Abstract
Dysosma pleiantha, an important threatened medicinal plant species, is restricted in distribution to southeastern China. The species is capable of reproducing both sexually and asexually. In this study, inter-simple sequence repeat marker data were obtained and analyzed with respect to genetic variation and genetic structure. The extent of clonality, together with the clonal and sexual reproductive strategies, varied among sites, and the populations under harsh ecological conditions tended to have large clones with relatively low clonal diversity caused by vegetative reproduction. The ramets sharing the same genotype show a clumped distribution. Across all populations surveyed, average within-population diversity was remarkably low (e.g., 0.111 for Nei’s gene diversity), with populations from the nature reserves maintaining relatively high amounts of genetic diversity. Among all populations, high genetic differentiation (AMOVA: ΦST = 0.500; Nei’s genetic diversity: G ST = 0.465, Bayesian analysis: ΦB = 0.436) was detected, together with an isolation-by-distance pattern. Low seedling recruitment due to inbreeding, restricted gene flow, and genetic drift are proposed as determinant factors responsible for the low genetic diversity and high genetic differentiation observed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Information on genetic diversity patterns can provide insight into evolutionary and demographic history of a taxon (Milligan et al. 1994). Understanding the relative importance of processes that structure diversity within and among populations (specifically inbreeding, gene flow, genetic drift, and selection) can provide both a means to assess future risk of erosion of diversity and a means for designing effective conservation strategies for rare taxa (Neel and Ellstrand 2003). The distribution of genetic variation in space is also a prime factor to consider in the conservation and management of natural populations (McCue et al. 1996). For example, developing sample strategies for recovery and, where appropriate, reintroduction may greatly benefit from this information (Neel and Cummings 2003).
The world trade figures suggest that China is first with exports of medicinal raw materials (Lange 1997). The current trend toward increased commercialization has resulted in overharvesting of some economically important medicinal plants, many of which have become threatened. Threatened medicinal plant species have become the focus of world attention because they represent a vanishing and decreasing flora in need of protection and conservation and because of their role as an essential commodity for health care (Kala 2005). The Podophyllaceae [formerly considered as a separate family but now included in Berberidaceae (APG II 2003)] comprises six genera: Achlys, Diphylleia, Dysosma, Jeffersonia, Ranzania, and Podophyllum. Among the six genera, Dysosma, consisting of seven species, is restricted to China. Dysosma pleiantha (Hance) Woodson (Berberidaceae), a perennial herb, is restricted in distribution to southeastern China (Ying et al. 1993). This region has been recognized as an important center of origin for threatened medicinal plants (Kala 2005). The rhizomes of Dysosma pleiantha, also known as guijiu, are well known in traditional Chinese medicine. The active constituent of guijiu is podophyllotoxin, which has cytotoxic and antitumor properties and has been used for cancer treatment (Jackson and Dewick 1985). In recent years, the species has been subject to a rapid demographic decline. Based on census estimates (H. L. Liu, unpubl. data), the population size of D. pleiantha is generally small, ranging from 150 to 800 individuals (see Table 1). The two populations within Tianmushan Nature Reserve (TM) and Sanqing Nature Reserve (SQ) are the largest known populations of the species, comprising approximately 800 individuals. The other populations were estimated to be fewer than 300 individuals (Table 1). As to its overall conservation status, D. pleiantha has been ranked as “threatened” on the China Species Red List (Wang and Xie 2004).
Many perennial plants combine sexual reproduction and clonal propagation as population regeneration mechanisms (Richards 1997). In some clonal species, the success of sexual versus clonal recruitment often varies geographically in response to ecological and genetic factors that limit one regeneration mechanism or the other (Eckert 2002). The demographic balance between sexual and clonal recruitment is likely to have important consequences for the clonal diversity and genetic structure of plant populations (Ellstrand and Roose 1987; Eckert and Barrett 1993). Higher rates of sexual reproduction will increase heterozygosity and decrease population differentiation (Balloux et al. 2003). As clonal recruitment may reduce the number of genetically distinct individuals within a population, an understanding of clonality is critical for the implementation of the most appropriate conservation management of threatened clonal plants (Young et al. 2002). Clonal reproduction is thought to be extensive for the species in Dysosma, and rates of germination and seedling emergence and establishment in the field tend to be low (Qiu et al. 2005). Our previous isozyme study on the clonal diversity of five populations of D. versipellis and one population of D. pleiantha showed that D. pleiantha appears to consist of multiple genets compared with its allopatric congener D. versipellis. Qiu et al. (2005) speculated that sexual reproduction is likely more important than clonal reproduction in D. pleiantha; however, only one D. pleiantha population was included in a previous isozyme analysis. Again, a problem with the application of enzyme electrophoresis for clonal identification and detecting genetic variation is the low number of polymorphic loci available in many studies (Ellstrand and Roose 1987; Qiu et al. 2005; Clark-Tapia et al. 2005). DNA type markers are able to detect the genetic variation beyond coding loci and to provide broader information on the amount of genetic variation and the genetic divergence among populations. Dominantly expressed multilocus DNA markers such as inter-simple sequence repeats (ISSRs) have been successfully used to assay the levels of clonal diversity and apportionment of genetic diversity within plant species and populations (Culley and Wolfe 2001; Qiu et al. 2006). Moreover, ISSR is useful in the estimation of genetic diversity when compared with other types of neutral DNA markers (Nybom 2004).
