III) reductases from Paracoccus denitrificans. Eur J Biochem 271:553–562
Miranda AT, González MV, González G, Vargas E, Campos-García J, Cervantes C (2005) Involvement of DNA helicases in chromate resistance by Pseudomonas aeruginosa PAO1. Mutat Res 578:202–209
Moshtaghie AA, Ani M, Bazrafshan MR (1992) Comparative binding study of aluminum and chromium to human transferrin. Effect of iron. Biol Trace Elem Res 32:39–46
Nicholson ML, Laudenbach DE (1995) Genes encoded on a cyanobacterial plasmid are transcriptionally regulated by sulfur availability and CysR. J Bacteriol 177:2143–2150
Nies A, Nies DH, Silver S (1990) Nucleotide sequence and expression of a plasmid-encoded chromate resistance determinant from Alcaligenes eutrophus. J Biol Chem 265:5648–5653
Nies DH, Koch S, Wachi S, Peitzsch N, Saier MH (1998) CHR, a novel family of prokaryotic proton motive force-driven transporters probably containing chromate/sulfate antiporters. J Bacteriol 180:5799–5802
Ohtake H, Fuji E, Toda K (1990) Bacterial reduction of hexavalent chromium: Kinetic aspects of chromate reduction by Enterobacter cloacae HO1. Biocatalysis 4:227–235
Ohtake H, Silver S (1994) Bacterial detoxification of toxic chromate. In: Chaudhry GR (ed) Biological degradation and bioremediation of toxic chemicals. Dioscorides, Portland, OR, pp 403–415
Pao SS, Paulsen IT, Saier MH (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34
Park CH, Keyhan M, Wielinga B, Fendorf S, Matin A (2000) Purification to homogeneity and characterization of a novel Pseudomonas putida chromate reductase. Appl Environ Microbiol 66:1788–1795
Pimentel BE, Moreno-Sánchez R, Cervantes C (2002) Efflux of chromate by cells of Pseudomonas aeruginosa expressing the ChrA protein. FEMS Microbiol Lett 212:249–254
Plaper A, Jenko-Brinovec S, Premzl A, Kos J, Raspor P (2002) Genotoxicity of trivalent chromium in bacterial cells. Possible effects on DNA topology. Chem Res Toxicol 15:943–949
Puzon GJ, Petersen JN, Roberts AG, Kramer DM, Xun L (2002) A bacterial flavin reductase system reduces chromate to a soluble chromium(III)-NAD(+) complex. Biochem Biophys Res Commun 294:76–81
Rai D, Sass BM, Moore DA (1987) Chromium(III) hydrolysis constants and solubility of chromium(III) hydroxide. Inorg Chem 26:345–349
Riley RG, Zachara JM, Wobber FJ (1992) Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. Report DOE/ER-0547T. US Department of Energy, Washington, DC
Riveros-Rosas H, Pfeifer GD, Lynam DR, Pedroza JL, Julián-Sánchez A, Canales O, Garfias J (1997) Personal exposure to elements in Mexico City air. Sci Total Environ 198:79–96
Riveros-Rosas H, Julián-Sánchez A, Villalobos-Molina R, Pardo JP, Piña E (2003) Diversity, taxonomy and evolution of medium-chain dehydrogenase/reductase superfamily. Eur J Biochem 270:3309–3334
Saier MH Jr (2003) Tracing pathways of transport protein evolution. Mol Microbiol 48:1145–1156
Salnikow K, Zhitkovich A, Costa M (1992) Analysis of the binding sites of chromium to DNA and protein in vitro and in intact cells. Carcinogenesis 13:2341–2346
Shanker AK, Cervantes C, Loza-Tavera H, Avudainayagam S (2005) Chromium toxicity in plants. Environ Int 31:739–753
Shi X, Dalal NS (1990) On the hydroxyl radical formation in the reaction between hydrogen peroxide and biologically generated chromium (V) species. Arch Biochem Biophys 277:342–350
Silver S, Phung LT (1996) Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol 50:53–89
Sparla F, Tedeschi G, Pupillo P, Trost P (1999) Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT_diaphorase. FEBS Lett 463:382–386
Sumner ER, Shanmuganathan S, Sideri TC, Willets SA, Houghton JE, Avery SV (2005) Oxidative protein damage causes chromium toxicity in yeast. Microbiology 151:1939–1948
Suzuki Y, Yoda T, Ruhul A, Sugiura W (2001) Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1–2 isolated from soil. J Biol Chem 276:9059–9065
Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41
Tauch A, Schluter A, Bischoff N, Goesmann A, Meyer F, Puhler A (2003) The 79,370-bp conjugative plasmid pB4 consists of an IncP-1β backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene bla NPS-1, and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol Gen Genomics 268:570–584
US Environmental Protection Agency (1998) Toxicological review of hexavalent chromium. CAS No. 18540–29–9. Washington, DC, 77 pp
Vincent JB (2004) Recent developments in the biochemistry of chromium (III). Biol Trace Elem Res 99:1–16
Wang P, Mori T, Toda K, Ohtake H (1990) Membrane-associated chromate reductase activity from Enterobacter cloacae. J Bacteriol 172:1670–1672
Wong PT, Trevors JT (1988) Chromium toxicity to algae and bacteria. In: Nriagu JO, Nieboer E (eds), Chromium in the natural and human environments. Wiley, New York, pp 305–315
Zayed A, Terry N (2003) Chromium in the environment: factors affecting biological remediation. Plant Soil 249:139–156
Zhitkovich A, Voitkun V, Costa M (1996) Formation of the amino acid-DNA complexes by hexavalent and trivalent chromium in vitro: importance of trivalent chromium and the phosphate group. Biochemistry 35:7275–7282
Acknowledgments
Research in our laboratories was supported by grants from CIC (Universidad Michoacana) and CONACYT (No. 41712-Q).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ramírez-Díaz, M.I., Díaz-Pérez, C., Vargas, E. et al. Mechanisms of bacterial resistance to chromium compounds. Biometals 21, 321–332 (2008). https://doi.org/10.1007/s10534-007-9121-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-007-9121-8