Abstract
Invasive populations of green iguanas (Iguanidae: Iguana iguana) are widely established beyond their native Central, South American, and Lesser Antillean range in various islands of the Pacific, Florida USA, and in the Greater Caribbean Region. Although widespread, information about these invasions is scarce. Here we determine the origin of invasive populations of green iguanas in Puerto Rico, Fiji, The Caymans, Florida USA, The Dominican Republic, the US Virgin Islands (USVI) of St. Thomas and St. Croix, and a U.S.A pet store. We sampled 120 individuals from these locations and sequenced one mitochondrial (ND4) and two nuclear (PAC and NT3) loci. We also include a preliminary characterization of population structure throughout Puerto Rico using six microsatellite loci to genotype individuals across 10 sampling sites. Comparing the genealogical relationships of all our samples to published sequencing data from the native range, we found that sampled populations were largely a product of populations from Colombia and El Salvador; two countries with multiple, industrial-size pet iguana farming operations. Notably, we found that haplotypes detected exclusively in the USVI and Puerto Rico’s outlying island of Vieques are closely linked to green iguanas native to Saba and Montserrat (Lesser Antilles); a clade not reported in the pet trade. Our population genetic analyses did not reveal isolation among sampling sites in Puerto Rico, rather the evidence supported admixture across the island. This study highlights the roles of the pet trade and lack of regulation in the spread of green iguanas beyond their native range.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human trade has greatly facilitated the movement of species (Wilson et al. 2009), though not all that travel prosper. Only a small percentage of introduced species survive and become invasive (Kolar and Lodge 2001). The introduction and proliferation of invasive species outside their native ranges has led to problems for human health, the economy, and the environment (Pimentel et al. 2005; Reaser et al. 2007; White et al. 2008), which has resulted in a plethora of work conducting risk assessments of, and assessing the impacts by, invasive species. Moreover, ecologists view invasive species as a unique opportunity to explore the ecological and evolutionary processes underlying successful establishment in real time, as opposed to the millennia it may take under scenarios of natural range expansion (Sax et al. 2007; Betancur-R. et al. 2011; Lawson Handley et al. 2011). By studying invasive species during their different invasion stages (i.e., introduction, establishment, and spread), ecologists are able to examine factors that facilitate invasion success and introduction pathways, which lead to useful information for the management of invasive species (Lockwood et al. 2007).
An array of vertebrate species are now introduced due to human traffic and trade (Kraus 2003), these include lizards, birds (Russello et al. 2008), snakes (Bushar et al. 2015), mammals (Lippens et al. 2017) and frogs (Meshaka 2011). In south Florida, the Nile monitor (Varanus nilotucus) was introduced as a pet and has formed three separate breeding populations (Dowell et al. 2016). A study using mitochondrial (mtDNA), nuclear sequence, and microsatellite data to determine their source, found the monitor populations to be the product of multiple introductions from several pet trade countries throughout the southern coastal regions of West Africa. The Florida populations were nevertheless isolated, each from a single putative origin, thus highlighting the role of the pet trade in the foundation and persistence of each of the three invasive populations. Moreover, the collateral effect of human trade has also perpetuated invasions. In the case of the house mouse (Mus musculus domesticus), its spread and genetic diversity has a close link to colonial activities in Senegal. Lippens et al. (2017) used mtDNA markers together with 16 nuclear microsatellites to elucidate the invasion history and spatial expansion of this species. Using mtDNA haplotype data together with population STRUCTURE analysis data, Lippens et al. (2017) suggest there was one putative origin and subsequent spread in-land starting from the first founded port of the country. The authors demonstrate the utility of genetic tools to determine the origin and introduction history of the introduced house mouse, an invasion that occurred hundreds of years earlier. This link between the colonial history of the country and the spread of the species highlight the effect of trade on species’ distribution.
Initial population size and continued influx (i.e., propagule pressure) of individuals are among several factors that may influence the probabilities of invasion success (Sinclair and Arnott 2016). In conjunction with the role of genetic variability, these factors may aid colonizing individuals in their establishment (Le Roux and Wieczorek 2009). Introduction events that suffer severe and persistent bottlenecks are hypothesized to lead to decreased genetic diversity and, with it, fewer opportunities for invasion success (Nei et al. 1975; Dlugosch et al. 2015). Nevertheless, populations across multiple taxa made up of only a few founding individuals (Betancur-R. et al. 2011), or those that have gone through bottleneck events (e.g., Tsutsui et al. 2000), have had low genetic diversity yet have become highly invasive (Sakai et al. 2001; Bock et al. 2015). Some species, like the invasive argentine ant (Linepithema humile), whose reduction in genetic variability has led to reduced intraspecific aggression, have even benefited from this lower genetic variation (Tsutsui et al. 2000). Conversely, decreased genetic diversity may not even occur in invasive populations. Some founding populations may carry with them a robust representation of the genetic diversity of their source population (Collins et al. 2017; Foote et al. 2019). Even when diversity loss does occur, this can be offset by multiple founder events or admixture from multiple sources leading to invasive populations with higher genetic diversity than their native range sources (e.g., Kolbe et al. 2007; Tonione et al. 2011). A crucial first step towards determining the role of genetic variability in establishment success is to reconstruct the invasion history by identifying the origin of introduced populations (Simberloff 2009; Guillemaud et al. 2010; Lawson Handley et al. 2011).
Propagule pressure may also contribute to increased invasion success (Von Holle and Simberloff 2005; Simberloff 2009). However, the occurrence of multiple introductions, propagule size, and frequency of introductions are seldom recorded in the early stages of invasion making it difficult to quantify the role of these factors in successful establishment (Brockerhoff et al. 2014). Multiple studies suggest increased propagule size and frequency aid in successful invasion by increasing genetic variability and decreasing the effects of environmental and demographic stochasticity (reviewed in Simberloff 2009). As with understanding the role of genetic diversity in introduction success, identifying the origin of invasive populations is a crucial step to assessing the potential role of propagule pressure in species colonization events. In research by Ficetola et al. (2008), the use of the mitochondrial Cytochrome b locus allowed researchers to identify the native range origin of invasive European populations of the American bullfrog (Rana catesbeina), as well as estimate the number of founding individuals through demographic simulations. Their work showed that in some species, fewer than a dozen individuals may be able to establish new invasive populations, further highlighting the importance of genetic data in understanding invasion success.
The green iguana (Iguana iguana, Linnaeus 1758; Fig. 1), one of two species in the genus Iguana, is a generalist herbivore that is threatened due to overexploitation and habitat loss in localized regions of its native range of Central and South America and the Lesser Antilles (for a review on the biology of green iguanas, see Falcón et al. 2013). Human consumption and exports for the pet trade have lowered population numbers to such an extent that efforts are currently underway to protect native populations (e.g., Escobar et al. 2010). Paradoxically, this species is highly invasive where established outside of its native range (Lazell 1973; Rivero 1998; Townsend et al. 2003; Krysko et al. 2007; Falcón et al. 2012; Iguana Specialist Group 2017; Kwak et al. 2019). These areas include the mainland United States (southern Florida) and multiple islands including the Dominican Republic (Pasachnik et al. 2012), the Cayman Islands (Moss et al. 2018), Puerto Rico (Rivero 1998), and more recently Dominica (van den Burg et al. 2020a). In many areas of the Greater Caribbean Region (GCR; sensu Olsen et al. 2004), the introduction of green iguanas has raised concerns about regional biosecurity (Falcón et al. 2012). The GCR boast ample suitable habitat for future establishment, where the added possibility of biodiversity loss through hybridizations with congeneric species exists (Falcón et al. 2012; Vuillaume et al. 2015; Van Wagensveld and Van Den Burg 2018; Moss et al. 2018). Green iguanas are reported on some islands in the Pacific, including Hawaii, Japan, Fiji (Falcón et al. 2013), and more recently in Taiwan (Chin 2016) as well. Its far-reaching invasive distribution makes a regional approach to characterizing these invasive green iguana populations necessary.