In the present study, we used ISSR markers to (1) investigate the levels and distribution of genetic variability within and among populations of D. pleiantha, and (2) detect the possible factors that might explain the patterns and levels of genetic variation observed. Additionally, we present measures of ISSR variation within and among populations of D. pleiantha and compare them with data published for its congener D. versipellis (Qiu et al. 2006). Overall, such information can also serve as a guide to preserving the genetic resources of this medicinally important, severely threatened species.
Materials and Methods
Study System and Population Sampling
Dysosma pleiantha is a diploid, herbaceous perennial, with a reported chromosome number of 2n = 2× =12 (Zhang et al. 1991). It grows from the rhizomes and typically reaches 10–30 cm in height. The nonbranching shoots bear opposite, centrally peltate, rounded leaves with 5–9 lobes and finely dentate margin. Plants remain in a juvenile phase for 4–5 years. When mature, they produce a terminal cyme with 5–8 drooping red-purple flowers in April. The most frequent pollinators are Chrysomya megacephala and Musca domestica (Y. X. Qiu, pers. obs.). Each plant produces 1–3 large berries (about 4–5 cm long and 2–3 cm wide), each with 20–30 minute seeds. The fruit ripens in late June and is dispersed by gravity (Li and Wang 2006). Fruit set is typically low and varies widely from year to year (Ying et al. 1993; Qiu et al. 2005). D. pleiantha grows in rocky and humous soils on hillsides and occurs primarily in mixed evergreen and deciduous forests, at altitudes between ca. 500 and 800 m. The forest habitats of D. pleiantha in southeast China are dominated by subtropical and temperate woody species such as Quercus acutissima Carruth, Cunninghamia lanceolata (Lamb.) Hook., Liquidambar formosana Hance, Alangium chinense (Lour.) Harms., and Cyclobalanopsis glauca (Thunb.) Oerst. (Y. X. Qiu, pers. obs.). Some D. pleiantha populations, however, survive in artificial bamboo forests due to deforestation, and these populations are characterized by very low density and patchy distribution (e.g., populations DQ and JN in Table 1) (Li and Wang 2006). D. pleiantha is distributed only in fragmented populations within the Zhejiang, Jiangxi, Anhui, and Fujian provinces of southeast China (Ying et al. 1993). Most populations in southeast China are small and isolated (Qiu et al. 2005). This may be caused in part by (1) destruction of natural habitat, (2) narrow-niche habitats, and (3) historical factors (e.g., historical fragmentation).
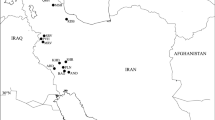
Collections of silica-dried leaf material of D. pleiantha were made from five natural populations, representing the overall distribution of the species in China (Fig. 1). For ISSR polymorphism, 15–23 individuals were assayed per population, with 105 putative genets in total (Table 1). Because the species can spread vegetatively via rhizomes, care was taken to collect leaf material from putative genets at intervals of more than 15 m.
Location of the five natural populations of Dysosma pleiantha in China. Abbreviations as in Table 1
The mode of reproduction (sexual vs. asexual) is likely to have important effects on genetic variation and its spatial distribution within plant populations. Thus, it is necessary to investigate the extent of clonality in Dysosma pleiantha. However, few highly clumped patches were found for D. pleiantha in the field, because of habitat destruction and overexploitation of the natural population for medicinal use (Qiu et al. 2005). Finally, two small plots (TT and JN, approximately 25 m2) and one large plot (TM, approximately 60 m2) were selected for clonal study. Leaf materials from all ramets within these plots were sampled (N = 113), and the exact locations of all ramets were recorded by measuring their distance from the plot margins. Voucher specimens representative of all the populations sampled (Table 1) are stored at the Herbarium of Zhejiang University.
