Abstract
Shrub encroachment frequently occurs in arid and semi-arid grasslands worldwide and affects the regional carbon balance. Many previous studies have revealed the effects of shrub encroachment on bulk carbon content of grasslands, but molecular evidence is surprisingly lacking. In this study, we examined the chemical composition of plant tissues and soil organic carbon (SOC), and soil microbial communities to identify the effects of shrub (Caragana microphylla) encroachment on SOC storage in the top layer (0–10 cm) along a gradient of natural shrub cover in the grasslands of Inner Mongolia. We found that SOC in the shrub patches was derived mainly from leaves, whereas SOC in the grassy matrix was composed of a mixture of fresh root- and leaf-derived compounds. Compared with pure grassland, the SOC decreased by 29% in the shrub-encroached grasslands (SEGs), and this decrease was enhanced by increasing shrub cover. We also found that free lipids and lignin-derived phenols increased while the ratios of ω-C18/∑C18 and suberin/cutin decreased with increasing shrub cover. In addition, the ratios of fungal to bacterial phospholipid fatty acids (PLFAs) and gram-negative to gram-positive bacterial PLFAs decreased with increasing shrub cover. These results indicate that the encroachment of nitrogen-rich legume shrubs can lead to carbon loss by altering the chemical composition of plant inputs as well as the soil microbial community in grassland ecosystems.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Soil organic carbon (SOC) is the most important carbon stock in terrestrial ecosystems (Schmidt et al. 2011), and even small changes in soil carbon could have large effects on the global terrestrial carbon cycle (Scharlemann et al. 2014). Over the past 150 years, shrub encroachment resulting from climate change and land use (e.g., overgrazing and reduced fire frequency) has become a major issue in arid and semi-arid ecosystems, which cover approximately 45% of Earth’s land surface (Bailey 1996; Van Auken 2009). Shrub encroachment has substantial effects on the ecosystem functioning because of changes in habitat heterogeneity, species composition, aboveground biomass and SOC, thereby affecting the regional carbon balance (Eldridge et al. 2011; Sutherland et al. 2013).
The evaluation of the influence of shrub encroachment on global biogeochemical cycles via effects on soil carbon storage has been a major topic (Liu et al. 2011). SOC has been reported to increase (Springsteen et al. 2010; Barger et al. 2011; Zhou et al. 2017), remain unchanged (Lett et al. 2004) or decrease (Jackson et al. 2002; Oelofse et al. 2016) after shrub encroachment. These inconsistencies might be attributable to differences in climate, vegetation type, soil properties, historical land use patterns and scales (Jackson et al. 2002; Barger et al. 2011; Li et al. 2016). Shrub encroachment usually creates “fertility islands” in grasslands (Schlesinger et al. 1996), and increases in SOC with shrub size and decreases along the shrub core-to-edge line gradient have been observed (Throop and Archer 2008; Bai et al. 2013). These studies reveal that shrub encroachment changes the within-site spatial distribution of soil resources and alters the resource flow (Li et al. 2008; Eldridge et al. 2011), which makes it difficult to accurately evaluate the effects of shrub encroachment on SOC due to the limited micro scale (Zhou et al. 2017). Shrub encroachment usually results in a landscape where shrub patches (shrub canopy) are embedded in the grassy matrix (natural grasslands), which is defined as a shrub-encroached grassland (SEG) (Fig. 1a; Chen et al. 2015). Shrub encroachment usually alters light, water, nutrient and plant–plant interactions (Asner and Heidebrecht 2005; Ratajczak et al. 2012; Peng et al. 2013), which makes SEG different from pure grasslands (D’Odorico et al. 2012). Many studies have assessed these effects by comparing SEG and pure grasslands at the plot level. For example, Eldridge et al. (2011) concluded that shrub encroachment increases SOC globally in drylands. Similarly, Li et al. (2016) observed an increase in SOC content in wetter regions and a greater rate of increase in grassland encroached by leguminous shrubs. However, molecular-level evidence to support these results is surprisingly lacking.
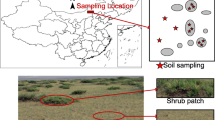
Illustration of sampling location and design. a Sampling location and photos; b twelve plots that represent a natural shrub cover gradient and five-pointed stars represent composite soil samples; c plant samples collected in the study area. Except for S. grandis and C. korshinskyi, which were collected in both the shrub patch and grassy matrix, the other herbs were collected only in the grassy matrix
SOC is mainly derived from aboveground litter, root litter and exudates and is metabolized by microorganisms (Armas-Herrera et al. 2016). The quality and quantity of plant litter together with soil microbial biomass are important drivers of SOC accumulation (Hibbard et al. 2001; Carbone and Trumbore 2007). Plant litter consists of polysaccharides (such as cellulose), aromatics (such as lignin and tannic acid) and aliphatics (such as waxes, cutin and suberin) (Crow et al. 2009), and these molecular components remain intact in the soil. Fontaine et al. (2007) and Kuzyakov (2010) reported that recalcitrant plant inputs led to higher carbon stocks, whereas labile plant inputs led to lower carbon stocks. Similarly, carbon stocks are also regulated by the decomposition and resynthesis of plant inputs by the microbial community (Kuzyakov 2010; Schmidt et al. 2011; Tamura and Tharayil 2014). Therefore, understanding the changes in plant inputs and the chemical composition of SOC as well as the soil microbial community after shrub encroachment should greatly clarify the effects of shrub encroachment on the carbon cycle (DeMarco et al. 2016). Previous studies have shown that shrub encroachment could alter the quality and quantity of plant inputs (Filley et al. 2008; Knapp et al. 2008; Throop and Archer 2008) and microbial communities (Li et al. 2017). However, how the input chemistry and microbial community respond to the shrub cover gradient remain unknown.