Invasive green iguanas represent a problem for both the general public and wildlife managers (Townsend et al. 2003; Krysko et al. 2007). Following hurricane Andrew in 1992, the number of green iguana populations in Florida increased, likely aided by canopy openings, planting of additional food sources, and the inadvertent creation of nesting sites. They are now considered a pest due to their consumption of residential and commercial vegetation (Krysko et al. 2007). Green iguanas are documented to interact with endemic wildlife such as use of the burrows of the Florida Burrowing Owl (Anthene cunicularia floridana), raising concern over possible competition or disruptions to the owl’s life history (McKie et al. 2005; Krysko et al. 2007). In the Cayman Islands, where green iguanas are considered an invasive pest after establishing in residential areas (Seidel and Franz 1994; Echternacht et al. 2011; Ledger 2015; Haakonsson 2016) and causing infrastructure damage (Rivera-Milán and Haakonsson 2020), a culling program was put in place and led to the removal of 874,252 individuals in the span of 11 months. Moreover, in Puerto Rico green iguanas are considered an invasive nuisance due to its impact on human agriculture, horticulture, and travel (Engeman et al. 2005; Falcón et al. 2012, 2013). In addition, although little is known about its interactions with native iguanid species, hybridization with the congeneric Lesser Antillean iguana I. delicatissima (Vuillaume et al. 2015), and even with the Sister Islands Rock Iguana (Cyclura nubila caymanensis; Moss et al. 2018) was reported. As both I. delicatissima and C. nubila caymanensis are considered critically endangered, the added threat of extirpation through hybridization is an alarming concern (Moss et al. 2018; Van Wagensveld and van den Burg 2018).
In this study, we investigated the origin of green iguana populations throughout their invasive range to assess possible introduction pathways. One plausible introduction pathway for islands in the Caribbean Region is over-water dispersal, which was documented for the species (Censky et al. 1998). A second possible pathway is the pet trade which, given the species’ popularity in the pet trade, coupled with the large amounts of reported exports from several countries during the last two decades (i.e., Colombia and El Salvador, see Hoover 1998; Stephen et al. 2011), is cited as the likely source of invasions (Rivero 1998; Krysko et al. 2007; Iguana Specialist Group 2017). In our study, we expected species to have origins in the major pet trade exporting countries without ruling out the possibility of introduction via natural dispersal for islands in the Antilles. To evaluate the source of invasive populations and introduction pathways, we analyzed invasive populations from the GCR (South Florida USA, the Dominican Republic, the Cayman Islands, the US Virgin Islands, and Puerto Rico) and the Pacific Island of Qaemea, Fiji. We sought to distinguish the origin of each invasive population from among the four discrete geographically defined clades identified by Stephen et al. (2012). To identify these clades, the authors used data from the mtDNA locus ND4 and nuclear loci PAC and NT3 loci from pet trade, captive, and wild caught individuals with known country of origin. In our work, we collected genetic data for those same markers from wild and pet trade individuals and combined these data with historical import and export records of pet green iguanas. In our study, if an invasive population originated from native range populations and was the result of natural dispersal, we expected it to have a closer genotypic relationship to those sources (e.g., Saba and St. Lucia) as opposed to documented exporting countries (i.e., Colombia, where a pet trade individual was sequenced at the ND4 marker by Stephen et al. 2012 and El Salvador, see Hoover 1998; Stephen et al. 2011). Additionally, we amplified and analyzed six microsatellite loci to explore the presence of possible population structure in the invasive green iguana introduction in Puerto Rico. With these data we also explored possible invasion scenarios (i.e., multiple introductions or single introductions). Using the sequence and microsatellite data we begin to infer the role that propagule pressure might have played in the green iguana’s successful invasion of Puerto Rico.
Methods
Sample collection
Our collaborators collected samples for this study throughout subtropical and tropical low elevation habitats (Falcón et al. 2012) at elevations below 400 m including mangroves, parks, zoos, farms, and tropical dry forests. Seven invasive green iguana introductions were sampled from sites in the GCR, including Miami (MIA; n = 5) and Davie (DAV; n = 4) in Florida (continental USA), the USA Virgin Islands (USVI) of St. Thomas (ANR; n = 5) and St. Croix (STX; n = 5), the Dominican Republic (RD, n = 4), the Cayman Islands of Grand Cayman (CY; n = 11) and Little Cayman (CYL, n = 2). In Puerto Rico (PR), a more intensive sampling scheme was completed to conduct population structure analysis. We acquired samples from Puerto Rico through collaborations with local hunters, wildlife managers, and private landowners who captured and euthanized green iguanas in an effort to control population growth. In total, 124 green iguanas from 10 sampling sites in Puerto Rico are included in this study. Moreover, we acquired samples from outside of the GCR from an introduced population on the Pacific island of Qamea, Fiji (FIJ; n = 3). Additionally, to consider haplotypes present in the pet trade, we sampled two green iguana pets purchased in Salt Lake City (Utah, USA). We stored blood or tissue samples (i.e., toe and tail clips, liver, dorsal or crest scales) in 2.0 mL tubes in 90% EtOH or in a solution of 20% DMSO, 0.25 M EDTA, NaCl to oversaturation and ddH2O at a pH of 7.5 to 8.0 at − 20.0 °C.
DNA extraction and sequencing
To identify the origins of green iguana populations, we collected DNA sequence data from a maximum of six tissue samples for all localities in the GCR and Fiji. In the case of Puerto Rico in particular, and due to the extensive sampling conducted, we collected samples from up to 10 individuals per site throughout the main island and Vieques (an island municipality located off the eastern coast of Puerto Rico). We extracted DNA from blood and tissue using the Qiagen DNEasy blood and tissue kit following the manufacturer’s protocol (Qiagen, Valencia, CA). If the tissue had scales, we extended digestion time to 24-h and added 40.0 µl of proteinase K. We assessed the DNA quality by gel electrophoresis in 1% agarose gel and measured DNA concentration using NanoDrop (Thermo Fisher). We diluted all samples above 30.0 ng/µl to 20.0 ng/µl.
We sequenced three loci for which Stephen et al. (2012) published haplotype data from green iguanas in their native range. Based on their variability, the authors showed these markers to be useful for distinguishing individuals from within four phylogeographic regions in the native range. We amplified a 825 bp fragment of the mitochondrial (mtDNA) NADH dehydrogenase subunit 4 (ND4) locus, a 563 bp fragment of the 3′ untranslated region of the nuclear locus polymerase alpha catalytic subunit (PAC) and a 489 bp region of the nuclear locus neurotrophin-3 (NT3) loci and locus using published primer sequences (Noonan and Chippindale 2006; Pasachnik et al. 2008) by polymerase chain reaction (PCR). The ND4 fragment was amplified using primers ND4 (5′-CAC CTA TGA CTA CCA AAA GCT CAT GTA GAA GC-3′) and LEU (5′-CAT TAC TTT TTA CTT GGA TTT GCA CCA-3′). The PAC fragment was amplified using primers PAC AF (5′-CCC AGT GAG AGT TGC TGG A-3′) and PAC S2 (5′-CTT TCC CCT CCC AAA CAA AC-3′). The NT3 fragment was amplified using NT3-F3 (5′ ATA TTT CTG GCT TTT CTC TGT GGC-3′) and NT3-R4 (5′-GCG TTT CAT AAA AAT ATT GTT TGA CCG G-3′) primers. We amplified all fragments in a total volume of 10.0 µl using 5.0 µl Qiagen’s Taq PCR master mix kit, 0.5 µl primer forward, 0.5 µl primer reverse, 0.3 µl BSA, 2.7 µl ddH2O, and 1.0 µl of DNA template.