DNA Extraction and ISSR-PCR Amplification
Total genomic DNA was extracted using the modified CTAB method of Doyle (1991). DNA concentrations were determined on ethidium bromide-stained agarose gels by comparison with known amounts of DNA, and by spectrophotometry. Working stocks of DNA were then prepared based on both estimates, and stored in 0.1× TE buffer. ISSR-PCR reactions were carried out in 25 μl total volume containing 60 ng genomic DNA, 10 mM Tris–HCl (pH 9.0), 25 mM KCl, 1.5 mM MgCl2, 0.25 mM dNTPs, 2% formamide, 1.0 U Taq polymerase, and 0.3 μM primers. Amplifications were performed using a GeneAmp 9700 DNA Thermal Cycler (Perkin-Elmer, USA). The PCR program included an initial denaturing step for 5 min at 94°C, followed by 45 cycles of 1 min at 94°C, 45 s at 52°C, and 2 min at 72°C. A final extension step of 5 min at 72°C was performed after the cycles. Amplified products were separated on a 2.0% agarose gel at 120 V for 3 h, along with a GeneRuler 100-bp ladder (Fermentas, Hanover, MD), visualized with ethidium bromide, and photographed with an Epson PhotoPC 850Z digital camera (Seiko Epson, Japan). One hundred primers (UBC primer set no. 9, Biotechnology Laboratory, University of British Columbia, Canada) were screened initially in 10 samples of D. pleiantha in two replicate trials to identify primers that amplified well and produced polymorphic bands for all accessions on both occasions. These 12 primers were finally selected for the full survey of all 211 individuals: UBC815 = (CT)8G, UBC823 = (TC)8C, UBC822 = (TC)8A, UBC845 = (CT)8RG, UBC857 = (AC)8YG, UBC852 = (TC)8RA, UBC853 = (TC)8RT, UBC854 = (TC)8RG, UBC866 = (CTC)6, UBC874 = (CCCT)4, UBC895 = AGAGTTGGTAGCTCTTGATC, UBC900 = ACTTCCCCACAGGTTAACACA.
Data Analysis
Statistical analysis of ISSR patterns was based on three assumptions. (1) ISSR fragments in D. pleiantha behave as diploid, dominant markers with alleles being either present (amplified) or absent (nonamplified). (2) Comigrating fragments represent putatively homologous loci. (3) Fragments are of nuclear origin and inherited biparentally (Arafeh et al. 2002).
Clonal diversity was detected at fine-scale levels and evaluated by the following indices: (1) number of genotypes, G; (2) the mean clone size, Nc = N/G, where N represents total number of ramets; (3) proportion of distinguishable genotypes, PD = G/N; and (4) a modified version of the Simpson diversity index, D = 1 − Σ[n i (n i − 1)/N(N − 1)] ,where n i is the number of ramets of the ith genet.
To characterize the levels of genetic diversity of D. pleiantha, the following parameters were calculated for each population: (1) percentage of polymorphic fragments (PPF) using PopGene version 1.31 (Yeh et al. 1997) and TFPGA version 1.3 (Miller 1997); (2) Shannon’s index (I) of phenotypic diversity (Lewontin 1972), calculated across all loci as I = −ΣP ilog2 (P i), where P i is the relative frequency of a given ISSR fragment, using PopGene version 1.31 (Yeh et al. 1997) and TFPGA version 1.3 (Miller 1997); and (3) Nei’s (1973) expected heterozygosity or gene diversity (Hpop), calculated from allele frequencies based on the square root of the frequency of the null (recessive) allele, or the unbiased estimator of Lynch and Milligan (1994), using PopGene version 1.31 (Yeh et al. 1997) and TFPGA version 1.3 (Miller 1997). Given that the above estimation of allele frequencies from dominant markers requires the assumption of Hardy–Weinberg equilibrium, within-population gene diversity was also estimated using a Bayesian approach (Zhivotovsky 1999; parameter H B) employing the Hickory program, version 1.0 (Holsinger and Lewis 2003). We used default values for burn-in (50,000), sampling (250,000), and thinning (50).
To characterize genetic differentiation among D. pleiantha populations, we adopted three different approaches to explore any possible bias induced by the Hardy–Weinberg assumption. First, we calculated Nei’s (1973) coefficient of population differentiation (G ST) using PopGene version 1.31 (Yeh et al. 1997). Second, a Bayesian estimate of population structure (ΦB) was estimated using the f-free analysis option in Hickory, which allows for uncertainties in the level of within-population inbreeding (Holsinger et al. 2002). Finally, nonhierarchical AMOVA was used to partition the total phenotypic variance of the entire data set into within-population and among-population components using Arlequin version 2.000 (Schneider et al. 2000). Statistical significance of fixation indices was tested with 10,000 permutations (Excoffier et al. 1992). The same software was used to calculate values for pairwise ΦST (analogous to F ST) between populations. To visualize the genetic relationships among all ISSR phenotypes, their Euclidean distance matrix was subjected to a principal coordinates analysis using the program MVSP, version 1.3 (Kovach 1999). In addition, a matrix of Nei’s D (1972) was calculated between all pairs of populations and subjected to UPGMA clustering as implemented in TFPGA version 1.3 (Miller 1997). Bootstrap values were obtained by resampling with replacement over loci (5,000 replicates). The correlation between pairwise ΦST values and geographic distances (Mantel’s r M) was tested for all natural populations by using Mantel’s test with 3,000 permutations (Mantel 1967).