Previous studies have hypothesized that the impacts of encroachment increase with woody cover and woody biomass (Ratajczak et al. 2012). Many previous studies have examined SOC changes with the shrub encroachment process using natural shrub cover gradients. For example, Blaser et al. (2014) found that total soil carbon stocks increased along a natural cover gradient from 0% (open grassland) to 100% (shrubland) in a vast floodplain ecosystem in central Zambia. Similarly, Gonzalez-Roglich et al. (2014) found that SOC stocks increased along a woody plant cover gradient (0–94%) in savannas of central Argentina. However, these studies focused on a large grassland-shrubland transition zone and did not eliminate the effect of disturbances due to differences in climate and environment. Therefore, a study focused on the SEG along a natural shrub cover at the same location could distinguish plant–microbe interactions from climatic and soil factors.
Mongolian grassland is the most typical temperate grassland in the world and has experienced increasing temperature and livestock densities over the past several decades (Fang et al. 2015). A previous study showed that the area of SEG has reached 5.1 × 106 ha in Inner Mongolia (Peng et al. 2013). In this study, we explored the effects of shrub encroachment on SOC in the top layer (0–10 cm) and its chemical composition along a natural shrub cover gradient in the Xilinhot grassland. The chemical composition of plant organic carbon (OC) and the soil microbial community were also examined. Specifically, the objectives of this study were to (1) investigate the chemical composition of plant-derived OC in the top layer of soil of shrub patches and grassy matrix, (2) demonstrate the contribution of aboveground and belowground plant inputs to SOC in the shrub patch and grassy matrix, and (3) reveal the effects of the chemical composition of plant inputs and the microbial community on the SOC accumulation in the top layer with increasing shrub cover when treating SEG as a vegetation type.
Materials and methods
Study site and sampling
Our study was conducted in a SEG near Xilinhot City in the Inner Mongolia Autonomous Region (43.93°N, 116.20°E; Fig. 1a). The mean annual temperature is 1.8 °C and ranges from − 20.1 °C in January to 20.2 °C in July. The mean annual precipitation is 293.6 mm, and precipitation is concentrated in the period from June to August. The natural community is Leymus chinensis + Stipa grandis, and the common herbs include Cleistogenes squarrosa, Agropyron cristatum, Artemisia frigida, Kochia prostrata, Allium ramosum and Iris tenuifolia. The dominant shrub is a nitrogen-fixing leguminous shrub, C. microphylla. The soil is classified as dark chestnut soil with a loamy clay texture (Chen et al. 2015).
Twelve plots with a size of 20 × 20 m were established along a transect with a natural shrub cover gradient in August 2015. The long and short crown diameters of all shrub canopy patches were measured to calculate the shrub cover at the plot scale. Then, we set up 5 subplots of 6 × 6 m at the quadrangles and centre of each plot. In each subplot, we collected soil samples from the top layer (0–10 cm) using a stainless-steel core from a randomly selected shrub patch and the neighbouring grassy matrix (Fig. 1b). To avoid heterogeneity in terms of the canopy size in each subplot, 1, 3 or 5 core samples under the shrub canopy were mixed into one sample (Fig. 1b). We sampled only in the grassy matrix in the plot without shrubs. All soil samples were kept frozen at − 80 °C until being freeze dried. The plant roots were carefully removed from the soil using tweezers, and the soil were ground thoroughly using a grinder. Then, composite samples of soil from the same plot and same position (in the shrub patch or the grassy matrix) (n = 23) were prepared.
The leaves and roots of the shrub (C. microphylla) and dominant herbs (S. grandis, Carex korshinskyi, A. frigida, Melilotoides ruthenica and I. tenuifolia) were collected in July 2016. We established three 20 × 20 m plots in the soil sampling location. Three repeats of specific species samples were collected from each plot, and each repeat was collected from different individuals. With the exception of S. grandis and C. korshinskyi, which were collected in both the shrub patch and grassy matrix, the other herbs were collected only in the grassy matrix. Moreover, we established paired 1-m2 quadrats in both the shrub patch and the neighbouring grassy matrix in each plot and then clipped the roots and leaves of herb plants in 0.25 m2 subquadrats to make mixed herb samples (Fig. 1c). Plant samples were frozen at − 20 °C until drying at 40 °C. The samples were ground thoroughly using a grinder. Composite leaves and roots samples for the specific species (n = 16) and for mixed herb samples within shrub patches and the grassy matrix (n = 4) were prepared.
Soil carbon and nitrogen measurements, chemical extraction and quantification
The soil total carbon and nitrogen content were measured using an elementary analyser (Vario EL III, Elementar, Germany), and the inorganic carbon content was measured by a carbonate content analyser (Eijkelkamp 08.53, Netherlands). The SOC was calculated by subtracting the inorganic carbon from the total carbon content.