We performed our PCR cycling for mtDNA ND4 and for nuclear PAC fragments using the following conditions: an initial 2 min denaturation step at 94.0 °C for 1 cycle, followed by 35 cycles of 94.0 °C for 20 s, 55.0 °C 30 s, 72.0 °C for 1.5 min and a final extension at 72.0 °C for 10 min. The nuclear NT3 fragment PCR cycling was performed at 94.0 °C for one initial denaturation cycle, followed by 35 cycles of 94.0 °C for 20 s, 50.0 °C 30 s and 72.0 °C for 1.5 min. We verified the PCR products by gel electrophoresis on a 1% agarose gel. PCR products of successful amplifications were purified using PCR ExoSAP-IT® (Affymetrix, USA) using 0.45 µl shrimp alkaline phosphate, 0.3 µl, 2.45 µl and 6.0 µl of PCR product, for a total volume of 9.0 µl. We sequenced the PCR products using an ABI 3130xl with the original primers. We edited and aligned sequence data for all loci using Sequencher v4.8. We verified electropherograms for ambiguous base calls by examining forward and reverse sequences. In all, we sequenced 107 individuals for the mtDNA ND4 locus and 81 and 84 individuals for the nuclear markers PAC and NT3, respectively. The Genotyping and Sequencing Facility of the University of Puerto Rico, Río Piedras (http://www.sgf.hpcf.upr.edu) conducted all the sequencing in this study.
Microsatellite data extraction and genotyping
Due to their high levels of polymorphism, microsatellite markers are used to compare genetic variability between populations and remain an important tool in understanding the changes in allelic frequencies that may occur during invasion (Allendorf and Luikart 2007; Dlugosch and Parker 2008; Montarry et al. 2010). To represent the population found in Puerto Rico, we sampled 173 individuals from 10 sampling sites, nine on the main island of Puerto Rico and one on Vieques. To determine the population structure of the I. iguana population in Puerto Rico, we collected data from six microsatellite loci (IgdL12, IgdL14, IgdL17, IgdL19, IgdL20, IgdL24) found to be polymorphic in I. iguana and its sister species I. delicatissima (Valette et al. 2013). We used PCR to amplify each locus in two separate PCR rounds. We carried out both rounds in 10 µl total volume using: 5.0 µl Qiagen’s Taq PCR master mix kit, 3 µl BSA, 2.7 µl ddH2O, and 1.0 µl of DNA template and 0.5 µl primer forward, 0.5 µl primer reverse. In the second reaction the forward primer was substituted by 0.5 µl fluorescently-labeled FAM or HEX. Both rounds of PCR cycling for all loci were performed using the following conditions: an initial 2 min denaturation step at 94.0 °C for 1 cycle, followed by 35 cycles of 94.0 °C for 20 s, 56.0 °C 35 s and 72.0 °C for 1.5 min and a final extension at 72.0 °C for 10 min. We verified PCR products by gel electrophoresis on a 1% agarose gel. Following successful amplifications, we genotyped samples in an ABI 3130xl at the University of Puerto Rico, Río Piedras Genotyping and Sequencing Facility. Subsequent allele scoring was completed in GeneMapper 4.08. We used Micro-Checker (Van Oosterhout et al. 2004) to check for evidence of scoring errors due to stuttering, allele dropouts and excess homozygosity in our data set.
Haplotype network analysis
To determine the relationships among haplotypes sampled in the invasive and native ranges of green iguanas, we inferred the genealogical relationships of the mtDNA ND4 and nuclear PAC and NT3 loci. We obtained haplotype data for native range populations and a pet-trade individual from Colombia published by Stephen et al. (2012) from GenBank, as well as for the outgroup species Iguana delicatissima (Malone et al. 2000). We phased haplotypes for the two nuclear markers (PAC and NT3), haplotypes using DnaSP 5.0 (Librado and Rozas 2009). This program was also used to count the number of different haplotypes (h) in our ND4, NT3, and PAC loci. We then employed the Median-Joining Networks (MJN) method (Bandelt et al. 1999), a type of network analysis is used to illustrate the mutational relationships among haplotypes (e.g., Zarza et al. 2008; McCartney-Melstad et al. 2012; De Busschere et al. 2016), to visualize the relationships between green iguana haplotypes from the native and the invasive range in PopART (Leigh and Bryant 2015) for each locus.
Population structure analyses in the invasive range of Puerto Rico
To describe the population structure of invasive green iguanas across Puerto Rico, we measured the level of allelic diversity, inferred the levels of genetic variation from the average number of alleles per locus (A), the observed heterozygosity (Ho), unbiased expected heterozygosity (He), and the number of private alleles (Na) using GenAlex 6.5 (Peakall and Smouse 2006, 2012). To determine how variation is partitioned in our sample set, we used GenAlex to perform an AMOVA (Excoffier et al. 1992). We then determined the Allelic Richness (Rs) for each sampling site by locus using FSTAT 2.9.3 (Goudet 1995, 2001). Finally, we assessed population structure using the Bayesian inference program STRUCTURE 2.3.2 (Pritchard et al. 2000). Because the origin of each sample and the occurrence of admixture were identified by our ND4, PAC and NT3 loci, we used both the USEPOPINFO and the admixture model within STRUCTURE. We performed 10 runs with 500,000 Markov chain Monte Carlo iterations after a 250,000 burn-in period with cluster (K) estimates between 1 and 10. We estimated the optimal number K using the ∆K method described by Evanno et al. (2005) and implemented in Structure HARVESTER 0.6.94 (Earl and VonHoldt 2012). We then generated and stylized STRUCTURE plots in POPHELPER 1.0.10 (Francis 2017).
We further investigate the potential population structure of green iguanas on Puerto Rico using Discriminant Analysis of Principal Components (DAPC) (Jombart et al. 2010) in the adegenet package (Jombart and Bateman 2008) in R version 4.0.2 (R Development Core Team 2013). This multivariate analysis is used to study complex genetic structures in green iguanas (Vuillaume et al. 2015) and shown to be useful in scenarios with high levels of admixture. We first transformed our data into a DAPC object using the genclone2genind function. We performed a Principal Component Analysis (PCA) and K- means clustering to determine the best number of clusters K using the function find.clusters. We evaluated up to K = 40 clusters and retained 100 principal components (PCs) based on eigen values that would encapsulate over 90% of the cumulative variance. We then chose the K with the lowest BIC. Following group assignment, we used the function dapc to transform the data using a PCA and then complete the Discriminant Analysis (DA). We used the optim.a and optim.a.score functions to determine the number of PCs to retain so as to avoid overfitting the model. Using this method, we retained 15 PCs and 9 discriminant functions. We used a scatterplot to visualize the relationships between individuals and clusters returned from the DAPC.
Results
Haplotype network analysis
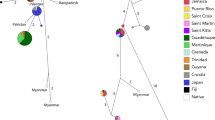
A Median Joining Network (MJN) used to infer relationships among ND4 haplotypes sampled from the native range, the invasive range, and I. delicatissima shows shared haplotypes between the Utah pet shop (pet trade) and the invasive population from Puerto Rico, Fiji, Dominican Republic, St. Croix, and Davie, Florida (Fig. 2). The network for ND4 revealed clusters consistent with the four major geographic clades previously described for the native range of green iguanas (Fig. 2). Individuals from the Dominican Republic, both Florida populations, the Cayman Islands, Fiji, and the pet store in Utah clustered within the Central American clade, while only one individual sampled in Miami clustered with the Northwest of Andes (particularly Colombia) clade (Fig. 2). With the exception of one individual that clustered within the Northwest of the Andes clade, green iguanas from St. Thomas and St. Croix were the only ones to cluster within the Caribbean clade.