Results
Clonal Study
Estimates of clonal diversity for three plots of D. pleiantha are given in Table 2. Twelve selected ISSR primers identified 45 genotypes in 113 individuals, and all the plots examined were composed of more than one genet. The plot in population TM contained 31 multilocus genotypes for 52 individuals sampled. The two plots from populations TT and JN, however, had only 6 and 8 multilocus genotypes, respectively. The average number of genotypes was 14.7. The average size of genotypes ranged from 1.677 (TM) to 4.833 (TT). The largest clone from population TT consisted of 21 samples, and each of these multisample clones spread at a maximum of 3.5 m long (Fig. 2, TT). The mean clonal diversity (measured as Simpson’s diversity index) was 0.734 (Table 2). More distinct multilocus genotypes were detected in the plot from TM than in the plots from TT and JN, which parallels the relatively higher genetic diversity measured in population TM. A dendrogram based on genetic distances between individuals showed that all individuals from the same plots formed a group (data not shown), indicating that none of the 45 genotypes was found at more than one location. Therefore, they were all local genotypes and no widespread genotype occurred. The spatial distribution of genets is shown in Fig. 2.
Distribution of multilocus genotypes in three plots from three Dysosma pleiantha populations (TM, from Tianmushan Nature Reserve; TT, from Mount Tiantai; JN, from Mount Shiyan). Each dot represents a sampled individual. Genotyped ramets sharing the same multilocus genotype are included within a single encircled group, and each dot outside the groups represents a distinct multilocus genotype
Genetic Diversity Within Populations
The survey of 105 individuals from 5 populations of D. pleiantha with 12 ISSR primers generated a total of 165 fragments, 98 (59.39%) of which were polymorphic. The band sizes ranged from 300 to 2500 bp (Fig. 3). All individuals tested produced different ISSR profiles. Levels of ISSR variation within populations varied widely across populations (Table 3). Assuming Hardy–Weinberg equilibrium, the average gene diversity (Hpop) ranged from 0.075 (JN) to 0.134 (SQ) for D. pleiantha, with an average of 0.111 ± 0.026 at the population level. Shannon’s index (I) ranged from 0.115 to 0.207, with an average of 0.170 ± 0.039 at the population level. Among the five populations, SQ and TM from nature reserves exhibit the greatest variability (PPF 48.48% and 39.39%, I 0.207 and 0.197, Hpop 0.134 and 0.131, H B 0.247 and 0.229, respectively). By contrast, genetic diversity was lowest in population JN, with PPF 26.06%, I 0.115, Hpop 0.075, and H B 0.145 (Table 3). Considering groups of populations, those from the nature reserves (SQ and TM) had on average higher levels of diversity (PPF 43.94%, I 0.202, Hpop/H B 0.133/0.238) than those from outside nature reserves (DQ, TT, and JN: PPF 31.92%, I 0.148, Hpop/H B 0.097/0.180) (Table 3).
Population Structure of ISSR Variation
Across the five populations of D. pleiantha surveyed for ISSR variation, Nei’s (1973) estimator of population substructure (G ST) indicated a fairly high level of population differentiation (G ST = 0.465). These G ST values translated into correspondingly low levels of gene flow (Nm), with 0.575 migrants exchanged between populations (on average) each generation. The AMOVA also revealed highly significant genetic differences among the five populations of D. pleiantha. Of the total genetic diversity, 49.99% of the variance occurred among populations (ΦST = 0.500, P < 0.001) and 50.01% occurred among individuals within populations. In the Bayesian approach to infer population structuring, we used the f-free model because it had the smallest DIC value (1614.2). The ΦB was 0.436, similar to the ΦST values from AMOVA.
The plot of the first and second principal coordinates from a principal coordinates analysis (PCoA) (accounting for 24.67% and 10.64% of the variation, respectively) is depicted in Fig. 4. PCoA revealed three plots, which were defined according to their geographic origin. Estimates of genetic distance, in terms of Nei’s (1972) D, between all pairs of populations ranged from 0.0757 (between TM and DQ) to 0.2201 (between TM and JN). Subjecting the genetic distance matrix to UPGMA clustering (Fig. 5) also revealed that populations spatially near each other tended to be genetically similar. Neighbor-joining clustering of the same distance matrix resulted in an identical tree topology (not shown). Corroborating this, Mantel’s test revealed that the genetic distances between these populations were significantly and positively correlated to their geographic distances (r M = 0.833; P = 0.027) as expected under Wright’s (1943) isolation-by-distance model of population structure.