The chemical extractions included solvent extraction (for free lipids), base hydrolysis (for bound lipids) and copper(II) oxide (CuO) oxidation (for lignin-derived phenols) (Feng et al. 2010; Zhao et al. 2014).
Samples (~ 6 g soil or ~ 0.2 g plant) were extracted with 15 mL (4 mL for plant samples) of chromatography-grade dichloromethane (DCM): methanol (MeOH) (1:1; v:v) by ultrasonication (15 min) and centrifugation (3000 rpm for 10 min) three times. The combined solvent extracts were filtered through glass fibre columns, concentrated by rotary evaporation and transferred to 2 mL glass vials.
After the solvent extraction, the air-dried residues were subjected to base hydrolysis and oxidized with CuO. Approximately 3 g soil residues (~ 0.1 g plant residues) was sealed in Teflon-lined bombs and heated at 100 °C for 3 h with 15 mL of 1 mol/L KOH/MeOH. After cooling, the suspension was transferred into a 100-mL round-bottom flask and acidified to pH 1 with 6 mol/L HCl. The bound lipids were recovered by liquid–liquid extraction with 20 mL ethyl acetate and concentrated by rotary evaporation three times after removing the remaining water with anhydrous Na2SO4. Then, the product was dried under N2 gas and methylated for 90 min at 70 °C with 1 mL of 14% BF3:MeOH. The bound lipids were obtained by lipid–lipid extraction with 2 mL hexane three times and transferred to 2 mL glass vials.
The air-dried residues (soil and plant) remaining after solvent extraction were sealed in Teflon-lined bombs and heated at 170 °C for 2.5 h with 1 g CuO, 100 mg ammonium iron(II) sulphate hexahydrate and 15 mL of 2 mol/L NaOH after oxygen exhaustion with N2 gas. The mixture was centrifuged at 3000 rpm for 10 min, and the supernatant was then transferred into another centrifuge tube and acidified to pH 1 with 6 mol/L HCl. The extracts were kept at room temperature in the dark for 1 h to prevent the reaction of cinnamic acids. After centrifugation (3000 rpm for 10 min), the lignin-derived phenols were recovered by liquid–liquid extraction with 20 mL ethyl acetate three times, concentrated by rotary evaporation and transferred to 2 mL glass vials.
Products from the solvent extraction, base hydrolysis and CuO oxidation were dried under N2 gas, and all biomarkers were trimethylsilyl (TMS)-derivatized by reaction with 100 µL N,O-bis-(trimethylsilyl)trifluoroacetamide (BSTFA) and 100 µL pyridine for 2 h at 60 °C. Biomarkers from solvent extraction and base hydrolysis were diluted with DCM, whereas the biomarkers from CuO oxidation were diluted with ethyl acetate.
All biomarkers were identified by gas chromatography-mass spectrometry (GC-MS) on an Agilent 7890A gas chromatograph (Agilent Technologies, Palo Alto, CA, USA) coupled to an Agilent 5973 N quadruple mass selective detector and were quantified with a flame ionization detector (FID) coupled to the Agilent 7890A instrument. The biomarkers from the solvent extraction and base hydrolysis were quantified by comparing the peak areas of biomarkers to that of an internal standard (squalane for free lipids and 5α-cholestane for bound lipids), whereas the amounts of lignin-derived phenols were quantified using the calibration curves of eight lignin-phenol standards. Details of the instrument configuration are provided in Zhao et al. (2014).
Estimation of soil microbial phospholipid fatty acid (PLFA)
The microbial communities were evaluated based on the PLFAs extracted from freeze-dried soil samples (Bligh and Dyer 1959). Briefly, approximately 8 g of soil was extracted with a MeOH:chloroform:citric acid solution (2:1:0.8; v:v:v). Then, the phospholipids were recovered from the glycolipids and neutral lipids using silica columns and methylated by alkaline methanolysis. The phospholipids were converted into fatty acid methyl esters via mild acid methanolysis and identified and quantified using the Agilent 6890 instrument (Agilent Technologies, Palo Alto, CA, USA) and the MIDI Sherlock Microbial Identification System (MIDI Inc., Newark, DE, USA). The content of each PLFA was expressed as PLFA mg g−1 OC against an internal standard (methyl ester C19:0; Matreya Inc., State College, PA, USA). PLFAs specific to actinomycetes (10Me 16:0, 10Me 17:0 and 10Me 18:0), gram-positive bacteria (G+; i14:0, a15:0, i15:0, i16:0, a17:0 and i17:0), gram-negative bacteria (G−; 16:1 w7c, 16:1 w9c, cy 17:0 w7c, 17:1 w8c, 18:1 w7c, cy 19:0 w7c and cy19:0 w9c) and fungi (F; 18:1 w9c and 18:2 w6c) were quantified (Fierer and Jackson 2006; Willers et al. 2015; Chen et al. 2016).
