Abstract
Previously we reported that double disruption of the proteinase genes (tppA and pepE) improved heterologous protein production by Aspergillus oryzae (Jin et al. Appl Microbiol Biotechnol 76:1059–1068, 2007). Since A. oryzae has 134 protease genes, the number of auxotrophy in a single host is limited for multiple disruptions of many protease genes. In order to rapidly perform multiple gene disruptions in A. oryzae, we generated the marker recycling system in highly efficient gene-targeting background. A. oryzae ligD gene homologous to Neurospora crassa mus-53 gene involved in nonhomologous chromosomal integration was disrupted, followed by disruption of the pyrG gene for uridine/uracil auxotroph. We further performed successive rounds of gene disruption (tppA and pepE) by the pyrG marker with high gene-targeting efficiency allowed by the ΔligD background. After each disruption process the pyrG marker was excised by the direct repeats consisting of ~300 bp upstream flanking region of the target gene, resulting in no residual ectopic/foreign DNA fragments in the genome. Consequently, we succeeded to breed the double proteinase gene disruptant (ΔtppA ΔpepE) applicable to further sequential gene disruptions in A. oryzae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aspergillus oryzae is an excellent host for heterologous protein production due to its high protein productivity and its safety guaranteed by its use in the manufacture of Japanese fermented foods for over a thousand years (Kitamoto 2002). In order to enhance the ability of protein production it is important to breed the host applicable to multiple rounds of genetic manipulations. Our breeding of A. oryzae quadruple auxotrophic host (niaD − sC − ΔargB adeA −) has enabled us to manipulate as many as four genes in a single host strain (Jin et al. 2004).
In general, the production level of heterologous proteins by A. oryzae is much lower compared to the homologous (fungal) proteins (Tsuchiya et al. 1994; Nakajima et al. 2006; Jin et al. 2007; Ito et al. 2007). We reported that double proteinase gene disruption (tppA and pepE) improved the heterologous protein production (Jin et al. 2007). The genome sequencing project revealed that A. oryzae has 134 protease genes (Machida et al. 2005). For multiple disruptions of protease genes, however, the number of selective markers in a single host is limited in A. oryzae.
The pyrG gene encoding orotidine-5′-phosphate (OMP) decarboxylase has been used for marker recycling, allowing multiple gene disruptions in Aspergillus fumigatus and Aspergillus nidulans (d’Enfert 1996; Nielsen et al. 2006), since positive selection for pyrG-excised strains can be done by using 5-fluoro-orotic acid (5-FOA) which is converted to the toxic intermediate 5-fluoro-UMP (Boeke et al. 1984). The Cre/loxP recombinase system of bacteriophage P1 also allows consecutive gene disruptions in A. nidulans (Forment et al. 2006). Based on these reports, however, multiple gene disruptions led to accumulation of many copies of ectopic/foreign DNA fragments in the genome. In A. oryzae, no attempts for marker recycling have been reported.
In order to perform multiple gene disruptions rapid and efficient methods are required. Ninomiya et al. (2004) developed a highly efficient gene-targeting system in Neurospora crassa by disruption of the genes encoding Ku70 and Ku80 that play a role in nonhomologous chromosomal integration. Ku-deficient strains have been employed for highly efficient gene-targeting in A. oryzae (Takahashi et al. 2006). Moreover, it was reported that all pathways of nonhomologous chromosomal integration in N. crassa are under the control of MUS-53 (human Lig4 homolog) (Ishibashi et al. 2006). The mus-53 gene disruptant shows 100% gene-targeting frequency even with short (100-bp) flanking sequences, providing a remarkably efficient system for gene targeting. Very recently, it was shown that gene-targeting efficiency was improved by defect of the homolog of MUS-53 (LigD) in A. oryzae (Mizutani et al. 2008).
In this study we performed multiple gene disruptions by developing a marker-recycling system in A. oryzae. In order to rapidly carry out multiple gene disruptions, high targeting frequency was achieved by disruption of the gene homologous to N. crassa mus-53. Finally, we generated the double proteinase gene disruptant (ΔtppA ΔpepE) applicable to further multiple gene disruptions in A. oryzae.