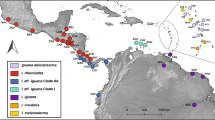
Distribution of and relationships among haplotypes recovered from native and invasive populations of green iguanas (Iguana iguana). The figure shows a the native and invasive distribution of green iguanas in the Americas, color coded by the major geographic clades found by Stephen et al. (2012) and the geographic origin of haplotypes (indicated by three-letter codes), b the distribution of ND4 and PAC haplotypes, with multiple haplotypes from a geographic clade represented by tonal variations of colors among invasive sampling sites in the Greater Caribbean Region and Fiji, and c the Median Joining Network of the mitochondrial ND4 and nuclear PAC loci for I. iguana haplotypes from invasive and native populations in relation to the major clades found in the native range (panels), color coded by invasive population. The size of the circles in panel C indicates the number of sequences
Network analysis also revealed that novel, unique haplotypes found within Puerto Rico had an average of 3 bp differences (out of a maximum of 8 bp) between them and the most closely related haplotypes from the native range. In Puerto Rico, invasive individuals grouped within the Central America and Northwest of the Andes clades. The Central American haplotypes described by Stephen et al. (2012) were only found in samples from the west coast of Puerto Rico where an exact match to the CA4 haplotype, unique to Honduras and El Salvador populations, was detected. Although not an exact match, a Colombian haplotype, SA5, was the least divergent in the Northwest of the Andes clade from haplotypes found in Puerto Rico (0.36% nucleotide divergence).
Our MJN analysis for nuclear data supported geographic clades described for native range green iguanas, but revealed relationships that differ from those inferred by the ND4 data. Analysis of the PAC locus clustered invasive range green iguanas from the Utah pet store, both Florida sites, Cayman Islands, the Dominican Republic, Fiji, and Puerto Rico together with haplotypes of Central American and North West of the Andes. Three iguanas collected in St. Croix also contained haplotypes found within the Central American and West and Southeast of the Andes Clades, whereas the remaining two clustered with St. Thomas individuals into the Caribbean haplotype clade. The NT3 data lacked much of the resolution of the other two loci and clustered all invasive range individuals within either Central or South American clades (Figure S1) with no distinction among East or West of the Andes (Table S2). Moreover, MJN for both PAC and NT3 loci showed haplotypes from all four native range green iguana geographic clades to be distributed throughout the population in Puerto Rico.
Haplotype alignment and novel haplotype identification
Mitochondrial ND4
Previous work using the mtDNA ND4 locus revealed 11 haplotypes distributed among four major phylogeographic regions within the native distribution of green iguanas (Table 1), dividing the range into Central American (CA), Caribbean (CAR), North West (NWA) and North East of the Andes clades (NEA) (Stephen et al. 2012). We aligned 109 sequences of the mtDNA ND4 locus recovered in this study from green iguanas of the invasive range and compared these to 11 of the published native-range haplotypes, using I. delicatissima as our outgroup. The alignment contained 15 novel haplotypes (not reported previously) identified from the invasive range for a total of 26 haplotypes with an average of 22 nucleotide differences among them. For each country we found the following: Florida samples (Davie and Miami, n = 6) had four novel haplotypes, three of which were unique (i.e., only found at this sampling site); Grand Cayman (n = 9) and Little Cayman (n = 2) had four novel haplotypes, three of which were unique; the Dominican Republic (n = 1) had one novel and unique haplotype; Puerto Rico (n = 76) had seven novel haplotypes out of which five were unique; and in the USVI St. Croix (n = 5) there was one novel haplotype.
Nuclear PAC and NT3
We aligned invasive range PAC sequences to published native range green iguana haplotypes. This alignment included 13 native range haplotypes, 86 invasive range samples, and the sequence of I. delicatissima as an outgroup. The alignment revealed a total of 25 haplotypes, with 12 only found in invasive populations. The native and invasive range haplotypes had an average of 3.6 nucleotide differences among them. The 12 novel haplotypes were distributed among our samples as follows: in the Dominican Republic (n = 5) we found two novel haplotypes, one of which was unique; in Puerto Rico (n = 48) we found 10 novel haplotypes with nine unique; and in Fiji (n = 2) we found one novel and one unique haplotype. We aligned 171 NT3 invasive range sequences to the only two published green iguana native range haplotypes and to I. delicatissima. We found three novel haplotypes for a total of five haplotypes in the NT3 alignment. Both the total number of haplotypes and the number of nucleotide differences among haplotypes, 1.5 nucleotide differences on average, were lowest in the NT3 locus. These three novel NT3 haplotypes were found within Puerto Rico (n = 54), which had two haplotypes, and Grand Cayman (n = 11) with one.
Within the PAC alignment, we found nine exact haplotype matches for haplotypes from Central and South America (Fig. 2). The CA3 haplotype, unique to Central America, was the most frequently found. This haplotype was recovered from green iguanas in the Utah pet store, Puerto Rico, Fiji, Davie, St. Croix, Cayman Islands, and the Dominican Republic. Moreover, the CA4 haplotype from Mexico was shared among individuals from Puerto Rico, St. Croix and the pet trade animal, while the CA5 haplotype from Panama was only present in Puerto Rico. The South American SA2 haplotype from Ecuador was also sequenced in multiple invasive iguanas from Puerto Rico, Miami, St. Croix, and the Dominican Republic. Finally, the SA7 haplotype from the Caribbean clade, was only identified within individuals from the USA Virgin Islands of St. Croix and St. Thomas (Fig. 2).
The NT3 locus had matches for both its reported haplotypes, corresponding to Central (CA1) and Southern America (SA1) clades. Individuals from Puerto Rico, St. Thomas, St. Croix and Davie, Florida shared the same SA1 haplotype as their native range counterparts. Whereas, the CA1 haplotype from Central America was matched by invasive individuals from Puerto Rico, the Dominican Republic, Cayman Islands, and Miami, Florida, as well as the pet trade animals in the Utah pet store.
Population structure in the invasive range of Puerto Rico
Our genotyping efforts produced unequal amounts of data among loci and among sampling sites. We analyzed six subsets of our data to determine if a difference in the outcome of our analysis would occur based on the number of individuals, the number of loci, or amounts of missing data. We built these subsets by either reducing the number of loci (i.e., six, five, four or three loci) or by selecting individuals with increasing amounts of genotyped loci (i.e., individuals with data in at least three, four, five or six loci). We found that all six subsets produced comparable results and here we present results of our analysis of the subset containing six loci (L6) which includes all the individuals genotyped and high levels of missing data. In our supplementary materials we provide the results of a subset of individuals with data at all six loci (W6 = constrained subset, fewer individuals and no missing data). The L6 subset had data for all 10 sampling sites in Puerto Rico (CAB, DOR, FJD, GUA, HUM, ISB, LIZ, MYZ, STI, VIQ) and a total of 169 individuals. Percent missing data varied by locus and sampling site; ranging from 15.43% in Igdl12 to 49.94% in Igdl17 and from 24.17% in the MYZ to 51.85% in LIZ (Table 2).
We found no evidence of scoring error due to stuttering or allele dropouts. Out of our six microsatellite loci, three (Igdl17, Igdl19, Igdl 20) were found to have a general excess of homozygotes and the possibility of null alleles if under Hardy–Weinberg equilibrium (HWE). The number of alleles (A) for the sampled loci ranged from 11 to 24 in our L6 subset (Table 2). At least one locus per sampling site deviated significantly from HWE in the L6 subset. However, when we added these values to determine mean deviation from HWE, only locus Igdl 24 was significant in the L6 subset (Table 2). Observed heterozygosity, Ho, ranged from 0.204 to 0.651 in our L6 subset and allelic richness, Rs, ranged from 1 to 4.465 across Puerto Rican sampling sites of green iguanas (Table S 2). The Rs in the L6 subset was highest in the STI site (Rs = 4.0). Expected heterozygosity (He) ranged between 0.427 and 0.744 in the L6 subset and was higher than the range of Ho (0.204–0.651).
The AMOVA analysis on our L6 subset showed that the greatest variation (51.0%) was explained by differences among individuals within sites, 45% of the variation was found within individuals, and among site variation was lowest with 4%.