Statistical analyses
The package “vegan” in the R 3.3.1 platform (R Foundation for Statistical Computing, Vienna, AT) was used for the statistical analyses. A paired t test was conducted to examine differences in the elemental contents, PLFAs and biomarkers between the shrub patch and the grassy matrix. In addition, principal component analyses (PCAs) were used to assess the contributions of SOC derived from aboveground and belowground inputs. All analyses were performed with a significance level of p < 0.05. To explore the effects of shrub encroachment along the shrub cover gradient, we calculated the variables at the plot scale (B p ) by the weighted method (Maestre et al. 2012, 2015). The formula was: Cover = ∑ (πab/4)/A′; B p = B g × (1 −Cover) + B s × Cover, where Cover represents the shrub cover at the plot scale; a and b represent the long and short crown diameter of the shrub patch, respectively; A′ represents the plot area; and B s and B g represent the response values in the shrub patch and grassy matrix, respectively.
Results
Chemical composition of plant inputs
Free lipids, which include odd-numbered n-alkanes (C27 to C33), even-numbered n-alkanoic acids (C16 to C30) and n-alkanols (C18 to C30), were identified in the solvent extracts (Fig. 2a–c). The mixed samples reflected the characteristics of the herb communities. The maximum abundance (Cmax) of n-alkanoic acids was at C16 and C18 in the leaves and roots, respectively, whereas the long-chain (> C20) alkanoic acids were detected mainly in the roots (Fig. 2a). The Cmax of n-alkanols was at C28 or C30, and n-alkanes displayed a Cmax at C29 in the shrub leaves, C31 in the herb leaves and C27 in the roots (Fig. 2).
Carbon number distribution of major biomarkers of plant leaves and roots. a, b, and c show the products of the total solvent extraction, and d, e, and f show the products of the base hydrolysis. Sl and Gl represent the mixed leaf samples of the shrub patches and grassy matrix, respectively; Sr and Gr represent the mixed root samples of the shrub patches and grassy matrix, respectively. CM, Caragana microphylla; SG, Stipa grandis; CK, Carex korshinskyi; IT, Iris tenuifolia; AF, Artemisia frigida; MR, Melilotoides ruthenica. All samples of specific herb species listed were collected in the grassy matrix
Homologous bound aliphatic lipids (alkanoic acids, alkanedioic acids and hydroxyalkanoic acids) were detected after base hydrolysis and displayed a strong even-over-odd predominance. The Cmax of homologous alkanoic acids (C14 to C30) was at C28 or C30 in the plant tissues (Fig. 2d). Alkanedioic acids (C20 to C26) were more abundant in the roots than in the leaves (Fig. 2e). The hydroxyalkanoic acids displayed a Cmax of C16 in leaves and C20–C24 in roots (Fig. 2f).
Eight lignin-derived phenols were identified in the CuO oxidation extracts, including the prevalent vanillyls (V; vanillin, acetovanillone and vanillic acid), syringyls (S; originating from angiosperms; syringaldehyde, acetosyringone and syringic acid) and cinnamyls (C; in the non-woody plant tissues; p-coumaric acid and ferulic acid).
Compared with the leaves and roots of the herbs, the leaves of C. microphylla had lower ratios of C/N, S/V and C/V and higher ratios of vanillic acid/vanillin ((Ad/Al)v) and syringic acid/syringaldehyde ((Ad/Al)s), which consisted of higher abundant free lipids (labile compounds) and lower abundant cutin and lignin (recalcitrant compounds), suggesting that C. microphylla produces more labile litter (Online Resource 1).
SOC content, microbial community and chemical composition of SOC
The SOC concentration of the shrub patches was significantly higher than that of the grassy matrix (0.74 vs. 0.59%, p = 0.03). The total, bacterial and fungal PLFAs and F/B ratio were slightly higher (p > 0.05) in the shrub patches, and the ratio of G−/G+ was significantly higher in the shrub patches (p < 0.01; Table 1). The concentrations of extractable long-chain lipids, bound lipids and lignin-derived phenols in the shrub patches were more abundant than those in the grassy matrix (p = 0.05; p < 0.01; p = 0.49; Table 1).
The major compounds of the solvent extraction were n-alkanoic acids (C16 to C30), n-alkanols (C18 to C32), n-alkanes (C23 to C33) and some sugars. The Cmax of n-alkanoic acids was at C18 in both the shrub patches and grassy matrix (Fig. 3a). In addition, the Cmax of n-alkanols and n-alkanes was at C28 and C31 in the shrub patches and C18 and C31 or C33 in the grassy matrix, respectively (Fig. 3b, c).
Homologous aliphatic lipids, including alkanoic acids (C12 to C32), alkanedioic acids (C16 to C26) and hydroxyalkanoic acids (C16 to C26), dominated in the base hydrolysis products (Fig. 3d–f). Among these lipids, the alkanoic acids were the most abundant component and ω-hydroxyalkanoic acids were the dominant hydroxyalkanoic acids, representing 7.14 ~ 27.53% of the base hydrolysis product. These two types of biomarkers showed a Cmax of C16 in both the shrub patches and the grassy matrix (Fig. 3d, f). Moreover, alkanedioic acids with a Cmax of C16 in the shrub patches and C24 in the grassy matrix represented 12.7 ~ 17.7% of the bound lipids (Fig. 3e).
