Abstract
Objective
To determine the effects of the extra N-terminal seven-amino-acid sequence on the function of chitosanase CsnA.
Results
Sequence and structure analysis indicated that the mature CsnA contains a seven-amino-acid extension in a disordered form at the N-terminus. To determine the function of this sequence, both mature CsnA and its N-terminus-truncated mutant, CsnAΔN, were expressed in Escherichia coli and characterized. Compared with CsnAΔN, CsnA exhibited a 15 °C higher temperature optimum, enhanced pH stability, thermostability and catalytic efficiency. The underlying mechanisms responsible for these changes were analyzed by circular dichroism (CD) spectroscopy. CD analysis revealed that the deletion of the N-terminal sequence resulted in a decrease in the Tm of 4.3 °C and this sequence altered the secondary structure of the enzyme.
Conclusions
The N-terminal sequence is essential for the stability and activity of chitosanase CsnA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chitosanase (EC 3.2.1.132) randomly attacks the β-1,4-linkages of the chitosan backbone, thereby producing bioactive chitooligosaccharides (COS). Based on structure and amino acid sequence similarities, chitosanases are mainly grouped into glycoside hydrolase (GH) families 5, 8, 46, 75 and 80 (http://www.cazy.org). Among these families, GH46 chitosanases have been extensively studied in terms of their catalytic features, enzymatic mechanisms and protein structures (Fukamizo and Brzezinski 1997; Saito et al. 1999). Most of characterized GH46 chitosanases consist only of a single catalytic module and the structure and function of this module have been intensively investigated. The enzymes are mostly α-helical proteins, dumbbell-shaped, and are composed of two globular lobes (a major lobe and a minor lobe) separated by a substrate-binding cleft (Fukamizo and Brzezinski 1997; Saito et al. 1999). Two strictly conserved residues (Glu and Asp) in GH46 enzymes are essential for catalysis (Boucher et al. 1995). These function, respectively, as a general acid and a nucleophile. Compared with the sequence of the catalytic module, terminal sequences are less conserved and even in a disordered form in chitosanases. Consequently, research on the role of the N- or/and C-terminal sequences in structure and function of the enzymes remains scarce. However, recent studies have proposed that the terminal regions have significant effects on protein soluble expression, folding and function (Hu et al. 2017; Li et al. 2014; Liu et al. 2011; Lu et al. 2016).
A GH46 chitosanase CsnA was identified from Renibacterium sp. QD1. It had maximum activity at pH 5.6 and stability over a wide pH range (Xing et al. 2014). Based on sequence alignment and homology modeling with the Streptomyces sp. N174 chitosanase (PDB code: 1CHK) as the template, the mature CsnA has an extra seven-amino-acid sequence (Ser-1-Ala-7) that did not form a regular secondary structure at the N-terminus and its function was unknown. The objective of this study was to examine the effect of this sequence on the enzyme function: the N-terminal segment has a distinct impact on optimal temperature, pH stability, thermostability and catalytic activity. To the best of our knowledge, this is the first report about the effects of terminal sequence on chitosanase. This study provides more insights into the significance of terminal region and the integration of structure and function of the enzyme.
Materials and methods
Strains, plasmids, and chemicals
E. coli DH5α and BL21(DE3) were used for plasmid amplification and expression, respectively. The plasmids pEASY-T1 (TransGen, China) and pET-22b(+) (Invitrogen, USA) were used for gene cloning and heterologous gene expression, respectively. Chitosan (≥90% deacetylated) was purchased from Qingdao MdBio, Inc. (China).
Gene cloning and sequence analysis
Genomic DNA of Renibacterium sp. QD1 was used as the template for amplification of CsnA gene (CsnA). Primers used for gene cloning are shown in Supplementary Table 1. Comparison of the amino acid sequence of CsnA with known sequences was conducted with the BLASTP programs at NCBI. The Swiss-model server (http://swissmodel.expasy.org/) was used to built the three-dimensional structure of CsnA with the chitosanase from Streptomyces sp. N174 (PDB code: 1CHK) as the template, which showed the highest percentage identity (67.4%) among the available amino acid sequences. A Ramachandran plot (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) was used for structure validation. The molecular masses of the enzymes were predicted using Compute pI/Mw tool on the ExPASy Server (http://web.expasy.org/compute_pi/).