Our STRUCTURE analysis resolved K = 4 for our L6 (∆K = 16.76) as the optimal number of clusters (see Table S 4 and Figure S 2). For our data, the model did not reveal any clear geographically defined groups, rather individuals from all predefined sampling sites were represented across all genotypes (Fig. 3). Our DAPC analysis resolved K = 6 (Fig. 4) as the number of inferred genetic clusters based on the lowest BIC value (acquired with the K-means algorithm) when K = 1 to K = 40 was tested. As with our STRUCTURE analysis, individuals from the multiple sampling sites had memberships across all 6 genetic partitions and showed no evidence of geographic isolation (Fig. 4).
a Bayesian Information Criterion (BIC) value of inferred number of clusters (K) for microsatellite subset L6. This method yielded K = 6 as the most likely number of genetic clusters. b Distribution of the individuals in each sampling site in their inferred cluster. c Structure like assignment of individuals into each of the six genetic clusters. d Distribution of individuals along DAPC axes where ellipses represent the inferred cluster assignment
Discussion
Exploring the origin(s) and frequency of introductions of a non-native species can lead to insights into the invasion process and understanding the factors facilitating invasion success. Multiple introductions from different geographic regions can, for instance, generate founding populations with high genetic variation and, thus, with a high adaptive potential (Lockwood et al. 2005). Understanding the geographic origin of invasive species and propagule pressure can provide crucial information for future prevention campaigns. In this paper, we assessed the origin of many invasive green iguana populations by performing network distance-based analyses, and further looked at the population structure in the invasive range of Puerto Rico. We confirmed that invasive green iguanas originated from the pet trade, often via multiple introductions, a finding supported both by our network analysis of mitochondrial (ND4) and nuclear (PAC and NT3) loci. Our STRUCTURE analysis revealed all populations on Puerto Rico are admixed and without detectable isolation, and further, that the invasion success there was likely mediated by high propagule pressure. Overall, our findings are consistent with the previous reports of origin in green iguana populations in the Lesser Antilles (St. Eustatius, Saba, Guadeloupe, Martinique and St. Lucia) documented by multiple authors (Vuillaume et al. 2015; van den Burg et al. 2018) who found potential origins in Florida, Greater Antilles, Central America, Caribbean and NE South America; like us, they base their findings on Stephen et al. (2012)’s published haplotype data.
Network analysis
Our network analysis results are consistent with historic trade data from the native range of the green iguana, which shows Colombia (EOA) and El Salvador (CA) as the two main countries farming and exporting green iguanas into the pet trade (Hoover 1998; Stephen et al. 2011). A reported 3,680,301 green iguanas were exported from Central and South America from 1983 to 1994, while between 2004 and 2009 a reported 1,976,891 individuals were exported from El Salvador alone (Hoover 1998; Stephen et al. 2011). During the same periods, the USA imported and re-exported a large number of green iguanas, receiving 2,979,820 individuals between 1983 to 1994 (Hoover 1998) and 971,602 from 2001 to 2008 (Stephen et al. 2011). In general, invasive green iguana populations from sampled localities have clear origins in the reported pet trade countries (Hoover 1998; Stephen et al. 2011). This holds true for green iguanas in Fiji, with origins in Honduras and El Salvador and for samples localities in the GCR. For individuals introduced into the Dominican Republic and Puerto Rico, export records from the CITES database (https://trade.cites.org/) further support our findings, with individuals imported live from El Salvador to both the Dominican Republic and Puerto Rico and additional individuals imported from Colombia to the Dominican Republic.
Interestingly, in the offshore island of Vieques in Puerto Rico, and in the US Virgin Islands of St. Thomas and St. Croix (where the origin of the green iguanas is debated) we found mixed results indicative of both pet trade origin, and origins from much older native populations (from which no recorded recent transport has occurred). Resident scientist and wildlife officials assume that green iguanas were introduced to the USVI, and dispute whether the introduction occurred in pre-Columbian times by the indigenous peoples of these islands, or recently through the pet trade (see discussions in Platenberg and Boulon 2006; Platenberg 2007; Akin 2012). These islands have green iguanas that appear to be morphologically distinct from typical pet trade animals in terms of their coloration (Fig. 1), a trait that was included in the classification of subspecies in the Caribbean (Breuil et al. 2019).
Our data do not seem to support the notion that green iguanas in St. Thomas originated through the modern pet trade. Haplotypes for both mitochondrial and nuclear markers on St. Thomas originated in populations native to the Caribbean clade (i.e., Montserrat and Saba), not yet described as occurring commonly (or legally) within the pet trade. In St. Croix, where both pet trade and Caribbean clade haplotypes were detected, only one of five clustered together with St. Thomas into the Caribbean clade while the remaining four grouped with the Central America clade. Here, green iguana were reported as early as 1859 (Günther 1859), pre-dating the pet trade boom of the 1980s and 1990s by over 100 years. Similar reports by Davis, Olasee (pers. comm to CDJV in 2016) affirm Günther (1859)’s observation that green iguanas were only present on eastern St. Croix in the nineteenth century. Today, these lizards are abundant throughout the island. While the presence of pet trade individuals is clear, the possibility of introduction by indigenous people or by natural dispersal (e.g., Censky et al. 1998) cannot be disregarded. Efforts should be made to prevent further imports of green iguanas to St. Thomas and St. Croix to retain possible remnant populations introduced by indigenous peoples or of natural arrival while their origin is resolved. These efforts should follow a regional approach, and particularly focus on preventing intentional and/or accidental introductions from Puerto Rico (unintended introductions by boat are possible; see Falcón et al. 2012). It is worth mentioning that until 2013, the green iguana was protected on St. Thomas and St. Croix. However, that year, the sale, hunt and consumption of green iguana was made legal through Bill No. 30–0277 (Barshinger et al. 2013). Our sampling in St. Thomas was limited to one location. Therefore, sampling should be expanded throughout the island to adequately represent the phylogenetic relationship between green iguanas on St. Thomas and its native range conspecifics. In the meantime, we propose a revision to the amendment of the Bill No. 30–0277 while origin status is researched.
In Puerto Rico, the green iguana was first reported in 1964 (both of these specimens were deposited in the University of Puerto Rico’s Museum of Zoology, V5403 and V5404) and by the 1990s, it had established localized populations in the western Mayagüez Zoo, in the northeastern mangrove forest of Carolina and Loíza, and in the Metropolitan Area of San Juan (Rivero 1998, 2006). Our data did not identify these early reported populations as founders, rather a strong level of admixture was detected throughout. Based on our findings, it would appear that continued admixture, be it during transportation of green iguanas from the U.S mainland to Puerto Rico or on island, masked the possibility of detecting a single introduction and rather reflects that these reptiles were imported from the same sources as those in the contiguous U.S (El Salvador and Colombia).
The green iguana in Puerto Rico was declared an invasive pest in 2004 (Departamento de Recursos Naturales y Ambientales 2004) and its import and sale became illegal. To our knowledge, the only record of green iguanas imported to Puerto Rico comes from the CITES database, when in 1993 a shipment for the pet trade of 600 live captive-bred specimens went directly from El Salvador to Puerto Rico. Haplotypes from captive bred individuals from El Salvador identified by Stephen et al. (2011) are present in Puerto Rico, providing evidence for the introduction of green iguanas to Puerto Rico through the pet trade. As for Colombia, their influence in the pet trade is also evident in Puerto Rico where sampling sites from the island clustered within the Colombian SA5 mitochondrial haplotype group suggesting an ancestral origin in Colombia. However, green iguana origin in Puerto Rico was not limited to Colombia and El Salvador, as haplotypes from six other countries were also identified on the island. It is worth noting that native range iguana farms may begin the process of admixture prior to exportation, as they may add individuals from wild populations into their stock, potentially crossing border lines (Stephen et al. 2011, 2012). Nevertheless, in Puerto Rico, haplotypes from areas in the native range that would otherwise be separated by geographic barriers interacted (e.g., the Andes), resulting in a novel admixed population.
