Abstract
To control the genetic quality during the whole process of tissue culture of the traditional Chinese medicinal plant, Saussurea involucrate Kar. et Kir., DNA polymorphisms and genetic variations were investigated using randomly amplified polymorphic DNA (RAPD) and inter-simple sequence repeats (ISSR) markers. The genetic stability/variation in tissue-cultured products, including three calli, three adventitious shoots, regenerated plantlets and 2 year-old regenerated plantlets cultivated in the planting base in Tianshan Mountain, were assessed compared with 1 year-old and 2 year-old seedlings cultivated in the same planting base using aseptic seedlings as reference. Apparent genetic variation was detected in the 11 type of plant materials. The percentages of polymorphic bands in the RAPD and ISSR analysis were, respectively, 35% and 33%. Cluster analysis indicated that the genetic similarity values calculated on the basis of RAPD and ISSR data among the 11 type of plant materials were respectively ranged from 0.823 to 0.995 with a mean of 0.878 and 0.825 to 0.974 with a mean of 0.885, which classified the samples into three groups. The similarity coefficient also revealed that differences among three calli were not remarkable by both RAPD and ISSR analysis, and only chemical components and growth properties needed consideration in the screening of callus used for the next redifferentiation studies. But there are remarkable differences among three adventitious shoots analyzed by ISSR markers. Therefore, RAPD and ISSR markers are efficient tools in genetic variation assessment and quality control in plant tissue culture process.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Saussurea involucrata Kar. et Kir. is a valuable traditional Chinese herb of the family Compositae. The whole plant is usually used for treating rheumatoid arthritis, mountain sickness, easing of pain and anti-inflammation. In recent years, this plant received much concern on terminating early pregnancy, inhibiting ageing and growth of cancer cells and enhancing the immunity of the body (Xie et al. 2007). S. involucrata grows on high mountains and in severely cold areas. The inhabitation is thus very rigorous. Driven by commercial incentives, the wild population of this plant has been threatened with depletion in recent years due to excessive harvesting. It has been listed as a protected plant by Chinese government. In addition, large-scale cultivation of S. involucrata is hampered by its extremely vigorous inhabitation. Therefore, a method for rapid and efficient multiplication of the S. involucrata plants is highly desired.
Plant tissue culture might be a prospective technique not only in the protection of natural plant resources, but also in its potential commercial interest. It has been successfully used for large-scale propagation of a large number of plant species, including S. involucrata. Several studies focused on tissue culture of S. involucrata has been reported in China since 1990, and regenerated plantlets were established from different explants and culture methods (Wa et al. 1990, Piao et al. 2003, Fu et al. 2004, Wu et al. 2005, Zhao and Wang 2008). In our laboratory, we have established rapid propagation using tissue culture together with a large-scale artificial planting method, which improves the preservation of S. involucrata, protects the ecological environment and overcomes the disadvantages including feasibility of degeneration by multi-generation planting and suffering from plant diseases and pests (Zhao et al. 2005a, b).
To commercially propagate a crop by tissue culture, it is important to know the genetic stability of the propagates produced. A major problem associated with in vitro culture is the occurrence of somaclonal variation amongst sub-clones of one parental line, arising as a direct consequence of in vitro culture of plant cells, tissues or organs (Larkin and Scowcroft 1981, Gould 1986). Such variation has been observed among regenerants from a large number of species and aspects of this process (Karp 1991, Peschke and Phillips 1992). This variation is often heritable and is therefore unwanted in clonal propagation. Thus, detection of this genetic variation is important to avoid the process becoming economically disastrous. However, in the tissue culture process of a medicinal plant, the screening of different calli, adventitious shoots and regenerated plantlets mainly depends on their chemical components, the genetic stability of the tissue-cultured products has seldom been considered. To our knowledge, no report is available on the genetic stability of tissue culture of S. involucrata.
Molecular marker techniques are at present powerful and valuable tools used in analysis of genetic fidelity of in vitro propagated plants, and are the subject of many publications and reviews. Among the markers, RAPD and ISSR have been mostly favored because of their sensitivity, simplicity and cost-effectiveness. Both RAPD and ISSR markers have been successfully applied to detect genetic similarities or differences in tissue-cultured materials of various plants (Yang et al. 1999, Claudete et al. 2002, Guo et al. 2006, Thomas et al. 2006). The use of two types of markers, which amplify different regions of the genome, allows better analysis of genetic stability/variation of the plantlets.