Principal coordinates analysis of 165 ISSR phenotypes from five populations of Dysosma pleiantha. The first and second axes extracted 24.67% and 10.64% of the total genetic variance, respectively. Abbreviations as in Table 1
Discussion
Fine-Scale Genetic Structure
For species that reproduce sexually, populations usually contain many genets (Ellstrand and Roose 1987; Eriksson and Bremer 1993). In this study, clonal diversity in three small plots for D. pleiantha tended to be relatively high (G/N = 0.351, Simpson’s D = 0.734, on average, Table 2), when compared with values presented in reviews of clonal diversity among more than 20 primarily obligate clonal species by Ellstrand and Roose (1987) (G/N = 0.17, Simpson’s D = 0.62, on average). Our data on the distribution of genotypes suggested that, in general, the ramets sharing the same genotype show a clumped distribution (<3.5 m). Without seedling recruitment, clonal diversity is expected to decline rapidly and populations are expected to become dominated by a few large genets (Eriksson 1993). Thus, the above results provide evidence that sexual recruitment is a very important mechanism of regeneration in D. pleiantha populations, and most individual plants with identical or similar genotypes were located within the small spatial range. Clonal ability can contribute to propagating or perpetuating adapted genotypes (Salemaa and Sievanen 2002) and thus enhance genet survival under suboptimal conditions (Kudoh et al. 1999). For instance, clonal spread was reported to be higher under harsh ecological conditions (Eckert and Barrett 1993; Stenström et al. 2001; Young et al. 2002). In our study, clonal diversity among three plots varies greatly. Clonal spread occurs more readily in plots TT and JN from outside the nature reserves than in plot TM from the nature reserve (Table 2), which could be interpreted as a strategy for propagating or perpetuating adapted genotypes under the suboptimal ecological conditions resulting from human disturbance.
Within-Population Variation and Population Divergence
The levels of within-population genetic diversity in D. pleiantha are relatively low when compared with other seed plants, with either a similar life history or various breeding system attributes, that have been screened with a comparable (i.e., dominant) marker system, such as amplified polymorphic DNA (RAPD), as most recently reviewed by Nybom (2004). Thus, if for comparison we focus on the genetic diversity index Hpop, the total average of within-population ISSR diversity in D. pleiantha (Hpop/H B = 0.111/0.203; Table 3) is much lower than the corresponding average of RAPD diversity reported in long-lived perennial species (Hpop = 0.25) and outcrossers (Hpop = 0.27), whereas it is comparable to selfers (Hpop = 0.12) (Nybom 2004; see also Nybom and Bartish 2000, for almost identical estimates). When compared with its congener species, these values of genetic diversity for D. pleiantha are higher than that observed from ISSR analysis in D. versipellis (Hpop/H B = 0.084/0.177) (Qiu et al. 2006). The results are also in good agreement with that obtained by our previous allozyme analysis (Qiu et al. 2005).
The low levels of heterozygosity observed in this species could result from significant amounts of selfing occurring within D. pleiantha populations. This interpretation would be in conflict with a predominantly outcrossing mating system of this species (Qiu et al. 2005). Genetic diversity, however, may be structured in neighborhoods, and mating may mainly take place among genetically related and geographically close individuals and/or intraclone ramets. Field observations of Chrysomya megacephala, the main pollinator of this species, have shown that its flight distance is less than 2 m, and it flies to the nearest plant to collect pollen and nectar (Y. X. Qiu, pers. obs.). No specialized structures were observed either on the fruit or seed surface that would facilitate dispersal. In natural conditions, the mature berries of D. pleiantha usually drop in the vicinity of their mother plant during the rainy season, and small seeds are released following decay of the fruit pulp (Li and Wang 2006). The pollinator behavior, coupled with the lack of specialized seed dispersal mechanisms, could be favoring the establishment of neighborhoods of related individuals. Thus, inbreeding resulting from these pollinations could be one of the major factors responsible for the low genetic variation within the populations of these species. The proportion of its sexual and asexual propagation may determine the level of genetic diversity (Ayres and Ryan 1997). Relatively low levels of genetic variation and small numbers of distinct multilocus genotypes observed in the JN and TT populations may indicate that vegetative reproduction and spreading predominate in these populations. The relatively high level of genetic variation observed within the TM and SQ populations suggests that the balance between vegetative reproduction and sexual reproduction is more in favor of sexual reproduction in the large populations than in the small populations (e.g., JN and TT); indeed, the relatively high fruit production observed in TM and SQ confirms this hypothesis.