Population structure in the invasive range of Puerto Rico
Although the role of genetic diversity in this species’ invasion success warrants further study, invasive biology theory would suggest high levels of diversity to be advantageous in overcoming the challenges that invasive species face during establishment and range expansion (Sakai et al. 2001). We consider the results from our population level analyses of Puerto Rico’s introduced green iguanas as support for the hypothesis that the invasion is the result of multiple introduction events from several locations. We do so with the caveat that our results suffer from the possibility of elevated estimates of genetic admixture that can be caused by missing data (Reeves et al. 2016). We did not observe population isolation or evidence of a single introduction event, but rather signs of admixture throughout the island of Puerto Rico. The number of haplotypes found and high levels of variability do suggest, however, that there remains much undescribed genetic diversity in the native range. The exact number of propagules and their size will remain unknown, though the levels of genetic variability described here could indicate that a large number of individuals founded the populations throughout the island of Puerto Rico. The variable number of K clusters found in our data set may also support the possibility that multiple founding populations, from at least three of the four geographic clades described by Stephen et al. (2012) led to the current levels of variability. Future work should focus on deeply examining the relative levels of genetic diversity of invasive populations when compared to populations in the native range.
Conclusions
Using network-based analyses, we were able to link the origin of invasive green iguana populations to the pet trade and to major pet trading countries. This finding contributes to the growing body of literature on biological invasions that are aided by the human pet trade (Bushar et al. 2015). Our work also highlights, albeit indirectly, the role of increased propagule pressure, as seen in the post 1990′s expansion and increase of the populations of green iguana on the island of Puerto Rico. This is supported by the close haplotypic relationship between iguanas exported from El Salvador for the pet trade and the iguanas found in the wild in Puerto Rico that further demonstrates the influential role of the pet trade in species invasion. Moreover, Stephen et al. (2012) highlighted the need to adequately sample throughout the range of the green iguana in order to detect possible cryptic species or lineages. Our research supports the necessity espoused by Stephen et al. (2012), whose work stirred up the conversation amongst green iguana experts about the need for a taxonomic revision of the genus Iguana. Because we found novel haplotypes not previously described for green iguanas, a more thorough sampling is needed across the native range of green iguanas prior to any taxonomic reassessment. Regions where green iguanas were protected from exploitation and where undetected haplotypes may persist should be prioritized because, although this species is not endangered, localized extirpation in unstudied native range populations could lead to the loss of green iguanas with significantly distinct evolutionary histories. Finally, we concur with the recommendations of Falcón et al. (2012, 2013) and the IUCN Iguana Specialist Group (2017) in the need to adopt laws, regulations, and management plans to prevent further spread and invasions by green iguanas outside their native range. As the green iguana continues to expand into new regions, most recently into southeast Asia (van den Burg et al. 2020b), it is important that a regional management approach is adopted.
Availability of data and materials
Sequences obtained in this study were submitted to GenBank (ND4: MH150962-MH150975, NT3: MH150947-MH150949 and PAC: MH150950-MH150961). Microsatellite data files are available in the supplementary material.
Code availability
Code used in DAPC analysis in R, and associated data files can be found https://github.com/chri360/DAPC_GreenIguana.
References
Akin B (2012) Undercurrents: Icon or Pest, the Great Iguana Debate. St. Croix Source 4–7.
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell Publishing, Malden, MA
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036
Barshinger CW, Malone S-M, Capehart D, et al (2013) Bill No. 30–0277. Thirtieth Legislature of the Virgin Islands, United States Virgin Islands
Betancur-R R, Hines A, Acero PA et al (2011) Reconstructing the lionfish invasion: insights into Greater Caribbean biogeography. J Biogeogr 38:1281–1293. https://doi.org/10.1111/j.1365-2699.2011.02496.x
Bock DG, Caseys C, Cousens RD et al (2015) What we still don’t know about invasion genetics. Mol Ecol 24:2277–2297. https://doi.org/10.1111/mec.13032
Breuil M, Vuillaume B, Schikorski D et al (2019) A story of nasal horns: two new subspecies of Iguana Laurenti, 1768 (Squamata, Iguanidae) in Saint Lucia, St Vincent & the Grenadines, and Grenada (southern Lesser Antilles). Zootaxa 4608:201–232. https://doi.org/10.11646/zootaxa.4608.2.1
Brockerhoff EG, Kimberley M, Liebhold AM et al (2014) Predicting how altering propagule pressure changes establishment rates of biological invaders across species pools. Ecology 95:594–601. https://doi.org/10.1890/13-0465.1
Bushar LM, Reynolds RG, Tucker S et al (2015) Genetic characterization of an invasive boa constrictor population on the caribbean island of Aruba. J Herpetol 49:602–610. https://doi.org/10.1670/14-059
Censky EJ, Hodge K, Dudley J (1998) Over-water dispersal of lizards due to hurricanes. Nature 395:556–556. https://doi.org/10.1038/26886
Chin J (2016) Former pet iguanas become nuisance around Kaohsiung. In: Taipei Times. http://www.taipeitimes.com/News/taiwan/archives/2016/04/16/2003644093. Accessed 15 Apr 2019
Collins CJ, Chilvers BL, Osborne A et al (2017) Unique and isolated: population structure has implications for management of the endangered New Zealand sea lion. Conserv Genet 18:1177–1189. https://doi.org/10.1007/s10592-017-0969-z
De Busschere C, Courant J, Herrel A et al (2016) Unequal contribution of native South African phylogeographic lineages to the invasion of the African clawed frog, Xenopus laevis, in Europe. PeerJ 4:e1659. https://doi.org/10.7717/peerj.1659
Departamento de Recursos Naturales y Ambientales (2004) Reglamento para regir la conservación y el manejo de la vida silvestre, Las Especies Exotica y la Caza en el estado libre asociado de Puerto Rico. Puerto Rico
Dlugosch KM, Anderson SR, Braasch J et al (2015) The devil is in the details: Genetic variation in introduced populations and its contributions to invasion. Mol Ecol 24:2095–2111. https://doi.org/10.1111/mec.13183
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449
Dowell SA, Wood JP, Campbell TS et al (2016) Combining genetic and distributional approaches to sourcing introduced species: a case study on the Nile monitor (Varanus niloticus) in Florida. R Soc Open Sci 3:150619. https://doi.org/10.1098/rsos.150619
Earl DA, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Echternacht AC, Burton FJ, Blumenthal JM (2011) The amphibians and reptiles of the Cayman islands: conservation issues in the face of invasions. In: Conservation of Caribbean Island Herpetofaunas Volume 2: Regional Accounts of the West Indies. BRILL, pp 129–147
Engeman RM, Smith HT, Constantin B (2005) Invasive Iguanas as airstrike hazards at Luis Munoz Marin international airport, San Juan, Puerto Rico. J Aviat Educ Res 14:45–50
Escobar RA, Besier E, Hayes WK (2010) Evaluating headstarting as a management tool: post-release success of green iguanas (Iguana iguana) in Costa Rica. Int J Biodivers Conserv 2:204–214. https://doi.org/10.5897/IJBC.9000053
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Falcón W, Ackerman JD, Daehler CC (2012) March of the green iguana: Non-native distribution and predicted geographic range of Iguana iguana in the greater Caribbean region. IRCF Reptil Amphib 19:150–160
Falcón W, Ackerman JD, Recart W, Daehler CC (2013) Biology and impacts of Pacific island invasive species. 10. Iguana iguana, the green iguana (Squamata: Iguanidae). Pac Sci 67:157–186. https://doi.org/10.2984/67.2.2
Ficetola GF, Bonin A, Miaud C (2008) Population genetics reveals origin and number of founders in a biological invasion. Mol Ecol 17:773–782. https://doi.org/10.1111/j.1365-294X.2007.03622.x
Foote I, Godfrey SS, Robertson BC (2019) Mate choice explains high genetic diversity in a small founding population of the New Zealand sea lion (Phocarctos hookeri). Aust J Zool 66:343–351. https://doi.org/10.1071/ZO19023
Francis RM (2017) Pophelper: an R package and web app to analyse and visualize population structure. Mol Ecol Resour 17:27–32. https://doi.org/10.1111/1755-0998.12509
Goudet J (1995) FSTAT (Version 1.2): A Computer Program to Calculate F-Statistics. J Hered 86:485–486. https://doi.org/10.1093/oxfordjournals.jhered.a111627
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3).