This present study is the first assessment of the genetic stability of tissue cultures of S. involucrata. Genetic stability/variation in tissue-cultured products (calli, adventitious shoots, regenerated plantlets and 2 year-old regenerated plantlets cultivated in the planting base in Tianshan Moutain) compared with those in 1 year-old and 2 year-old seedlings cultivated in the planting base using aseptic seedlings as reference, were investigated by RAPD and ISSR markers. The information gained on genetic stability/variation will be valuable for screening of the tissue-cultured products and quality control of the large-scale propagation of S. involucrata from tissue culture.
Materials and methods
Callus initiation and maintenance
Seeds of Saussurea involucrata Kar. et Kir. were collected from Hejing County, Xinjiang Province. They were soaked in 70% (v/v) ethanol for 1 min and then rinsed three times in sterile distilled water, surface disinfected with 2% (w/v) sodium hypochlorite for 10 min and then rinsed five times in sterile distilled water and aseptically germinated on MS medium (Murashige and Skoog 1962) containing 3% (w/v) sucrose, solidified with 0.6% agar, at pH 5.8.
When the aseptical seedlings were 3–5 cm long, hypocotyls, cotyledon, and root were cut into sections of 8 mm in length and used as explants for all in vitro experiments. Three kinds of explants were respectively inoculated on the surface of the MS medium supplemented with 2 mg α-naphthaleneacetic acid (NAA/l), 0.5 mg 6-benzylaminopurine (6-BA/l), 3% (w/v) sucrose and 0.6% agar, pH 5.8. The cultured explants were subcultured every 20 days. The experiments were conducted at 25°C under a 16-h photoperiod regime of fluorescent light (30 mmol m−2 s−1).
Plantlet regeneration, hardening and transplant to field
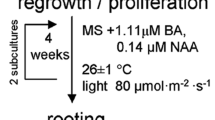
Calli subcultured within three generations were used to induce adventitious shoots. Adventitious shoot induction medium was MS medium supplemented with 0.05 mg NAA/l and 0.5 mg 6-BA/l, 3% (w/v) sucrose and 0.6% agar, pH5.8. Elongated shoots (5–7 cm long) were transferred onto the root induction medium [1/2 MS medium supplemented with 0.2 mg Indole-3-acetic acid (IAA/l), 1 mg activated carbon/l, 2% (w/v) sucrose and 0.6% agar, pH 5.8]. The culture conditions were the same as above. The rooted shoots (regenerated plantlets) were removed from the medium and, after being washed thoroughly to remove any trace of medium, transferred to nursery bed containing soil of Tianshan Mountain and vermiculite. The nursery bed were maintained under greenhouse conditions with temperatures of 25–30°C, disperse light and moderate humidity. The nursery bed was covered with plastic membrane for the first week and increased ventilation gradually at the second week in order to let the regenerated plantlets adapt to the outer environment. The plastic membrane was taken away from the third week. Survived plantlets were transplanted to the planting base in Tianshan Mountains in Xinjiang province after 40–50 days. The planting base located at the 1,600–2,000 m high altitude. The suitable transplanting time is June. After three months growing from June to September, the survival ratio was higher during the whole winter.
Plant materials of S. involucrata used in the genetic analysis
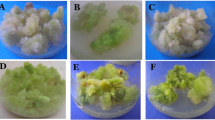
Three kinds of yellowish-green calli were induced from hypocotyls, cotyledon, and root of the aseptical seedlings. Accordingly, three kinds of adventitious shoots induced from the above calli were acquired respectively. Roots germinated from the adventitious shoots induced from hypocotyls. 2 year-old regenerated plantlets, as well as the 1 year-old and 2 year-old seedlings growing in the same planting base in Tianshan Mountain were collected and used as plant materials. The symbols of above 11 types of plant materials (eight tissue-cultured products and three seedlings) are listed in Table 1.
Ten samples of each type of plant materials were used in the following genetic analysis. Proper amount of the 10 samples of each type were mixed and the mixed sample represented the whole genetic information of this type of plant material.