Materials and methods
Strains and growth media
Aspergillus oryzae wild type strain, RIB40 (Machida et al. 2005) and quadruple auxotrophic strain, NSAR1 (Jin et al. 2004) (Supplementary Table 1) were used as a DNA donor and for transformation, respectively. The strains generated in this study are listed in Supplementary Table 1. DPY (20 g/l dextrin, 10 g/l polypeptone, 5 g/l yeast extract, 5 g/l KH2PO4, and 0.5 g/l MgSO4 · 7H2O (pH 5.5)) and potato/dextrose/agar (PDA) media were used for growth. M + Met medium (2 g/l NH4Cl, 1 g/l (NH4)2SO4, 0.5 g/l KCl, 0.5 g/l NaCl, 1 g/l KH2PO4, 0.5 g/l MgSO4 · 7H2O, 0.02 g/l FeSO4 · 7H2O, 20 g/l glucose, 1.5 g/l methionine pH 5.5) with required supplements (0.1 g/l adenine, 10–40 mM uridine, and 1–4 g/l uracil) was used for transformation and growth of A. oryzae strains. Escherichia coli DH5α (supE44 ΔlacU169 (Φ80 lacZ ΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1) was used for DNA manipulation.
Transformation of A. oryzae
Transformation of A. oryzae was done according to the method of Kitamoto (2002).
Molecular techniques
The recombination reactions for plasmid construction in the MultiSite Gateway system were carried out as instructed by the manufacturer (Invitrogen, San Diego, CA, USA). DNA fragments were amplified with the PrimeSTAR HS DNA Polymerase (TaKaRa, Otsu, Japan) and their nucleotide sequences were confirmed with ABI PRISM 310NT Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). All primers used in this study are listed in Supplementary Table 2. Gene disruption methods in A. oryzae are described in Supplementary Methods.
Excision of the pryG marker
Conidia (approx. 106–3 × 106) of the gene disruptants with the pyrG marker were spread on PD agar medium containing 1 mg 5-FOA/ml and 20 mM uridine, and then incubated at 30°C. After 5–8-day cultivation, colonies were observed to grow and transferred onto another 5-FOA agar medium supplemented with uridine. The 5-FOA resistant strains were examined for uridine/uracil auxotrophy. In order to verify the pyrG excision PCR analysis was done using the primers (tpp-1340_F and tpp + 237_R for tppA, pEp-488_F and pEt + 162_R for pepE) and genomic DNAs of transformants as template.
Southern analysis
The strains were identified by Southern analysis. After electrophoresis, the digested genomic DNAs were transferred onto Hybond N+ membrane (GE Healthcare, Piscataway, NJ, USA). ECL (enhanced chemiluminescence) direct nucleic acid labeling and detection system (GE Healthcare) and LAS-100plus luminescent image analyzer (Fuji Photo Film, Tokyo, Japan) were used for detection.
Results
Disruption of A. oryzae ligD gene homologous to N. crassa mus-53 gene
We searched the gene homologous to N. crassa mus-53 gene in A. oryzae Genome database DOGAN (http://www.bio.nite.go.jp/dogan/MicroTop?GENOME_ID=ao). The gene with the gene ID AO090120000322 (designated as ligD (Mizutani et al. 2008)) was predicted to encode the 1,006 amino acid protein sharing 49.4% homology with MUS-53. For disruption of the ligD gene the flanking regions of the ligD ORF were cloned and connected with the selective marker, argB. The DNA fragment for disruption of the ligD gene was introduced into the A. oryzae quadruple auxotrophic strain, NSAR1 (Jin et al. 2004; Supplementary Table 1). PCR analysis using the genomic DNAs revealed that in 6 of the 16 transformants the ligD gene was disrupted, which showed 38% disruption efficiency (Table 1). Southern analysis was also performed to verify disruption of the ligD gene (Supplementary Fig. 1).
The ligD disruptants grew and conidiated comparably to the non-disrupted transformants (Fig. 1a). This suggests that the strains can be normally used for further experiments such as protein production. On the other hand, the ΔligD strain reduced the growth in the presence of methyl methanesulfonate (MMS), a chemical mutagen (Fig. 1a). These phenotypes of the ΔligD strain generated in our study were consistent with the other recent report of the ligD gene disruption in A. oryzae (Mizutani et al. 2008).