Guillemaud T, Beaumont M, Ciosi M et al (2010) Inferring introduction routes of invasive species using approximate Bayesian computation on microsatellite data. Heredity (Edinb) 104:88–99. https://doi.org/10.1038/hdy.2009.92
Günther A (1859) On the reptiles from St. Croix, West Indies, collected by messrs, A. and B. Newton Ann Mag Nat Hist 4:209–217
Haakonsson J (2016) Green Iguana Invasion. FLICKER Bull. Cayman Islands Dep. Environ Terr Resour unit 23:1–5
Hoover C (1998) The U.S. Role in the international live reptile trade: Amazon tree boas to Zululand dwarf chameleons.
Iguana Specialist Group (2017) Position Statement of the IUCN SSC Iguana Specialist Group on Non-native Invasive Iguanas.
Jombart T, Bateman A (2008) Genetics and population analysis adegenet: a R package for the multivariate analysis of genetic markers 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94. https://doi.org/10.1186/1471-2156-11-94
Kolar CS, Lodge DM (2001) Progress in invasion biology: predicting invaders. Trends Ecol Evol 16:199–204. https://doi.org/10.1016/S0169-5347(01)02101-2
Kolbe JJ, Glor RE, Schettino LR et al (2007) Multiple sources, admixture, and genetic variation in introduced anolis lizard populations. Conserv Biol 21:1612–1625. https://doi.org/10.1111/j.1523-1739.2007.00826.x
Kraus F (2003) Invasion pathways for terrestrial vertebrates. In: Ruiz GM, Carlton JT (eds) Invasive species: vectors and managment strategies, 2nd edn. Island Press, Washington, D.C., pp 68–92
Krysko KL, Enge KM, Donlan EM et al (2007) Distribution, natural history, and impacts of the introduced Green iguana (Iguana iguana) in Florida. Iguana 14:142–151
Kwak ML, Lee L, Okumura C, Da HC (2019) First Report of Co-invasion by the reptile nematode Ozolaimus megatyphlon (Nematoda: Pharyngodonidae) with invasive Green Iguanas (Iguana iguana) in the Asia-Pacific. Acta Parasitol 65:264–270. https://doi.org/10.2478/s11686-019-00144-7
Lawson Handley L-J, Estoup A, Evans DM et al (2011) Ecological genetics of invasive alien species. Biocontrol 56:409–428. https://doi.org/10.1007/s10526-011-9386-2
Lazell JD (1973) The lizard genus Iguana in the Lesser Antilles. https://www.biodiversitylibrary.org/item/25493#page/22/mode/1up 145:1–26.
Le Roux J, Wieczorek AM (2009) Molecular systematics and population genetics of biological invasions: Towards a better understanding of invasive species management. Ann Appl Biol 154:1–17. https://doi.org/10.1111/j.1744-7348.2008.00280.x
Ledger W (2015) ‘Green’ invasion project gets funding. In: Cayman News Serv. https://caymannewsservice.com/2015/06/green-invasion-project-gets-funding/. Accessed 24 Jun 2015
Leigh JW, Bryant D (2015) POPART: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210X.12410
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452. https://doi.org/10.1093/bioinformatics/btp187
Lippens C, Estoup A, Hima MK et al (2017) Genetic structure and invasion history of the house mouse (Mus musculus domesticus) in Senegal, West Africa: a legacy of colonial and contemporary times. Heredity (Edinb) 119:64–75. https://doi.org/10.1038/hdy.2017.18
Lockwood JL, Cassey P, Blackburn TM (2005) The role of propagule pressure in explaining species invasions. Trends Ecol Evol 20:223–228. https://doi.org/10.1016/j.tree.2005.02.004
Lockwood JL, Hoopes MF, Marchetti MP (2007) Invasion ecology. Blackwell Publishing, Malden, MA
Malone CL, Wheeler T, Taylor JF, Davis SK (2000) Phylogeography of the Caribbean rock iguana (Cyclura): implications for conservation and insights on the biogeographic history of the West Indies. Mol Phylogenet Evol 17:269–279. https://doi.org/10.1006/mpev.2000.0836
McCartney-Melstad E, Waller T, Micucci P, a, et al (2012) Population structure and gene flow of the yellow anaconda (Eunectes notaeus) in Northern Argentina. PLoS ONE 7:e37473. https://doi.org/10.1371/journal.pone.0037473
McKie AC, Hammond JE, Smith HT et al (2005) Invasive green iguana interactions in a burrowing owl colony in Florida. Florida F Nat 33:125–127
Meshaka WE (2011) A runaway train in the making: The exotic amphibians, reptiles, turtles, and crocodilians of Florida: Monograph 1. Herpetol Conserv Biol 6:1–101
Montarry J, Andrivon D, Glais I et al (2010) Microsatellite markers reveal two admixed genetic groups and an ongoing displacement within the French population of the invasive plant pathogen Phytophthora infestans. Mol Ecol 19:1965–1977. https://doi.org/10.1111/j.1365-294X.2010.04619.x
Moss JB, Welch ME, Burton FJ et al (2018) First evidence for crossbreeding between invasive Iguana iguana and the native rock iguana (Genus Cyclura) on Little Cayman Island. Biol Invasions 20:817–823. https://doi.org/10.1007/s10530-017-1602-2
Nei M, Maruyama T, Chakraborty R et al (1975) The bottleneck effect and genetic variability in populations. Evolution (N Y) 29:1–10. https://doi.org/10.2307/2407137
Noonan BP, Chippindale PT (2006) Vicariant origin of Malagasy reptiles supports late Cretaceous Antarctic land bridge. Am Nat 168:730–741
Olsen A, Triñanes JA, Wanninkhof R (2004) Sea–air flux of CO2 in the Caribbean Sea estimated using in situ and remote sensing data. Remote Sens Environ 89:309–325. https://doi.org/10.1016/J.RSE.2003.10.011
Pasachnik SA, Carreras De Leon R, Reynoso VH et al (2012) Green iguanas (Iguana iguana) in the Dominican Republic. IRC 19:132–134
Pasachnik SA, Fitzpatrick BM, Near TJ, Echternacht AC (2008) Gene flow between an endangered endemic iguana, and its wide spread relative, on the island of Utila, Honduras: When is hybridization a threat? Conserv Genet 10:1247–1254. https://doi.org/10.1007/s10592-008-9692-0
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research–an update. Bioinformatics 28:2537–2539. https://doi.org/10.1093/bioinformatics/bts460
Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pimentel D, Zuniga R, Morrison D (2005) Update on the environmental and economic costs associated with alien-invasive species in the United States. Ecol Econ 52:273–288. https://doi.org/10.1016/j.ecolecon.2004.10.002
Platenberg RJ (2007) Impacts of introduced species on an island ecosystem: non-native reptiles and amphibians in the US Virgin Islands. Manag Vertebr invasive species Proc an Int Symp Fort Collins, Color August 7–9, 2007 168–174.