DNA extraction and PCR amplification
Total DNA extraction following a cetyltrimethylammonium bromide (CTAB) protocol was described by Zou et al. (2001). Quality and quantity of DNA was monitored by both gel inspection and spectrometric measurements.
Eighty arbitrary, 10-mer RAPD primers (Sangon Biotech. Inc., Shanghai) were screened for the RAPD analysis. The optimized PCR was performed in a total volume of 25 μl containing 30–50 ng template DNA, 0.6 μM decamer primer, 1 × reaction buffer, 2.3 mM MgCl2, 0.35 mM dNTP mix and 1.8 U Taq polymerase (Promega). The amplification reaction consisted of an initial denaturation step at 95°C for 2 min, followed by 40 cycles of three steps: denaturation at 94°C for 40 s, annealing at 37°C for 1 min, extension at 72°C for 2 min, and a final extension for 7 min at 72°C.
A total of 41, 15–20 mer ISSR primers (Sangon Biotech. Inc., Shanghai) were screened for ISSR analysis. The optimized PCR was performed in a total volume of 25 μl containing 30–50 ng of template DNA, 0.8 μM primer, 1× reaction buffer, 2.5 mM MgCl2, 0.35 mM dNTP mix and 2.5 U Taq polymerase (Promega). The amplification reaction consisted of an initial denaturation step at 94°C for 5 min, followed by 40 cycles of three steps: denaturation at 94°C for 30 s, annealing at 53°C for 45 s, extension at 72°C for 2 min, and a final extension at 72°C for 7 min.
The detailed description of the RAPD and ISSR primers used in the present study were given in Table 2 and 3 respectively. Amplifications were carried out on a MG-96G (Longene, China) thermocycler. Amplification products in both the cases were mixed with loading buffer (0.005% each of Bromophenol Blue, as tracking dyes), run for 2 h on 2% agarose gels at constant voltage (100 V) at room temperature and detected using ethidium bromide.
Data analysis
Two independent amplifications were performed with all the selected RAPD and ISSR primers, and only clear and reproducible bands were scored. Data was scored as 1 for the presence and 0 for the absence of DNA band of each plant material. The similarity matrix and dendrogram were constructed using the NTSYSpc 2.10e (Exeter software, Setauket, N.Y.) software package (Rohlf, 2000). Genetic similarities between samples were measured by the Jaccard’s similarity coefficient (Jaccard, 1908) with SIMQUAL module. Similarity coefficients were used to construct dendrogram using the UPGMA and SHAN routine in NTSYS program.
Results and discussion
Analysis of genetic stability by RAPD
All the initial screening of 80 decamer RAPD primers resulted in selection of 27 oligonucleotides, which produced clear and reproducible amplification products. Each primer produced a unique set of amplification products ranging in size from 200 bp to 4,500 bp. The number of bands for each primer varied from 5 in primer S27 to 12 in primer S55. These 27 primers used in this analysis yielded 202 scorable bands with an average of 7.48 bands per primer. Of The 202 fragments scored from these primers, 132 were monomorphic and 70 were polymorphic (34.7%). The highest number (7) of polymorphic bands was obtained by primer S42, and lowest (0) by primer S26, S27 and S31. Total number of amplified products with all the primer calculated was 1926 (Tables 2, 4). RAPD amplification pattern of 11 mixed samples of S. involucrata was shown by representative gel profiles of primers S55 and S62 (Fig. 1). With primer S55, total number of bands scored was 12 and five were polymorphic (2200, 1900, 1400, 1100 and 700 bp). With primer S62, total number of bands scored was nine and one was polymorphic. One additional 400 bp band was present in Sa and R2.
Similarity values among 11 mixed samples ranged from 0.823 to 0.995 with a mean of 0.878. Among them, the highest similarity value was that between S1 and S2 (0.995). Sa was closely similar to R2 with the similarity value 0.921 and they were closely related to S1 and S2 (Table 5). As shown in the dendrogram (Fig. 2), generated by cluster analysis using the UPGMA method based on Jaccard’s coefficient, Sa and R2, S1 and S2 clustered together respectively and then they clustered into group 1. The three kind of calli were closest related and the similarity values among them ranged from 0.906 to 0.946. They clustered together and formed group 2. Similarly, the three adventitious shoots were closest related and the similarity values among them ranged from 0.931 to 0.961. Rp was more similar to the three adventitious shoots than the other materials. They clustered together and formed group 3.