Mutant construction, protein expression and purification
Sequence and structure analysis indicated that CsnA had an extra seven-amino-acid sequence that present in the form of random coil at the N-terminus. To verify the function of this fragment, we cloned the mature CsnA gene without the signal peptide-coding sequence and its truncated version (CsnAΔN) gene without the N-terminal sequence by PCR, respectively, with primers CsnA-F/CsnA-R and CsnAΔN-F/CsnAΔN-R (Supplementary Table 1). PCR products were digested by MscI and HindIII and ligated into the pET-22b(+) vector. The resulting plasmids, pET-CsnA and pET-CsnAΔN, were transformed into E. coli BL21(DE3). The positive transformants harboring pET-CsnA and pET-CsnAΔN were used for enzyme expression with IPTG induction (0.5 mM) at 25 °C for 60 h. The supernatants were harvested by centrifugation at 4 °C (10,000×g, 20 min) and then applied onto an Ni-Sepharose column equilibrated with 20 mM Tris/HCl buffer (pH 8) containing 0.5 M NaCl. Protein fractions were collected and analyzed by SDS-PAGE. The protein concentration was determined using a protein assay kit.
Enzymatic activity assay and characterization
The enzymatic activity was determined using the dinitrosalicyclic acid (DNS) method. One unit (U) was defined as the amount of enzyme that released reducing sugar equal to 1 µM glucosamine.HCl per min under standard conditions.
The pH optima and pH stability of the enzymes were measured from pH 3.6 to 10.6. The optimal temperature and thermostability were determined at 0–70 °C at the pH optimum. The T50 value, the temperature at which 50% of enzyme activity was retained, was calculated as described previously (Wulf et al. 2012).
Determination of kinetic parameters
The kinetic parameters were determined with chitosan from 1 to 10 mg/ml at 55 °C (CsnA) or 40 °C (CsnAΔN) for 1 min. K m and V max values were calculated based on Lineweaver–Burk plots using GraphPad Prism (version 5) software (GraphPad Software Inc., La Jolla, CA). The equation k cat = V max /[E] was used to calculate the k cat value, where [E] was the molar concentration of the enzymes used in this experiment. The experiments were carried out three times.
Analysis of reaction pattern and hydrolysis products
The hydrolysis products of CsnA and CsnAΔN were determined by TLC using silica gel plates 60F 254 (Merck). Time course analysis of enzymatic hydrolysates was conducted to determine the action pattern of the enzymes according to the method described previously (Xing et al. 2014).
Effects of metal ions and chemical reagents on enzyme activity
The effects of metal ions and chemical reagents on enzyme activity were determined by mixing the enzyme with 1 mM Mn2+, Al3+, Zn2+, Ba2+, Ni2+, Fe3+, Mg2+, Ca2+, Li+, SDS or EDTA under the standard conditions. The relative activity was calculated with respect to the control sample where the reaction was conducted without any additives.
Circular dichroism (CD) analysis
To investigate the structural difference between CsnA and CsnAΔN, the proteins were monitored by a Jasco J-810 spectropolarimeter. The enzymes (0.1 mg/ml) in 10 mM phosphate buffer (pH 7) were subjected to signal measurement in a 1 mm cell. All measurements were conducted three times.
Thermal denaturation curves of the proteins were obtained by monitoring the change in ellipticity [θ] at 222 nm as the temperature increased on a Jasco J-810 spectropolarimeter. The temperature of the midpoint of transition (Tm) was calculated by curve-fitting the resultant CD values versus temperature data on the basis of a least square analysis according to the method described previously (Liu et al. 2011).