Platenberg RJ, Boulon RH (2006) Conservation status of reptiles and amphibians in the U.S. Virgin Islands Appl Herpetol 3:215–235. https://doi.org/10.1163/157075406778116159
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team R (2013) R: A Language and environment for statistical computing. R Found Stat Comput 1:409
Reaser JK, Meyerson LA, Cronk Q et al (2007) Ecological and socioeconomic impacts of invasive alien species in island ecosystems. Environ Conserv 34:1–14. https://doi.org/10.1017/S0376892907003815
Reeves P, Bowker C, Fettig C, et al (2016) Effect of error and missing data on population structure inference using microsatellite data. bioRxiv 080630. doi: https://doi.org/10.1101/080630
Rivera-Milán FF, Haakonsson J (2020) Monitoring, modeling and harvest management of non-native invasive green iguanas on Grand Cayman. Biol Invasions, Cayman Islands. https://doi.org/10.1007/s10530-020-02233-5
Rivero J (1998) Amphibians and reptiles of Puerto Rico. Editorial de la Universidad de Puerto Rico, San Juan, Second
Rivero JA (2006) Guiá para la identificación de lagartos y culebras de Puerto Rico. La Editorial, Universidad de Puerto Rico Press, San Juan, First
Russello MA, Avery ML, Wright TF (2008) Genetic evidence links invasive monk parakeet populations in the United States to the international pet trade. BMC Evol Biol 8:217. https://doi.org/10.1186/1471-2148-8-217
Sakai AK, Allendorf FW, Holt JS et al (2001) The population biology of invasive species. Annu Rev Ecol Syst 32:305–332. https://doi.org/10.1146/annurev.ecolsys.32.081501.114037
Sax D, Stachowicz J, Brown J et al (2007) Ecological and evolutionary insights from species invasions. Trends Ecol Evol 22:465–471
Seidel ME, Franz R (1994) Amphibians and reptiles (exclusive of marine turtles) of the Cayman Islands. The Cayman Islands: Natrual History and Biogeography. Springer, Dordrecht, pp 407–433
Simberloff D (2009) The role of propagule pressure in biological invasions. Annu Rev Ecol Evol Syst 40:81–102. https://doi.org/10.1146/annurev.ecolsys.110308.120304
Sinclair JS, Arnott SE (2016) Strength in size not numbers: propagule size more important than number in sexually reproducing populations. Biol Invasions 18:497–505. https://doi.org/10.1007/s10530-015-1022-0
Stephen CL, Pasachnik S, Reuter A, et al (2011) Survey of Status, Trade, and Exploitation of Central American Iguanas.
Stephen CL, Reynoso VH, Collett WS et al (2012) Geographical structure and cryptic lineages within common green iguanas, Iguana iguana. J Biogeogr 40:50–62
Tonione MA, Reeder N, Moritz CC (2011) High genetic diversity despite the potential for stepping-stone colonizations in an invasive species of gecko on Moorea. French Polynesia PLoS One 6:e26874. https://doi.org/10.1371/journal.pone.0026874
Townsend JH, Krysko KL, Enge KM (2003) Introduced iguanas in southern Florida: a history of more than 35 years. Iguana 10:111–118
Tsutsui ND, Suarez AV, Holway DA, Case TJ (2000) Reduced genetic variation and the success of an invasive species. Proc Natl Acad Sci USA 97:5948–5953
Valette V, Filipová L, Vuillaume B et al (2013) Isolation and characterization of microsatellite loci from Iguana delicatissima (Reptilia: Iguanidae), new perspectives for investigation of hybridization events with Iguana iguana. Conserv Genet Resour 5:173–175. https://doi.org/10.1007/s12686-012-9761-z
van den Burg MP, Brisbane JLK, Knapp CR (2020a) Post-hurricane relief facilitates invasion and establishment of two invasive alien vertebrate species in the Commonwealth of Dominica, West Indies. Biol Invasions 22:195–203. https://doi.org/10.1007/s10530-019-02107-5
van den Burg MP, Meirmans PG, van Wagensveld TP et al (2018) The Lesser Antillean Iguana (Iguana delicatissima) on St. Eustatius: Genetically Depauperate and Threatened by Ongoing Hybridization. J Hered 109:426–437. https://doi.org/10.1093/jhered/esy008
van den Burg MP, Van Belleghem SM, De Jesús Villanueva CN (2020b) The continuing march of common Green Iguanas: arrival on mainland Asia. J Nat Conserv 57:125888. https://doi.org/10.1016/j.jnc.2020.125888
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. https://doi.org/10.1111/j.1471-8286.2004.00684.x
Van Wagensveld TP, Van Den Burg M (2018) First record on fecundity of an Iguana hybrid and its implications for conservation: evidence for genetic swamping of Iguana delicatissima populations by non-native iguanas. Herpetol Notes 11:1079–1082
Von Holle B, Simberloff D (2005) Ecological resistance to biological invasion overwhelmed by propagule pressure. Ecology 86:3212–3218. https://doi.org/10.1890/05-0427
Vuillaume B, Valette V, Lepais O et al (2015) Genetic Evidence of Hybridization between the Endangered Native Species Iguana delicatissima and the Invasive Iguana iguana (Reptilia, Iguanidae) in the Lesser Antilles: Management Implications. PLoS ONE 10:e0127575. https://doi.org/10.1371/journal.pone.0127575
White PCL, Ford AES, Clout MN et al (2008) Alien invasive vertebrates in ecosystems: pattern, process and the social dimension. Wildl Res 35:171. https://doi.org/10.1071/WR08058
Wilson JRUU, Dormontt EE, Prentis PJ et al (2009) Something in the way you move: dispersal pathways affect invasion success. Trends Ecol Evol 24:136–144. https://doi.org/10.1016/j.tree.2008.10.007
Zarza E, Reynoso VH, Emerson BC (2008) Diversification in the northern neotropics: mitochondrial and nuclear DNA phylogeography of the iguana Ctenosaura pectinata and related species. Mol Ecol 17:3259–3275. https://doi.org/10.1111/j.1365-294X.2008.03826.x
Acknowledgements
We would like to thank the following organizations and people for granting permits or for providing samples to this study: Departamento de Recursos Naturales y Ambientales de Puerto Rico, Francheska Ruiz Canino at the U.S Department of Fish and Wildlife-Vieques Wildlife Refuge, The Virgin Islands Department of Planning and Natural Resources Division of Fish and Wildlife, Jane Haakonsson at The Ministry of the Environment of the Cayman Islands, Nicole Angeli at the U.S Virgin Islands Division of Fish and Wildlife, Frank Ridgley at the Miami Zoo, Stesha Pasachnik at the Fort Worth Zoo, Rosanna Carreras at the Instituto Tecnológico de Santo Domingo, Los Iguaneros de Aguada, Carlos Andrés Rodríguez Gómez at the Puerto Rico Conservation Trust-Para la Naturaleza, Steven Flores, Efraín Martínez Cebollero and Alberto Puente Rolón at the University of Puerto Rico Mayagüez, Steve Connors, and Brian Layton. We thank Silvia Planas, Yadira Ortíz Ruíz and Dania Rodriguez for their guidance. We thank Dr. Steven Van Belleghem and Dr. Jason Kolbe for their helpful comments and edits. We also thank the editor and anonymous reviewers for their thoughtful comments and observations that helped to vastly improve this manuscript. All research at the International Institute of Tropical Forestry is done in collaboration with the University of Puerto Rico.
Funding
This work was funded by The National Science Foundation through The Louis Stokes Alliance for Minority Participation (LSAMP) Puerto Rico chapter—Bridge to the doctorate and Undergraduate Research Opportunities (HRD-1139888) to CDJV, The Department of Biology of University of Puerto Rico Rio Piedras and The office of the Dean of Graduate Studies and Research FIPI to DCJV and RP. The sequencing facility used in this study was supported by an Institutional Development Award Networks of Biomedical Research Excellence (INBRE) Grant P20GM103475 from the National Institute of General Medical Sciences, a component of the NIH; the Bioinformatics Research Core of the INBRE.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Ethics approval
The methods in this study were reviewed and approved by the Institutional Animal Care and use committee at the University of Puerto Rico Rio Piedras (IACUC 01,011–06-01–2015).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
De Jesús Villanueva, C.N., Falcón, W., Velez-Zuazo, X. et al. Origin of the green iguana (Iguana iguana) invasion in the greater Caribbean Region and Fiji. Biol Invasions 23, 2591–2610 (2021). https://doi.org/10.1007/s10530-021-02524-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-021-02524-5