Recent land uses and deforestation have resulted in population size reduction and habitat fragmentation of D. pleiantha, particularly for populations outside the nature reserves (DQ, JN, and TT) (Li and Wang 2006, see Table 1). All D. pleiantha populations are fewer than 800 individuals, based on our five-year field investigation (Qiu et al. 2005). From theoretical predictions and a number of studies, the smaller populations might be expected to show reduced levels of polymorphism and allelic richness (Young et al. 1996; Coates and Hansley 1999). Thus, we expected the populations outside the nature reserves (DQ, JN, and TT) to be genetically depauperate, compared with the populations in the nature reserves (TM and SQ). Diversity estimates obtained with ISSRs indicate that those from the nature reserves had on average higher levels of diversity (PPF = 43.94%, Hpop/H B = 0.133/0.238) than those from outside the nature reserves (DQ, TT, and JN: PPF = 31.92%, Hpop/H B = 0.097/0.180) (Table 3). Based on that analysis, inbreeding and random genetic drift could be responsible for the lower genetic diversity of this species.
Across the species’ range, natural populations were found to show an unexpectedly high level of genetic subdivision, with an estimated G ST value of 0.465 [Nei (1978) classified G ST < 0.05 as low, 0.05–0.15 medium, and >0.15 high]. There is a considerable amount of genetic differentiation among populations of D. pleiantha. Different analyses of the ISSR data all show a high between-population variation (AMOVA, ΦST = 0.500; Nei’s genetic diversity, G ST = 0.465; Bayesian analysis, ΦB = 0.436). The AMOVA-derived G ST analog, ΦST, is of comparable magnitude, 0.500. This value is much higher than the average reported in the RAPD literature for species with long-lived perennials (ΦST = 0.25) as well as outcrossers (ΦST = 0.27–0.28), but is still lower than generally found in predominant selfers (ΦST = 0.65; Nybom and Bartish 2000; Nybom 2004). A high level of population differentiation may be explained by several factors, including the species’ breeding system, genetic drift, or geographic isolation of populations (Hogbin and Peakall 1999). When the association between collection distance and among-population distance diversity was analyzed for outcrossing and selfing taxa separately, a strong positive relationship was found for outcrossing taxa, but not for selfing taxa (Nybom and Bartish 2000; Nybom 2004). Given that the mating system of the species was outcrossing and most populations of D. pleiantha were more than 250 km apart, we therefore believe that great geographic isolation between populations may have played an important role in promoting differentiation among these populations. When populations are small and isolated from one another, genetic drift also influences genetic structure and increases differentiation among populations (Ellstrand and Elam 1993). D. pleiantha seems to consist of a series of disjunct populations, more or less isolated from each other (Fig. 1). Restricted gene flow and genetic drift might have influenced the extent of differentiation among D. pleiantha populations. The G ST-derived Nm value of 0.575 is indicative of restricted gene flow among natural populations, and this value is actually below the level (Nm ≈ 1) needed to counteract genetic drift (Slatkin 1993). The significantly positive r M value observed in the isolation-by-distance analysis also indicates that gene exchange is largely restricted to nearest neighboring populations. Hence, based on the present data set, there is no evidence that serendipitous long-distance dispersal (or colonization) plays a large role in the population dynamics of D. pleiantha. These data strongly suggest that the major contemporary factor, in addition to breeding system attributes, determining the pronounced genetic structure of D. pleiantha is restricted gene flow due to limited pollen and seed dispersal with isolation by distance.
Conservation Implications
Knowledge of the levels and distribution of genetic diversity is important for designing conservation strategies for threatened and endangered species (Qiu et al. 2005). In the present study, the observed genetic differentiation among D. pleiantha populations is so great, and so little gene flow appears to exist among them, that management for the conservation of genetic variability in this species should aim to preserve not only large populations but also as many of the small populations outside nature reserves as possible. Reduced levels of genetic variation, especially in the smaller populations, will affect the species’ ability to adapt to changes in its habitat (Luijten et al. 2000). Positive correlations between population size, expected heterozygosity, and plant fitness were also found in Gentiana pneumonanthe (Oostermeijer et al. 1995) and a self-incompatible perennial (Arnica montana; Luijten et al. 2000). Vegetative reproduction in the small populations of D. pleiantha might postpone extinction, but it is essentially an evolutionary dead end. In the present situation, seedling recruitment in small populations (e.g., TT and DQ) is virtually absent (Y. X. Qiu, pers. obs.). Thus, policy plans should be developed to stimulate recruitment in the small populations. Artificial transplanting of individuals among different scattered plots in the same population may be advantageous to increase fruit set. Given that remarkably high levels of genetic differentiation existed among populations in D. pleiantha, crossing populations may bear a certain risk of outbreeding depression, which can be attributed to the disruption of local adaptation (i.e., extrinsic isolation), underdominance, or epistatic interactions (heterozygote–heterozygote interactions or interactions involving sex chromosomes) (Fischer and Matthies 1997; Edmands 2007).