Results
Sequence and structure analysis of CsnA, and protein expression
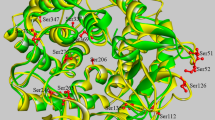
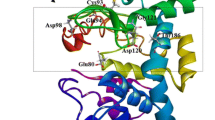
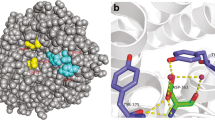
Analysis of the sequence of CsnA indicated that mature CsnA contained an extra N-terminal oligopeptide (7 residues), a catalytic motif of GH46 (215 residues) and an extra C-terminal oligopeptide (16 residues) (Fig. 1a). Three-dimensional structure of CsnA was built with Streptomyces sp. N174 chitosanase as the template. The Ramachandran plot showed that 96.5% of residues were in the favoured regions and 3.5% were in the allowed regions (Supplementary Fig. 1). These results indicated that side chain environments in the model of CsnA were acceptable. As shown in Fig. 1b, the 16-amino-acid extension at the C-terminus formed β-strands. However, the structural conformation of the first four amino acid residues (Ser-1-Asp-4) at the N-terminius was missing because the chitosanase template did not have the corresponding sequence and the retained three N-terminal amino acids (Leu-5-Ala-7) were present in the form of random coil. Many studies have reported that the regular secondary structures make stabilizing contributions to proteins (Sharma et al. 2008; Zhukovsky et al. 1994), thus we speculated that the C-terminal extension might be essential for enzyme function. Unlike the regular secondary structures, the biological significance of irregular segments still remains elusive. Therefore, in order to reveal the function of the N-ternimal random coil, we constructed the truncated mutant CsnAΔN (Fig. 1a). The theoretical molecular masses of CsnA and CsnAΔN were 28.2 and 27.6 kDa, respectively.
Schematic structures and three-dimensional structure. a Schematic structures of CsnA and its truncated mutant CsnAΔN; b Three-dimensional structure of CsnA. The α-helix is shown as a spiral ribbon in red, the irregular coil and β-turn are in green. The N-terminus and C-terminus are in gray and outlined with black ovals; c Illustration of the polar contacts connecting the N-terminus and the catalytic module. The N-terminal residues (Leu-5, Ser-6 and Ala-7) are shown as sticks and colored in gray. The residues involved in the polar contacts in the catalytic module are shown as sticks and colored in cyan. The polar contacts are represented by the yellow dotted lines
The enzymes were expressed in E. coli BL21(DE3) and purified to homogeneity. The molecular weights of CsnA and CsnAΔN estimated by SDS-PAGE were consistent with their theoretical molecular sizes (Fig. 2).
Enzyme properties of CsnA and CsnAΔN
Purified recombinant CsnA and its truncated derivative, CsnAΔN, had similar pH optima at 5.6 (Fig. 3a). However, CsnA was more stable over a broad pH range of 3.6 to 10.6 than CsnAΔN (Fig. 3b). Notebaly, CsnA retained 78–95% of its initial activity after incubation at pH 4.2–10, whereas no more than 45% of CsnAΔN activity was retained after incubation at pH below 5 and above 8.6. These results indicated that CsnA was more stable than CsnAΔN over a broad pH range.
Effects of pH and temperature on enzyme activity and stability. a The pH optima. The pH activity profiles were estimated over the pH range of 3.6-10.6; b pH stability. The enzymes were preincubated in the buffers at a pH range of 3.6–10.6 at 4 °C for 12 h, and residual activities were measured under conditions described above; c The optimal temperatures. The assays were conducted at 0–70 °C; d Thermostability. The thermostability of the enzymes was determined by measuring the residual activities after the enzymes were incubated at 0–70 °C for 1 h and then cooled at 4 °C for 10 min. Values shown are means of triplicate determinations ± standard error (SE)
The optimal temperature of CsnA was 55, 15 °C higher than that of CsnAΔN (Fig. 3c). Moreover, compared with CsnAΔN, CsnA was more stable over a temperature range of 10–70 °C (Fig. 3d). CsnA retained about 80% of activity after it was incubated at 40 °C for 1 h, whereas only 20% of CsnAΔN activity was remained under the same conditions. The T50 of CsnAΔN decreased by 18.7 °C, compared to WT. These results demonstrated that CsnA exhibited better thermostability than CsnAΔN.
Kinetic parameters
As shown in Table 1, the K m of CsnAΔN was comparable to that of CsnA, suggesting that the N-terminal deletion had little effect on the substrate affinity of the enzyme. However, CsnA had higher values of V max , k cat , catalytic efficiency (k cat /K m ) and specific activity than CsnAΔN. These results indicated that the deletion of the first 7 N-terminal amino acids decreased the catalytic efficiency and specific activity of the enzyme.