Analysis of genetic stability by ISSR
A total of fourteen of the forty-one ISSR primers were selected as suitable for the present study. The number of bands for each primer varied from 6 in primer 840 to 10 in primer 808. The 14 primers used in this analysis yielded 115 scorable bands with an average of 8.2 bands per primer. Of the 115 fragments scored from these primers, 77 were monomorphic and 38 were polymorphic (33%). The highest number (5) of polymorphic bands was obtained by primer 841 and 846, and lowest (0) by primer 868. Total number of amplified products with all the primer calculated was 1107 (Tables 3, 4). ISSR amplification pattern of 11 mixed samples of S. involucrata was shown by representative gel profiles of primers 848 and 873 (Fig. 3). With primer 848, total number of bands scored was 9 and 3 were polymorphic (900, 700 and 600 bp). With primer 873, total number of bands scored was 9 and 3 was polymorphic (2300, 1900 and 900 bp). One 2,300 bp band was missing in Sa and Ch; one additional 1,900 band was present in Sa; one 900 bp band was missing in Ch, Cr and Sr.
Similarity values among 11 mixed samples ranged from 0.824 to 0.974 with a mean of 0.885. Similar to the situation of RAPD, the highest similarity value was that between S1 and S2 (0.974), and they were closely related to R2 and Sa (Table 6). As shown in the dendrogram (Fig. 4), S1, S2, R2 and Sa clustered into group 1. The three calli were closest related and the similarity values among them ranged from 0.860 to 0.886. They clustered into group 2. Sc were closest related to Rp with the similarity values 0.947, which is different from the result in RAPD. They clustered together with Sh and Sr successively and formed Group 2. Different to that of RAPD analysis, Group 1 clustered together with group 2 and then clustered with group 3.
Genetic quality control and screening of tissue-cultured products
The similarity coefficients between three calli and Sa were closed revealed both by RAPD and ISSR markers (ranged from 0.842 to 0.868), indicating genetic differences among the three calli were not remarkable. Therefore, in the screening of calli used for the next redifferentiation studies, only chemical components and growth properties of the callous tissues needed consideration. The similarity coefficients between three adventitious shoots and Sa were also very closed assessed by RAPD marker (0.872–0.882) but ranged from 0.886 (between Sr and Sa) to 0.930 (between Sh and Sa) assessed by ISSR marker. Combined with the rooting ability of the three adventitious shoots, Rp induced from Sh was used to hardening and transplant to field. The similarity coefficient between Rp and Sa was lower than those between the adventitious shoots and Sa assessed by RAPD marker (0.823), but relatively higher assessed by ISSR marker (0.912). After two years’ cultivation in the planting base in Tianshan Mountain, R2 was closer to Sa both in RAPD and ISSR analyses (similarity coefficient was 0.921 and 0.930, respectively), and was much closer to S2 in ISSR analysis (similarity coefficient was 0.956) but not the same in RAPD analysis (similarity coefficient was 0.882).
In the current study, similar polymorphism within the tissue-cultured products and seedlings of S. involucrate obtained by RAPD (34.7%) and ISSR (33%) analysis. The DNA polymorphism in tissue-cultured products provides evidence for the occurrence of genetic variation produced in in vitro culture from hypocotyls, cotyledon and root explants of same seedlings. This seems to be quite alarming and further stresses the need for testing regenerated plantlets well before and after their actual outplanting. The presence or absence of genetic variation depends upon the source of explant and method of regeneration or on the source of the regenerants (callus, protoplast and cell) (Larkin et al. 1989). We have used callus for the rapid propagation of S. involucrata. This method is considered to be a high-risk method for genetic instability and, therefore, corroborate our findings of genetic variation among tissue-cultured products of S. involucrata.