Generation of the ΔligDΔpyrG strain in A. oryzae. (a) Growth of the ligD disruptant. Conidia (approximately 100 conidia/5 μl) were inoculated on the agar media and incubated at 30°C for 3 and 5 days in the absence or presence of MMS, respectively. (b) Growth of the ΔligDΔpyrG strain. Conidia (approximately 600 conidia/5 μl) were spotted on PD media with indicated supplements and 5-FOA (800 μg/ml). The agar plates were incubated at 30°C for 3 days. ΔligD::argB adeA −: the parental strain (NSR-ΔlD2), ΔligD::argB ΔpyrG::adeA: the pyrG disruptant (NSPlD1), ΔligD::argB ΔpyrG::adeA [pyrG]: the pyrG-complemented strains (Host: NSPlD1, Plasmid: pgEpG (See Supplementary Methods))
Isolation of the pyrG disruptant and its resistance to 5-FOA
For adding uridine/uracil auxotrophy in the ligD disruptant, the pyrG gene was disrupted. The 2 kb-flanking regions of the pyrG ORF were cloned and connected with the selective marker, adeA. The disruption fragment for the pyrG gene was introduced into the ligD disruptant (NSR-ΔlD2). Transformation frequency was not significantly influenced by disruption of the ligD gene. Transformants were obtained on the selective medium that contained uridine for growth of uridine/uracil auxotrophic strains. Out of the 13 transformants examined, 12 showed uridine/uracil auxotrophy, which showed the high efficiency (92%) for disruption of the pyrG gene (Table 1). Disruption of the pyrG gene was verified by Southern (Supplementary Fig. 2) and PCR analyses (data not shown).
The pyrG disruptant (NSPlD1) could be normally transformed with the plasmid harboring the wild type pyrG gene. The transformants were able to grow in the absence of uridine/uracil while the pyrG disruptants did not form colonies on the same medium (Fig. 1b). However, only the pyrG disruptant showed resistance against 5-FOA (Fig. 1b). These results suggested that positive selection using 5-FOA for uridine/uracil auxotrophs could be applied to marker recycling of the pyrG marker in A. oryzae.
Disruption of the tppA gene with the pyrG marker
The 1.3 kb flanking regions of the tppA ORF were cloned and connected with the pyrG gene. In this construct 3′-end of the upstream flanking region of the tppA ORF (~300 bp) was fused with the downstream flanking region of the tppA ORF so that the pyrG marker was flanked by the ~300 bp directed repeats (Fig. 2a; gray box). The disruption fragment for the tppA gene was introduced into the ΔligDΔpyrG strain (NSPlD1). PCR analysis revealed that out of the 9 transformants, 8 showed disruption of the tppA gene, indicating high disruption efficiency (89%) (Table 1). Disruption of the tppA gene was also verified by Southern analysis (Fig. 2b).
Disruption of the tppA gene in the ΔligDΔpyrG background. (a) Schematic model of disruption of the tppA gene. The boxes (1.3 kb) are the flanking regions used for disruption of the tppA gene. The 0.3 kb upstream flanking region of the tppA gene (boxed in gray) was attached at 5′-end of the downstream flanking regions, introducing direct repeats. (b) Southern analysis of the tppA disruptants The genomic DNAs were digested with HincII and SphI. All 3 strains analyzed in the panel (lanes 1–3) exhibited the expected band pattern for disruption of the tppA gene. “P” represents the parental strain (NSPlD1)
Excision of the pyrG marker by using 5-FOA
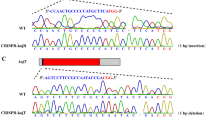
Since the uridine/uracil auxotrophs were resistant to 5-FOA in A. oryzae, positive selection for pyrG − strains was carried out using 5-FOA. It was expected that the pyrG inserted at the tppA locus would be excised out by homologous recombination with the direct repeats, in which the flanking regions of the tppA ORF were directly connected without leaving any ectopic/foreign DNA fragments (Fig. 3a). Conidia of the ΔtppA::pryG strains (NSlD-tA1~3) were spread onto the agar medium containing 5-FOA and uridine. One colony per approx. 106 conidia appeared after 5–8-day cultivation and the resulting 5-FOA resistant strains exhibited uridine/uracil auxotrophy (Fig. 3b). The strains were confirmed for pyrG marker excision by PCR (data not shown) and Southern analyses (Fig. 3c). These data indicate that the pyrG inserted at the tppA locus was successfully excised by homologous recombination with the flanking direct repeats.