PCA of the SOC chemical composition
PCA was performed to assess the correlations between the SOC and the biomarkers as well as the microbial PLFAs (Fig. 4). The first two principal components (PCs) together could explain 72.9% of the variation of the grassy matrix (Fig. 4a). PC1 was most positively influenced by SOC, cutin and suberin, and PC2 was most negatively correlated with the PLFAs and cutin∨suberin (the compounds in both cutin and suberin; Fig. 4a). SOC was strongly positively influenced by suberin and cutin but negatively correlated with PLFAs (Fig. 4a). Similarly, the first two PCs together could explain 60.7% of the variation in the shrub patches (Fig. 4b). PC1 was negatively influenced by cutin∨suberin as well as PLFAs and cutin, and PC2 was strongly correlated with suberin and lignin. SOC was strongly positively influenced by cutin, lignin and cutin∨suberin but weakly correlated with PLFAs (Fig. 4b).
Changes in the SOC chemical composition and microbial community with shrub cover
The SOC concentration in the top layer was highest in the pure grassland and decreased with increasing shrub cover in the SEG at the plot scale (R = 0.87) because the concentration decreased sharply in the grassy matrix (R = 0.92; Fig. 5a). From the perspective of chemical composition, the contents of free lipids and lignin-derived phenols as well as the ratios of S/V, (Ad/Al)v and ω-C16∑/C16 increased while cutin, the ratios of suberin/cutin and ω-C18∑/C18 decreased with increasing shrub cover (Fig. 5). Moreover, the suberin contents showed a unimodal trend with increasing shrub cover (Fig. 5d). Alternatively, decreasing suberin but increasing cutin and (Ad/Al)s trends were observed in the shrub patches, whereas a consistent trend was observed for the biomarkers of the grassy matrix (Fig. 5). Microbial PLFAs increased while the ratios of G−/G+ and F/B decreased with increasing shrub cover (Fig. 5).
Changes in the biomarker-based OC components, parameters and microbial community of the SOC in the top layer with increasing shrub cover. Shrub cover represents the shrub cover at plot scale. SOC was standardized to %. Free lipid, cutin, suberin, lignin and PLFAs were standardized to mg/g OC. The remaining ratios are unitless. Lines are cubic splines with associated Pearson correlation coefficients (R)
Discussion
Influences of plant chemistry on SOC composition
Plant-derived molecular compounds are valuable biomarkers of SOC sources and degradation processes (Otto and Simpson 2006; Andreetta et al. 2013). An advantage of this study is that the biomarkers from the leaf and root tissues of the dominant plant species in the study area were measured simultaneously, rather than citing plant characteristics from studies of different plant species or different areas (Paul et al. 2003; Pisani et al. 2013). Consistent with previous studies, the distribution of the carbon numbers of biomarkers indicated the different plant origins (Feng et al. 2010, 2015). The dominant C18 alkanoic acid, C16 ω-hydroxyalkanoic acid and C16 alkanedioic acid indicated leaf inputs, whereas the C24 alkanedioic acid indicated root inputs (Fig. 2; Otto and Simpson 2006). The bimodal dominant C31 and C33 n-alkanes in the grassy matrix are likely attributable to the higher herb richness (Chen et al. 2015), whereas the abundant C29 n-alkanes suggested an important contribution of shrub leaves in the shrub patches (Fig. 2c). However, the C18 n-alkanols and C16 hydroxyalkanoic acids may be derived from microbes because of low concentrations in the plant (Fig. 2b, f; Zhao et al. 2014).
Contributions of aboveground and belowground inputs to SOC
The PCA results indicated that the aboveground and belowground inputs to SOC were considerably different in the shrub patches and the grassy matrix (Fig. 4). The SOC in the shrub patches was derived mainly from leaves, as evidenced by a close positive correlation between SOC and cutin (Fig. 4b; Angst et al. 2016). However, the SOC in the grassy matrix was composed of a mixture of fresh root-derived (more important) and leaf-derived compounds, which was confirmed by the closer correlation between the SOC and suberin compared to that between the SOC and cutin (Fig. 4a; Kögel-Knabner 2002). This difference is likely because of the high density of herb roots in the shallow layer and a greater contribution of shrub leaves (Tamura and Tharayil 2014). The significantly higher ω-C16/∑C16 ratio in the grassy matrix than in shrub patch might be attributed to the larger number of root inputs in the grassy matrix because a significant difference was not observed in the ω-C18/∑C18 ratio with enhanced cutin oxidation (Goñi and Hedges 1990; Feng et al. 2010). Similarly, the suberin/cutin ratio, which is an indicator of the relative input of root (bark)/leaf (Goñi and Hedges 1990; Feng et al. 2010), was significantly higher in the grassy matrix (p = 0.01; Table 1). In addition, previous studies have demonstrated that the chemistry and input of the biopolymers incorporated into soil are controlled by soil animals and microbial communities (Filley et al. 2008; Dempsey et al. 2011). We observed a greater microbial contribution in the grassy matrix, which was supported by the close negative relationship between the microbial PLFAs and SOC. By contrast, the microbial PLFAs were weakly related to the SOC in the shrub patches, which might be attributed to the dilution of the fresh OC input (Tamura and Tharayil 2014).