Analysis of reaction pattern and hydrolysis products of CsnA and CsnAΔN
Both CsnA and CsnAΔN could only hydrolyze colloidal chitosan, and no activity was detected when chitosan powder was used as the substrate (data not shown). The predominant end products analyzed by TLC were chito-biose and -triose for both CsnA and CsnAΔN (Supplementary Fig. 2). Time course analysis of enzymatic hydrolysates indicated that both the enzymes acted in an endolytic manner (Supplementary Fig. 2). These results implied that the N-terminal deletion did not alter the degradation pattern of CsnA.
Effects of metal ions and chemical reagents on the activities of CsnA and CsnAΔN
The effects of metal ions and chemicals on the enzyme activity were not significantly affected by the N-terminal deletion (Supplementary Table 2).
CD analysis
The Tm of CsnAΔN was 36.06, 4.3 °C lower than that of CsnA (40.36 °C) (Fig. 4a). These results suggested that the N-terminal deletion certainly impaired the structural stability of the enzyme.
CD measurements. a The thermal unfolding curves of CsnA and CsnAΔN. The denaturation CD data were measured at 222 nm from 20 to 60 °C upon heating at a rate of 2 °C/min. b The CD spectra for CsnA and CsnAΔN were recorded between 190 and 250 nm with a band width of 1 nm and a scanning speed of 100 nm/min at 25 °C
CD spectroscopy was used to detect possible changes in the secondary structure of truncated mutant compared to the wild-type enzyme. As shown in Fig. 4b, the CD spectra of CsnAΔN were hardly altered by removal of the extra N-terminal oligopeptide. However, a detailed analysis of the CD spectra showed that the secondary structure content of CsnAΔN was different from that of CsnA (Supplementary Table 3). Thus, we assumed that this extra N-terminal oligopeptide had effects on the secondary structure of CsnA.
Discussion
Many studies have shown that terminal regions influence enzymes in different ways (Kim et al. 2011; Song et al. 2015). However, there has been no report on the relationship between terminal regions and chitosanase function. In this study, we identified an extra 7-residue N-terminal sequence that did not form regular secondary structure in chitosanase CsnA from Renibacterium sp. QD1. To explore the effect of this sequence on enzyme function and structure, we cloned the full-length CsnA gene and truncated CsnAΔN gene, expressed them in E. coli, and characterized their enzymatic properties.
The deletion of the N-terminal amino acids led to a significant decrease in optimum temperature, pH stability and thermal stability relative to CsnA. We speculated that this terminal sequence may help to keep enzyme structure stable. Indeed, the N-terminal Leu-5 and Ala-7 formed three polar contacts with the Gln-141, Lys-10 and Lys-11 in the catalytic module (Fig. 1c). The importance of polar contacts in protein structural stability has been reported (Bhardwaj et al. 2010), therefore, the polar contacts around N-terminus might contribute to the stability of CsnA. Furthermore, the effect of this extra sequence on the structural stability of the enzyme was confirmed by the decrease of the Tm at the time of deletion of this sequence. In addition, CD analysis revealed differences between CsnA and its truncated mutant, indicating that the N-terminal segment did affect the secondary structures of CsnA. It can be concluded from the above mentioned observations that the deletion of the extra N-terminal segment not only broke the polar contacts around N-terminus but also changed the secondary structure of the enzyme and thus made the local structure less compact and rigid. These results suggested that the N-terminal sequence confer the structural stability of CsnA, and hence, prevent the overall thermal unfolding of the enzyme at higher temperature and broader pH range.
The first seven amino acids are seated upstream of a helix and probably act as a major element for its folding. Therefore, the N-terminal deletion might disturb this folding. The decreased catalytic efficiency of the truncated mutant can probably be ascribed to the disturbed folding and unstable structure of the enzyme.
In summary, we identified an extra N-terminal sequence in disordered form in chitosanase CsnA. This oligopeptide simultaneously increased the optimal temperature, pH stability, thermostability and catalytic efficiency. Analysis of the mechanisms underlying these changes revealed that this N-terminal segment significantly contributed to the structural stability of the enzyme. The observations made here provide information about the biological significance of terminal regions in enzyme structure and function. In future studies, we plan to obtain the crystal structure of CsnA to get a better mechanistic understanding and then to manufacture the enzyme for better performance by engineering its N-terminus.