Excision of the pyrG gene targeted at the tppA locus. (a) Schematic model of disruption of the tppA gene. By homologous recombination of the direct repeats consisting of the 0.3 kb upstream flanking region of the tppA gene (boxed in gray), the pyrG gene targeted at the tppA locus is excised, and then the upstream and downstream flanking region of the tppA ORF are directly connected. Note that no ectopic/foreign DNA fragments are left in the genome after excision of the pyrG marker. (b) Growth of the pyrG-excised strains. The conidia (approximately 500 conidia/5 μl) of the strains were spotted onto the M+Met medium with or without uridine and uracil, and then incubated at 30°C for 3 days. “P1–P3” represent the parental strains (NSlD-tA1~3). “1–3” are the pyrG-excised strains derived from the NSlD-tA1~3 strains. (c) Southern analysis of the pryG-excised strains. The genomic DNAs were digested with EcoRV and SphI
Successive round of gene disruption (pepE) and marker recycling
Next, the 1.3 kb flanking regions of the pepE ORF were cloned and connected with the pyrG gene. In this construct 3′-end of the upstream flanking region of the pepE ORF (~300 bp) was fused with the downstream flanking region of the pepE ORF so that the pyrG marker was flanked by the ~300 bp directed repeats (Fig. 4a; hatched box). The disruption fragment for the pepE gene was introduced into the ΔligDΔpyrGΔtppA strain (NSPlD-tA3). PCR analysis revealed that out of the 10 transformants 9 showed uridine/uracil auxotrophy, which was 90% efficiency for disruption of the tppA gene (Table 1). Disruption of the pepE gene was verified by Southern (Fig. 4a) and PCR analyses (data not shown).
Successive round of gene disruption (pepE) and marker recycling. (a) Southern analysis of the pepE disruptants. The boxes (2.0 and 1.9 kb) are the flanking regions for disruption of the pepE gene. The 0.3 kb upstream flanking region of the pepE ORF (hatched box) was attached at 5′-end of the downstream flanking regions, introducing direct repeats. The genomic DNAs were digested with SphI and XbaI. All 4 strains analyzed in the panels (lanes 1–4) exhibited the expected band pattern for disruption of the pepE gene. “P” represents the parental strain (NSPlD-tA3). (b) Southern analysis of the pyrG-excised strains. The genomic DNAs were digested with SphI and XbaI. All 4 strains analyzed in the panels (lanes 1–4) exhibited the expected band pattern for excision of the pyrG gene from the pepE locus. “P” represents the parental strain (NSlD-tApE2)
For another round of marker recycling, pyrG − strains were positively selected from the ΔpepE::pyrG strain (NSlD-tApE2) using 5-FOA. One colony per approximately 2.3 × 105 conidia appeared after 5–8-day cultivation on the agar medium containing 5-FOA and uridine. The 5-FOA resistant strains showed uridine/uracil auxotrophy and confirmed for pyrG marker excision by PCR (data not shown) and Southern analyses (Fig. 4b). Finally, we generated the double disruptant of proteinase genes (tppA and pepE), which is still applicable to successive rounds of gene disruptions.
Discussion
In order to perform multiple gene disruptions for breeding of excellent host strains, many target genes should be disrupted rapidly and efficiently. Therefore, we generated the ligD disruptant for highly efficient gene-disruption frequency in A. oryzae. The genes (pyrG, tppA, and pepE) could be disrupted at very high frequency (~90%) in the ΔligD background (Table 1). The disruption rate in the ΔligD background is higher than the A. oryzae Ku70-deficient strain (Takahashi et al. 2006). This may be consistent with that N. crassa mus-53 disruptants shows higher gene-targeting rate than the Ku-deficient strain (mus-52 disruptant) (Ishibashi et al. 2006). Moreover, the ΔligD strain grew normally and conidiated comparably to the non-disrupted transformants (Fig. 1a). These data propose that use of the ΔligD strain is effective in multiple gene disruptions. However, the gene disruption efficiency could not always reach 100% in the ΔligD background of A. oryzae. In other recent paper of the ligD disruptant in A. oryzae, some of the genes could not be disrupted at 100% efficiency (Mizutani et al. 2008). In this study we used the 1.3–2.0 kb flanking regions of target genes for disruption, while the same length of the flanking fragments led to 100% disruption frequency in N. crassa mus-53 disruptant (Ishibashi et al. 2006). This suggests some distinct chromosomal recombination mechanism in A. oryzae from N. crassa.
This paper first describes that the multiple gene disruptions with maker recycling were done in the highly efficient gene-targeting background in filamentous fungi. We took advantage of the direct repeats consisting of the short upstream flanking region of the disrupted ORF, allowing multiple gene disruptions without leaving any ectopic/foreign DNA remnants in the genome during marker recycling. Recently, we reported that by monitoring global expression of protease genes in A. oryzae the nptB gene encoding a neutral protease was chosen from the protease genes induced during the cultivation and that its disruption improved the production level of the heterologous protein (Kimura et al. 2008). We are planning to breed additional gene disruptant such as the nptB gene using the double proteinase disruptant. It is expected that our technique of multiple gene disruptions would contribute to not only further improvement of heterologous protein production but also functional analyses of many genes of interest in A. oryzae.