Effects of input chemistry and microbial community on SOC along shrub cover
Consistent with previous studies, the SOC of the shrub patch was, on average, 28% higher (− 9 ~ 114%; p = 0.03) than that of the grassy matrix, indicating that fertile islands were prevalent in the SEG and increased the soil resource heterogeneity (Schlesinger et al. 1996; Archer et al. 2001). Compared with the pure grassland, the SOC decreased by 29% (4 ~ 48%) in the SEG with increasing shrub cover (R = 0.92; Fig. 5), consistent with the notion that “fertile islands” can create feedback that results in the degeneration of the ecosystem (Schlesinger et al. 1990). Maestre et al. (2009) observed interesting impacts of shrub patches on the grassy matrix in semi-arid Mediterranean grasslands and presumed that these impacts were likely attributable to biotic mechanisms, including altered litter inputs, microbial activity and microclimate. Similar to the shrubs in Mediterranean grasslands, C. microphylla, which is known as a rhizomatous clonal plant, in our studied grasslands can form a more horizontal, close-to-surface zone of plant inputs compared to the shrubs with elevated canopies and lower basal stems found in the Chihuahuan Desert (Xiong and Han 2005; Maestre et al. 2009). The decreased SOC observed in the grassy matrix might be affected by the discrete shrubs with sprawling canopies and root systems that had strong ability to extend several metres horizontally (Maestre et al. 2009).
Consistent with a study in kudzu-invaded soils, the distribution of the plant and microbial biomarkers reflected the effects of the dominant plant inputs on the composition of SOC in the top layer (Tamura and Tharayil 2014). In the pure grassland, a higher abundance of cutin and suberin and a lower concentration of free lipids were detected, suggesting the preservation of recalcitrant polymers derived from herb inputs (Fig. 5). In addition, this result was further supported by the low ω-C16/∑C16 ratio and PLFA concentration, which indicates a low degradation rate (Feng et al. 2010).
Compared with the leaves and roots of dominant herb species, the leaves of the legume shrub C. microphylla were labile plant inputs, as evidenced by the lower C:N ratio and chemical composition (Online Resource 1). Increased microbial activities were observed in the SEG, as supported by the increasing PLFAs in the grassy matrix, suggesting increased decomposition as a likely mechanism driving the carbon loss observed in the grassy matrix (Fig. 5; Nottingham et al. 2009). More recalcitrant substrates would increase the fungal community because of the high efficiency at which fungi use plant heteropolymers (Chigineva et al. 2009). The decreased F/B ratio with increasing shrub cover suggests a negative response of fungi, which was likely associated with less recalcitrant carbon allocation in the SEG (Feng et al. 2010).
Soil carbon loss because of co-metabolism of the recalcitrant substrates after the incorporation of fresh labile inputs has been reported (Fontaine et al. 2007; Kuzyakov 2010). Similarly, Whittinghill et al. (2012) found that carbon losses under nitrogen deposition might be caused by enhanced decomposition, which indicates that carbon loss may be induced by the encroachment of N-rich shrubs. The SOC remained unchanged in the shrub patches with the increasing shrub cover, which might have been caused by the replacement of prior carbon decompositions with the fresh input of shrub leaves (Fig. 5c; Fontaine et al. 2007; Filley et al. 2008). Similarly, the SOC decreased significantly with increasing shrub cover in the grassy matrix, which likely resulted from insufficient fresh plant inputs compared with the prior carbon decompositions (Fig. 5c, d; Knapp et al. 2008; Montané et al. 2010). Previous studies have shown that shrubs led to root litter decay by providing shadows and competition for moisture and nutrients (Casper and Jackson 1997; Alvarez et al. 2011). The decreased ratios of G−/G+, suberin/cutin and ω-C18/∑C18 indicated decreased root inputs as the shrub cover increased (Feng et al. 2010, 2015).
Conclusions
Shrub encroachment is expected to affect regional soil carbon dynamics. Previous studies have focused on changes in the bulk carbon contents. In this study, we demonstrated that the altered plant input chemistry and microbial community caused by shrub encroachment significantly affect SOC storage using a multiple biomarker approach. Our results provide molecular evidence for the biotic mechanisms by which SOC changes after shrub encroachment.