Change history
05 December 2017
In Table 1 as published, some of the data were wrong. The corrected Table 1 is shown below.
References
Bhardwaj A, Leelavathi S, Mazumdar-Leighton S, Ghosh A, Ramakumar S, Reddy VS (2010) The critical role of N- and C-terminal contact in protein stability and folding of a family 10 xylanase under extreme conditions. PLoS ONE 5:e11347
Boucher I, Fukamizo T, Honda Y, Willick GE, Neugebauer WA, Brzezinski R (1995) Site-directed mutagenesis of evolutionary conserved carboxylic amino acids in the chitosanase from Streptomyces sp. N174 reveals two residues essential for catalysis. J Biol Chem 270:31077–31082
Fukamizo T, Brzezinski R (1997) Chitosanase from Streptomyces sp. strain N174: a comparative review of its structure and function. Biochem Cell Biol 75:687–696
Hu H, Chen K, Li L, Long L, Ding S (2017) Characterization of the wild-type and truncated forms of a neutral GH10 xylanase from Coprinus cinereus: roles of C-terminal basic amino acid-rich extension in its SDS resistance, thermostability, and activity. J Microbiol Biotechnol 27:775–784
Kim YM et al (2011) Truncation of N- and C-terminal regions of Streptococcus mutans dextranase enhances catalytic activity. Appl Microbiol Biotechnol 91:329–339
Li Z et al (2014) A C-terminal proline-rich sequence simultaneously broadens the optimal temperature and pH ranges and improves the catalytic efficiency of glycosyl hydrolase family 10 ruminal xylanases. Appl Environ Microbiol 80:3426–3432
Liu L, Zhang G, Zhang Z, Wang S, Chen H (2011) Terminal amino acids disturb xylanase thermostability and activity. J Biol Chem 286:44710–44715
Lu X, Wang G, Feng Y, Liu S, Zhou X, Du G, Chen J (2016) The N-terminal alpha-helix domain of Pseudomonas aeruginosa lipoxygenase is required for its soluble expression in Escherichia coli but not for catalysis. J Microbiol Biotechnol 26:1701–1707
Saito J, Kita A, Higuchi Y, Nagata Y, Ando A, Miki K (1999) Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism. J Biol Chem 274:30818–30825
Sharma D, Feng G, Khor D, Genchev GZ, Lu H, Li H (2008) Stabilization provided by neighboring strands is critical for the mechanical stability of proteins. Biophys J 95:3935–3942
Song L, Tsang A, Sylvestre M (2015) Engineering a thermostable fungal GH10 xylanase, importance of N-terminal amino acids. Biotechnol Bioeng 112:1081–1091
Wulf H, Mallin H, Bornscheuer UT (2012) Protein engineering of a thermostable polyol dehydrogenase. Enzym Microb Technol 51:217–224
Xing P, Liu D, Yu W-G, Lu X (2014) Molecular characterization of an endo-type chitosanase from the fish pathogen Renibacterium sp. QD1. J Mar Biol Ass UK 94:681–686
Zhukovsky EA, Mulkerrin MG, Presta LG (1994) Contribution to global protein stabilization of the N-capping box in human growth hormone. Biochemistry 33:9856–9864
Acknowledgements
This work was supported by the National Natural Science Foundation of China (41376144, U1406402) and Key Technologies Research and Development Program of China (2013BAB01B02).
Supporting information
Supplementary Table 1—Primers used for gene cloning.
Supplementary Table 2—Effects of metal ions and chemical reagents on the activities of CsnA and CsnAΔN.
Supplementary Table 3—The secondary structure contents of CsnA and CsnAΔN.
Supplementary Fig. 1—Ramachandran plot of CsnA.
Supplementary Fig. 2—Time course analysis of enzymatic hydrolysates of CsnA (a) and CsnAΔN (b).
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at https://doi.org/10.1007/s10529-017-2444-9.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, Y., Gao, P., Yu, W. et al. N-Terminal seven-amino-acid extension simultaneously improves the pH stability, optimal temperature, thermostability and catalytic efficiency of chitosanase CsnA. Biotechnol Lett 40, 75–82 (2018). https://doi.org/10.1007/s10529-017-2436-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-017-2436-9