Considering that low seed set was observed in this species, a good strategy to encourage seed set, improved seed germination, and seedling recruitment needs to be considered. It is desirable to apply simple methods (e.g., enhancing seed germination or propagation via rhizome segments). These would be easy to perform in the field and cost effective. In vitro techniques such as tissue culture have proved to be an effective means for recovery of endangered species (Nadeem et al. 2000). At present, an effective protocol of in vitro propagation, involving multiple shoot formation from zygotic embryos and subsequent rooting, has been developed for D. pleiantha in our laboratory (Pan et al. 2006). In vitro propagation may well be used as a means to rescue zygotic embryos for this species. Currently efforts have been taken up along these lines. In addition to the demographic and genetic constraints, there are external threats to the species in the form of overexploitation of its rhizomes for medicinal use, which greatly reduces its chances of survival. In view of these investigations, we conclude that the external pressures on the species need to be stopped, and an integrated conservation strategy based on demographic, ecological, and genetic aspects should be prepared. In these ways, we hope that the future of this medicinally important, severely threatened species will be guaranteed.
References
APG II (Angiosperm Phylogeny Group) (2003) An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141:399–436
Arafeh R, Sapir Y, Shmida A, Iraki N, Fragman O, Comes HP (2002) Patterns of genetic and phenotypic variation in Iris haynei and I. atrofusca (Iris sect. Oncocyclus = the royal irises) along an environmental gradient in Israel and the West Bank. Mol Ecol 11:39–52
Ayres DR, Ryan FJ (1997) The clonal and population structure of a rare endemic plant, Wyethia reticulata (Asteraceae): allozyme and RAPD analysis. Mol Ecol 6:761–772
Balloux F, Lehmann L, De Meeûs T (2003) The population genetics of clonal and partially clonal diploids. Genetics 164:1635–1644
Clark-Tapia R, Alfonso-Corrado C, Eguiarte LE, Molina-Freaner F (2005) Clonal diversity and distribution in Stenocereus eruca, a narrow endemic cactus of the Sonoran Desert. Am J Bot 92:272–278
Coates DJ, Hansley VL (1999) Genetic divergence and the mating system in the endangered and geographically restricted species, Lambertia orbifolia Gardener (Proteaceae). Heredity 83:418–427
Culley TM, Wolfe AD (2001) Population genetic structure of the cleistogamous plant species Viola pubescens Aiton (Violaceae), as indicated by allozyme and ISSR molecular markers. Heredity 86:545–556
Doyle JJ (1991) DNA protocols for plants-CTAB total DNA isolation. In: Hewitt GM, Johnston A (eds) Molecular techniques in taxonomy. Springer-Verlag, Berlin, Germany, pp 283–293
Eckert CG (2002) The loss of sex in clonal plants. Evol Ecol 15:501–520
Eckert CG, Barrett SCH (1993) Clonal reproduction and patterns of genotypic diversity in Decodon verticillatus (Lythraceae). Am J Bot 80:1175–1182
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreedng and outbreeding for conservation and management. Mol Ecol 16:463–475
Ellstrand NC, Elam DR (1993) Population genetics consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242
Ellstrand NC, Roose ML (1987) Patterns of genotypic diversity in clonal plant species. Am J Bot 74:123–131
Eriksson O (1993) Dynamics of genets in clonal plants. Trends Ecol Evol 8:313–316
Eriksson O, Bremer B (1993) Genet dynamics of the clonal plant Rubus saxatilis. J Ecol 81:533–542
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: applications to human mitochondrial DNA restriction data. Genetics 131:479–491
Fischer M, Matthies D (1997) Mating structure and inbreeding and outbreeding depression in the rare plant Gentianella germanica (Gentianaceae). Am J Bot 84:1685–1692
Hogbin PM, Peakall R (1999) Evaluation of the conservation of genetic research to the management of endangered plant Zieria prostrata. Conserv Biol 13:514–522
Holsinger KE, Lewis PO (2003) Hickory: a package for the analysis of population genetic data. Version 0.8. Distributed by the authors, Department of Ecology and Evolutionary Biology, University of Connecticut, Storrs, CT
Holsinger KE, Lewis PO, Dey DK (2002) A Bayesian approach to inferring population structure from dominant markers. Mol Ecol 11:1157–1164
Jackson DE, Dewick PM (1985) Tumor-inhibitory aryltetralin lignans from Podophyllum pleianthum. Phytochemistry 24:2407–2409
Kala CP (2005) Indigenous uses, population density, and conservation of threatened medicinal plants in protected areas of the Indian Himalayas. Conserv Biol 19:368–378
Kovach WL (1999). MVSP – A multivariate statistical package for windows, version 3.1. Kovach Computing Services, Pentraeth, Wales