References
Alvarez LJ, Epstein HE, Li J et al (2011) Spatial patterns of grasses and shrubs in an arid grassland environment. Ecosphere 2:1–30
Andreetta A, Dignac MF, Carnicelli S (2013) Biological and physico-chemical processes influence cutin and suberin biomarker distribution in two Mediterranean forest soil profiles. Biogeochemistry 112:41–58
Angst G, John S, Mueller CW et al (2016) Tracing the sources and spatial distribution of organic carbon in subsoils using a multi-biomarker approach. Sci Rep 6
Archer S, Boutton TW, Hibbard KA (2001) Trees in grasslands: biogeochemical consequences of woody plant expansion. In: Schulze ED et al (eds) Global biogeochemical cycles in the climate system. A Harcourt Science and Technology Company, San Diego, pp 115–138
Armas-Herrera CM, Dignac MF, Rumpel C et al (2016) Management effects on composition and dynamics of cutin and suberin in topsoil under agricultural use. Eur J Soil Sci 67:360–373
Asner GP, Heidebrecht KB (2005) Desertification alters regional ecosystem–climate interactions. Glob Change Biol 11:182–194
Bai E, Boutton TW, Liu F et al (2013) 15N isoscapes in a subtropical savanna parkland: spatial-temporal perspectives. Ecosphere 4:1–17
Bailey RG (1996) Ecosystem geography. Springer, New York
Barger NN, Archer SR, Campbell JL et al (2011) Woody plant proliferation in North American drylands: a synthesis of impacts on ecosystem carbon balance. J Geophys Res 116:G4
Blaser WJ, Shanungu GK, Edwards PJ et al (2014) Woody encroachment reduces nutrient limitation and promotes soil carbon sequestration. Ecol Evol 4:1423–1438
Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–917
Carbone MS, Trumbore SE (2007) Contribution of new photosynthetic assimilates to respiration by perennial grasses and shrubs: residence times and allocation patterns. New Phytol 176:124–135
Casper BB, Jackson RB (1997) Plant competition underground. Annu Rev Ecol Syst 28:545–570
Chen L, Li H, Zhang P et al (2015) Climate and native grassland vegetation as drivers of the community structures of shrub-encroached grasslands in Inner Mongolia, China. Landscape Ecol 30:1627–1641
Chen D, Pan Q, Bai Y et al (2016) Effects of plant functional group loss on soil biota and net ecosystem exchange: a plant removal experiment in the Mongolian grassland. J Ecol 104:734–743
Chigineva NI, Aleksandrova AV, Tiunov AV (2009) The addition of labile carbon alters litter fungal communities and decreases litter decomposition rates. Appl Soil Ecol 42:264–270
Crow SE, Lajtha K, Filley TR et al (2009) Sources of plant-derived carbon and stability of organic matter in soil: implications for global change. Glob Change Biol 15:2003–2019
DeMarco J, Filley T, Throop HL (2016) Patterns of woody plant-derived soil carbon losses and persistence after brush management in a semi-arid grassland. Plant Soil 406:277–293
Dempsey MA, Fisk MC, Fahey TJ (2011) Earthworms increase the ratio of bacteria to fungi in northern hardwood forest soils, primarily by eliminating the organic horizon. Soil Biol Biochem 43:2135–2141
D’Odorico P, Okin GS, Bestelmeyer BT (2012) A synthetic review of feedbacks and drivers of shrub encroachment in arid grasslands. Ecohydrology 5:520–530
Eldridge DJ, Bowker MA, Maestre FT et al (2011) Impacts of shrub encroachment on ecosystem structure and functioning: towards a global synthesis. Ecol Lett 14:709–722
Fang J, Bai Y, Wu J (2015) Towards a better understanding of landscape patterns and ecosystem processes of the Mongolian Plateau. Landscape Ecol 30:1573–1578
Feng X, Simpson AJ, Schlesinger WH et al (2010) Altered microbial community structure and organic matter composition under elevated CO2 and N fertilization in the duke forest. Glob Change Biol 16:2104–2116
Feng X, Gustafsson Ö, Holmes RM et al (2015) Multi-molecular tracers of terrestrial carbon transfer across the pan-Arctic: comparison of hydrolyzable components with plant wax lipids and lignin phenols. Biogeosciences 12:4841–4860
Fierer N, Jackson RB (2006) The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103:626–631
Filley TR, Boutton TW, Liao JD et al (2008) Chemical changes to nonaggregated particulate soil organic matter following grassland-to-woodland transition in a subtropical savanna. J Geophys Res 113. doi:10.1029/2007jg000564
Fontaine S, Barot S, Barre P et al (2007) Stability of organic carbon in deep soil layers controlled by fresh carbon supply. Nature 450:277–280
Goñi MA, Hedges JI (1990) The diagenetic behavior of cutin acids in buried conifer needles and sediments from a coastal marine environment. Geochim Cosmochim Acta 54:3083–3093
González-Roglich M, Swenson JJ, Jobbágy EG et al (2014) Shifting carbon pools along a plant cover gradient in woody encroached savannas of central Argentina. For Ecol Manag 331:71–78
Hibbard K, Archer S, Schimel D et al (2001) Biogeochemical changes accompanying woody plant encroachment in a subtropical savanna. Ecology 82:1999–2011
Jackson RB, Banner JL, Jobbágy EG et al (2002) Ecosystem carbon loss with woody plant invasion of grasslands. Nature 418:623–626
Knapp AK, Briggs JM, Collins SL et al (2008) Shrub encroachment in North American grasslands: shifts in growth form dominance rapidly alters control of ecosystem carbon inputs. Glob Change Biol 14:615–623
Kögel-Knabner I (2002) The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol Biochem 34:139–162
Kuzyakov Y (2010) Priming effects: interactions between living and dead organic matter. Soil Biol Biochem 42:1363–1371