References
Boeke JD, LaCroute F, Fink GR (1984) A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346
d’Enfert C (1996) Selection of multiple disruption events in Aspergillus fumigatus using the orotidine-5′-decarboxylase gene, pyrG, as a unique transformation marker. Curr Genet 30:76–82
Forment JV, Ramón D, MacCabe AP (2006) Consecutive gene deletions in Aspergillus nidulans: application of the Cre/loxP system. Curr Genet 50:217–224
Ishibashi K, Suzuki K, Ando Y, Takakura C, Inoue H (2006) Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc Natl Acad Sci USA 103:14871–14876
Ito K, Asakura T, Morita Y, Nakajima K, Koizumi A, Shimizu-Ibuka A, Masuda K, Ishiguro M, Terada T, Maruyama J, Kitamoto K, Misaka T, Abe K (2007) Microbial production of sensory-active miraculin. Biochem Biophys Res Commun 360:407–411
Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K (2004) Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett 239:79–85
Jin FJ, Watanabe T, Juvvadi PR, Maruyama J, Arioka M, Kitamoto K (2007) Double disruption of the proteinase genes, tppA and pepE, increases the production level of human lysozyme by Aspergillus oryzae. Appl Microbiol Biotechnol 76:1059–1068
Kimura S, Maruyama J, Takeuchi M, Kitamoto K (2008) Monitoring global gene expression of proteases and improvement of human lysozyme production in nptB gene disruptant of Aspergillus oryzae. Biosci Biotechnol Biochem 72:499–505
Kitamoto K (2002) Molecular biology of the Koji molds. Adv Appl Microbiol 51:129–153
Machida M, Asai K, Sano M et al (2005) Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161
Mizutani O, Kudo Y, Saito A, Matsuura T, Inoue H, Abe K, Gomi K (2008) A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet Biol (in press)
Nakajima K, Asakura T, Maruyama J, Morita Y, Oike H, Shimizu-Ibuka A, Misaka T, Sorimachi H, Arai S, Kitamoto K, Abe K (2006) Extracellular production of neoculin, a sweet-tasting heterodimeric protein with taste-modifying activity, by Aspergillus oryzae. Appl Environ Microbiol 72:3716–3723
Nielsen ML, Albertsen L, Lettier G, Nielsen JB, Mortensen UH (2006) Efficient PCR-based gene targeting with a recyclable marker for Aspergillus nidulans. Fungal Genet Biol 43:54–64
Ninomiya Y, Suzuki K, Ishii C, Inoue H (2004) Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA 101:12248–12253
Takahashi T, Masuda T, Koyama Y (2006) Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol Genet Genomics 275:460–470
Tsuchiya K, Nagashima T, Yamamoto Y, Gomi K, Kitamoto K, Kumagai C, Tamura G (1994) High level secretion of calf chymosin using a glucoamylase-prochymosin fusion gene in Aspergillus oryzae. Biosci Biotechnol Biochem 58:895–899
Acknowledgements
We thank to Yukiko Oshima for experimental help. This study was supported by a Grant-in-Aid for Scientific Research (S) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplementary Fig. 1
Southern analysis of the ligD disruptants. The genomic DNAs were digested with BamHI and SmaI. The boxed regions are the flanking regions used for disruption of the ligD gene. Four strains were analyzed, confirming that the strains (lanes 1 and 3 in the panels) exhibited the expected band pattern for disruption of the ligD gene. “P” represents the parental strain (NSAR1) (PDF 79 kb)
Supplementary Fig. 2
Southern analysis of the pyrG disruptant. The genomic DNAs were digested with PstI and XhoI. The flanking regions used for disruption of the pyrG gene are boxed. All 4 strains analyzed in the panel (lanes 1 to 4) exhibited the expected band pattern for disruption of the pyrG gene. “P” represents the parental strain (NSR-ΔlD2) (PDF 182 kb)
Rights and permissions
About this article
Cite this article
Maruyama, JI., Kitamoto, K. Multiple gene disruptions by marker recycling with highly efficient gene-targeting background (ΔligD) in Aspergillus oryzae . Biotechnol Lett 30, 1811–1817 (2008). https://doi.org/10.1007/s10529-008-9763-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9763-9