References
Fu CX, Jin ZP, Yang R, Wu FY, Zhao DX (2004) Establishment of Saussurea involucrata hairy roots culture and plantlet regeneration. Chin J Biotechnol 20(3):366–371
Gould AR (1986) Factors controlling generation of variability in vitro. In: Vasil IK (ed) Cell culture and somatic cell genetics in plants. Academic Press, Orlando, pp 549–567
Guo WL, Gong L, Ding ZF, Li YD, Li FX, Zhao SP, Liu B (2006) Genomic instability in phenotypically normal regenerants of medicinal plant Codonopsis lanceolata Benth. et Hook. f., as revealed by ISSR and RAPD markers. Plant Cell Rep 25:896–906
Jaccard P (1908) Nouvelles recherches surla distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Karp A (1991) On the current understanding of somaclonal variation in plants. Oxford Surv Plant Mol Cell Boil 7:1–58
Larkin P, Scowcroft WR (1981) Somaclonal variation, a novel source of variability from cell cultures for plant improvement. Theor Appl Genet 60:197–214
Larkin PJ, Banks PM, Bhati R, Bretell RIS, Davis PA, Ryan SA, Scowcroft WR, Spindler LH, Tanner GJ (1989) From somatic variation to variant plants: mechanism and applications. Genome 31:705–711
Mangolin CA, Ottoboni LMM, Machado PS (2002) RAPD markers to evaluate callus tissue of Cereus peruvianus mill. (Cactaceae) maintained in different growth regulator combinations. Biochem Genet 40:351–358
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15:473–497
Peschke VM, Phillips RL (1992) Genetic implication of somaclonal variation in plants. Adv Genetics 30:41–75
Piao RZ, Cao HN, Chen YQ, Jin YS, Zong CW (2003) Study on the technology of rapid propagation in vitro for saussurea involucrata kar. et kir. Chin J Agr Sci Yanbian Uni 25(2):117–121
Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system. Exeter Software, New York
Thomas JB, Vijayan D, Joshi SD, Lopez SJ, Kumar RR (2006) Genetic integrity of somaclonal variants in tea (Camellia sinensis (L.) O Kuntze) as revealed by inter simple sequence repeats. J biotechnol 123:149–154
Wa G, Liu JL, Shi YH (1990) Tissue culture of Saussurea involucrata. Chin XinJiang Ind Sci 5:221–222
Wu LQ, Guo SX, Xiao PG (2005) Tissue culture and plantlet regeneration from embryo of Saussurea involucrata. China J Chin Materia Medica 30:814–816
Xie ZB, Jiang GF, Liao FS, Yue ZG (2007) Advances in studies of chemical composition and medicinal value of snowdrop. Chin Food Sci Technol 6:254–257
Yang H, Tabei Y, Kamada H, Kayano T, Takaiwa F (1999) Detection of somaclonal variation in cultured rice cells using digoxigenin-based random amplified polymorphic DNA. Plant cell Rep 18:520–526
Zhao HQ, Wang XJ (2008) Establishment of rapid propagation technique in tissue culture of Saussurea involucrata Kar. et Kir. from Tianshan Mountain. Chin J Anhui Agr Sci 36(7):2743–2744
Zhao B, Xu CM, Wang YC, Yuan XF, Lei L, Wang XD (2005a) Callus induction and rapid propagation technique of regenerated plantlets of Saussurea involucrata. Chin Invention Patent, Application No. 200510012283.2
Zhao B, Wang, XJ, Xu CM, Wang YC, Liu M, Bolati, Yuan XF, Zhao HQ, Wang XD (2005b) Large-scale artificial cultivation of regenerated plantlets of Saussurea involucrate. Chin Invention Patent, Application No. 200510012284.7
Zou YP, Ge S, Wang XD (2001) Molecular markers in systematic and evolutionary botany. Science Publisher, Beijing, pp 16–17
Acknowledgements
This study was supported by the National Natural Science Foundation of China (20506027) and Hi-tech Program of Western Plan of Chinese Academy of Sciences (KGCX2-YW-509).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yuan, X.F., Dai, Z.H., Wang, X.D. et al. Assessment of genetic stability in tissue-cultured products and seedlings of Saussurea involucrata by RAPD and ISSR markers. Biotechnol Lett 31, 1279–1287 (2009). https://doi.org/10.1007/s10529-009-9984-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-009-9984-6