Lett MS, Knapp AK, Briggs JM et al (2004) Influence of shrub encroachment on aboveground net primary productivity and carbon and nitrogen pools in a mesic grassland. Can J Bot 82:1363–1370
Li P, Wang N, He W et al (2008) Fertile islands under Artemisia ordosica in inland dunes of northern China: effects of habitats and plant developmental stages. J Arid Environ 72:953–963
Li H, Shen H, Chen L et al (2016) Effects of shrub encroachment on soil organic carbon in global grasslands. Sci Rep 6:28974
Li H, Zhang J, Hu H et al (2017) Shift in soil microbial communities with shrub encroachment in Inner Mongolia grasslands, China. Eur J Soil Biol 79:40–47
Liu F, Wu X, Bai E et al (2011) Quantifying soil organic carbon in complex landscapes: an example of grassland undergoing encroachment of woody plants. Glob Change Biol 17:1119–1129
Maestre FT, Bowker MA, Puche MD et al (2009) Shrub encroachment can reverse desertification in semi-arid Mediterranean grasslands. Ecol Lett 12:930–941
Maestre FT, Quero JL, Gotelli NJ et al (2012) Plant Species Richness and Ecosystem Multifunctionality in Global Drylands. Science 335:214–217
Maestre FT, Delgado-Baquerizo M, Jeffries TC et al (2015) Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc Natl Acad Sci USA 112:15684–15689
Montané F, Romanyà J, Rovira P et al (2010) Aboveground litter quality changes may drive soil organic carbon increase after shrub encroachment into mountain grasslands. Plant Soil 337:151–165
Nottingham AT, Griffiths H, Chamberlain PM et al (2009) Soil priming by sugar and leaf-litter substrates: a link to microbial groups. Appl Soil Ecol 42:183–190
Oelofse M, Birch-Thomsen T, Magid J et al (2016) The impact of black wattle encroachment of indigenous grasslands on soil carbon, Eastern Cape, South Africa. Biol Invasions 18:445–456
Otto A, Simpson MJ (2006) Evaluation of CuO oxidation parameters for determining the source and stage of lignin degradation in soil. Biogeochemistry 80:121–142
Paul EA, Morris SJ, Sixad J et al (2003) Interpretation of soil carbon and nitrogen dynamics in agricultural and afforested soils. Soil Sci Soc Am J 67:1620–1628
Peng H, Li X, Jiang Z et al (2013) Shrub encroachment with increasing anthropogenic disturbance in the semiarid Inner Mongolian grasslands of China. CATENA 109:39–48
Pisani O, Hills KM, Courtier-Murias D et al (2013) Molecular level analysis of long term vegetative shifts and relationships to soil organic matter composition. Org Geochem 62:7–16
Ratajczak Z, Nippert JB, Collins SL (2012) Woody encroachment decreases diversity across North American grasslands and savannas. Ecology 93:697–703
Scharlemann JPW, Tanner EVJ, Hiederer R et al (2014) Global soil carbon: understanding and managing the largest terrestrial carbon pool. Carbon Manag 5:81–91
Schlesinger WH, Reynolds JF, Cunningham GL et al (1990) Biological feedbacks in global desertification. Science 247:1043–1048
Schlesinger WH, Raikes JA, Hartley AE et al (1996) On the spatial pattern of soil nutrients in desert ecosystems. Ecology 77:364–374
Schmidt MW, Torn MS, Abiven S et al (2011) Persistence of soil organic matter as an ecosystem property. Nature 478:49–56
Springsteen A, Loya W, Liebig M et al (2010) Soil carbon and nitrogen across a chronosequence of woody plant expansion in North Dakota. Plant Soil 328:369–379
Sutherland WJ, Freckleton RP, Godfray HCJ et al (2013) Identification of 100 fundamental ecological questions. J Ecol 101:58–67
Tamura M, Tharayil N (2014) Plant litter chemistry and microbial priming regulate the accrual, composition and stability of soil carbon in invaded ecosystems. New Phytol 203:110–124
Throop HL, Archer SR (2008) Shrub (Prosopis velutina) encroachment in a semidesert grassland: spatial–temporal changes in soil organic carbon and nitrogen pools. Glob Change Biol 14:2420–2431
Van Auken OW (2009) Causes and consequences of woody plant encroachment into western North American grasslands. J Environ Manage 90:2931–2942
Whittinghill KA, Currie WS, Zak DR et al (2012) Anthropogenic N deposition increases soil C storage by decreasing the extent of litter decay: analysis of field observations with an ecosystem model. Ecosystems 15:450–461
Willers C, Jansen van Rensburg PJ, Claassens S (2015) Phospholipid fatty acid profiling of microbial communities-a review of interpretations and recent applications. J Appl Microbiol 119:1207–1218
Xiong X, Han X (2005) Spatial heterogeneity in soil carbon and nitrogen resources, caused by Caragana microphylla, in the thicketization of semiarid grassland, Inner Mongolia. Acta Ecolog ica Sinica 25:1678–1683
Zhao L, Wu W, Xu X et al (2014) Soil organic matter dynamics under different land use in grasslands in Inner Mongolia (northern China). Biogeosciences 11:5103–5113
Zhou Y, Boutton TW, Wu XB et al (2017) Soil carbon response to woody plant encroachment: importance of spatial heterogeneity and deep soil storage. J Ecol doi: 10.1111/1365-2745.12770
Acknowledgements
This study was funded by the National Natural Science Foundation of China (31330012, 31470525), the National Basic Research Program of China on Global Change (2014CB954001), and the China Postdoctoral Science Foundation (2016M591282).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Edith Bai.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, L., Li, H., Shen, H. et al. Shrub-encroachment induced alterations in input chemistry and soil microbial community affect topsoil organic carbon in an Inner Mongolian grassland. Biogeochemistry 136, 311–324 (2017). https://doi.org/10.1007/s10533-017-0396-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10533-017-0396-8