Kudoh H, Dhibaike H, Whigham DF, Kawano S (1999) Genet structure and determinants of clonal structure in a temperate deciduous woodland herb, Uvularia perfoloata. J Ecol 87:244–257
Lange D (1997) Trade figures for botanical drugs world-wide. Med Plant Conserv 3:16–17
Lewontin RC (1972) The apportionment of human diversity. Evol Biol 6:391–398
Li ZC, Wang WY (2006) Ecological and biological characteristics of endangered medicinal plant Dysosma versipellis. J Trop Subtrop Bot 14:190–195
Luijten SH, Dierick A, Oostermeijer JGB, Raijmann LEL, Den Nijs JCM (2000) Population size, genetic variation and reproductive success in the rapidly declining, self-incompatible perennial (Arnica montana) in the Netherlands. Conserv Biol 14:1776–1787
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
McCue KA, Buckler ES, Holtsford TP (1996) A hierarchical view of genetic structure in the rare annual plant Clarkia springvillensis. Conserv Biol 10:1425–1434
Miller MP (1997) Tools for Population Genetic Analysis (TFPGA), version 1.3. Department of Biological Sciences, Northern Arizona University, AZ, USA
Milligan BG, Leebens-Mack J, Strand AE (1994) Conservation genetics: beyond the maintenance of maker diversity. Mol Ecol 3:423–435
Nadeem M, Palni LMS, Purohit AN, Pandey H, Nandi SK (2000) Propagation and conservation of Podophyllum hexandrum Royle: an important medicinal herb. Biol Conserv 92:121–129
Neel MC, Cummings MP (2003) Effective of conservation targets in capturing genetic diversity. Conserv Biol 17:219–229
Neel MC, Ellstrand NC (2003) Conservation of genetic diversity in the endangered plant Eriogonum ovalifolium var. vineum (Polygonaceae). Conserv Genet 4:337–352
Nei M (1972) Genetic distance between populations. Am Nat 106:283–292
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Plant Ecol Evol Syst 3:93–114
Oostermeijer JGB, Van Eijck MW, Van Leeuwen NC, Den Nijs JCM (1995) Analysis of the relationship between allozyme heterozygosity and fitness in the rare Gentiana pneumonanthe L. J Evol Biol 8:739–757
Pan Q, Chen SY, Jiang WM, Gilbert CYS, Fu CX (2006) Study on the podophyllotoxin in the Dysosma versipellis (Hance) M. Cheng and its callus materials. J Zhejiang Univ (Agric & Life Sci) 32(1):56–59
Qiu YX, Zhou XW, Fu CX, Gilbert CYS (2005) A preliminary study of genetic variation in the endangered, Chinese endemic species Dysosma versipellis (Berberidaceae). Bot Bull Acad Sin 46:65–73
Qiu YX, Li JH, Liu HL, Chen YY, Fu CX (2006) Population structure and genetic diversity of Dysosma versipellis (Berberidaceae), a rare endemic from China. Biochem Syst Ecol 34:745–752
Richards AJ (1997) Plant breeding systems. Chapman and Hall, London, UK
Salemaa M, Sievanen R (2002) The effects of apical dominance on the branching architecture of Arctostaphylos uva-ursi in four contrasting environments. Flora 197:429–442
Schneider S, Roessli D, Excoffier L (2000) Arlequin ver 2.0. A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Geneva, Switzerland
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Stenström A, Jonsson BO, Jonsdottir IS, Fagerstrom T, Augner M (2001) Genetic variation and clonal diversity in four clonal sedges (Carex) along the Arctic coast of Eurasia. Mol Ecol 10:497–513
Wang S, Xie Y (2004) China species red list, vol 1. Higher Education Press, Beijing, pp 324
Wright S (1943) Isolation by distance. Genetics 28:114–138
Yeh FC, Yang RC, Boyle T, Ye ZH, Mao JX (1997) PopGene, the user friendly shareware for population genetic analysis. Molecular Biology and Biotechnology Center, University of Alberta, Edmonton, Canada
Ying TS, Zhang YL, Boufford DE (1993) The endemic genera of seed plants of China. Science Press, Beijing, China
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Young AG, Hill JH, Murray BG, Peakall R (2002) Breeding system, genetic diversity and clonal structure in the sub-alpine forb Rutidosis leiolepis F. Muell. (Asteraceae). Biol Conserv 106:71–78
Zhang D, Shao J, Li D (1991) A study on the karyotypes of Dysosma versipellis and D. pleiantha endemic to China. Guihaia 11(1):58–62
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgments
This research was supported by the National Basic Research Program of China, 973 Program (2007CB411600), the National Science Foundation of China (grant 30570177), and the Zhejiang Provincial Natural Science Foundation of China (grant M303092). The authors thank Mr. Zhang Hongwei and Mr. Gao Yuanlong for their help during fieldwork. We thank Hai Yan and Yu Jin for assistance in ISSR analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zong, M., Liu, HL., Qiu, YX. et al. Genetic Diversity and Geographic Differentiation in the Threatened Species Dysosma pleiantha in China as Revealed by ISSR Analysis. Biochem Genet 46, 180–196 (2008). https://doi.org/10.1007/s10528-007-9141-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-007-9141-7