Abstract
The aim of the present study was to examine the effects of ageing and training status on (1) markers of skeletal muscle mitochondrial content and (2) the ability to activate the acute signalling pathways associated with regulating exercise-induced mitochondrial biogenesis. Muscle biopsies were obtained from the vastus lateralis muscle of young untrained (24 ± 4 years, n = 6; YU), young trained (22 ± 3 years, n = 6; YT), old untrained (65 ± 6 years, n = 6; OU) and old trained (64 ± 3 years, n = 6; OT) healthy males before and after (3 h and 3 days post-exercise) completion of high-intensity interval cycling exercise. In resting muscle, lifelong training preserved mtDNA, PGC-1α and COXIV protein content such that muscles from OT individuals were comparable to muscles from both YU and YT individuals, whereas lifelong sedentary behaviour reduced such markers of mitochondrial content. Regardless of age or training status, acute exercise induced comparable increases in p38MAPK phosphorylation immediately post-exercise, PGC-1α and COXIV mRNA expression at 3 h post-exercise and COXIV protein at 3 days post-exercise. Data demonstrate that lifelong endurance training preserves skeletal muscle PGC-1α content and that despite the mitochondrial dysfunction typically observed with sedentary ageing, muscles from sedentary elderly individuals retain the capacity to activate the acute signalling pathways associated with regulating the early processes of mitochondrial biogenesis. We consider our data to have immediate translational potential as they highlight the potential therapeutic effects of exercise to induce skeletal muscle mitochondrial biogenesis persist late in adulthood, even after a lifetime of physical inactivity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ageing is a complex multifactorial biological process characterised by systemic decline (Kowald and Kirkwood 1996). This decline is particularly pronounced in skeletal muscle such that by the age of 70 muscle strength can be reduced by up to 40 % (Close et al. 2005). Age-related muscle frailty (termed sarcopenia) describes the deterioration of muscle mass and force generation capacity with age (McArdle et al. 2004). These age-related deficits increase frailty, disease risk, and decrease mobility to the extent that elderly individuals may become unable to perform everyday tasks independently (Carter et al. 2007). An expanding elderly population means the social and economic costs of combating muscle frailty are set to increase (Janssen et al. 2004).
With regards to the potential contributing mechanisms underpinning frailty, much attention has been given to the role of skeletal muscle mitochondria, an area frequently termed as the ‘mitochondrial theory of ageing’. Indeed, skeletal muscles from older adults exhibit electron transport chain (ETC) abnormalities (Lezza et al. 1994), oxidative stress (Gianni et al. 2004) and somatic mtDNA mutations (McKenzie et al. 2002). Furthermore, old adults display lower abundance and activity of ETC subunits, mitochondrial dysfunction, reduced mtDNA content and lower abundance of nuclear and mitochondrial transcription factors (Joseph et al. 2012; Safdar et al. 2010; Lanza et al. 2008). Interpretation of these data is complicated, however, as it is difficult to determine if the decline in mitochondrial content and function is due to ageing per se and/or a lifetime of physical inactivity. In this regard, it is noteworthy that many of the apparent aforementioned declines in mitochondrial content and function can indeed be attenuated by lifelong physical activity (Safdar et al. 2010; Lanza et al. 2008). Such data therefore suggest that lifelong physical activity or strategic exercise of sufficient intensity and duration may be a suitable strategy to preserve mitochondrial mass and function across the lifespan.
In considering the molecular mechanisms regulating the apparent preservation of mitochondrial mass and function with lifelong physical activity, recent research has focused on the proposed master regulator of mitochondrial biogenesis; peroxisome proliferator activated receptor gamma co-activator one alpha (PGC-1α). Indeed, data from rodents demonstrates that overexpression of PGC-1α in skeletal muscles prevents muscle wasting by reducing apoptosis, autophagy and proteasome degradation and also induces improvements in insulin sensitivity and insulin signalling (Wenz et al. 2009; Sandri et al. 2006). Despite the well-documented effects of muscle contraction on PGC-1α expression, aged rodents fail to up-regulate PGC-1α content in skeletal muscle in response to endurance training (Derbre et al. 2012), a finding which may be attributable to inability to activate signalling pathways (e.g., AMPK or p38MAPK phosphorylation etc.) upstream of PGC-1α (Ljubicic et al. 2009). Taken together, such data suggest that modulation of PGC-1α represents a potential avenue to combat age related disorders but that in the context of exercise, physical activity should be performed throughout the lifespan and not left until later life. In contrast, more recent data from humans demonstrates that elderly untrained individuals retain the ability to respond to an acute exercise challenge (when compared with trained elderly counterparts), as evidenced by increased phosphorylation of AMPK, p38MAPK and expression of PGC-1α mRNA in skeletal muscle (Iversen et al. 2011), thereby suggesting that aged muscle retains the ability to adapt to endurance training. However, it is not currently known whether aged muscle retains the ability to activate such signalling pathways in a manner comparable to young individuals.
With this in mind, the aim of the present study was to therefore examine the effects of ageing and training status on (1) markers of skeletal muscle mitochondrial content and (2) the ability to activate the acute signalling pathways associated with regulating exercise-induced mitochondrial biogenesis. To this end, we obtained muscle biopsies from the vastus lateralis muscle of young untrained (n = 6; YU), young trained (n = 6; YT), old untrained (n = 6; OU) and old trained (n = 6; OT) healthy males before and after (3 h and 3 days post-exercise) performing an acute high-intensity interval cycle training protocol (HIT). Although previous studies have compared skeletal muscle characteristics of old trained and untrained humans at rest (Safdar et al. 2010) or in response to exercise (Iversen et al. 2011), this study represents the first known attempt to simultaneously compare young and old individuals in the both the resting and exercised state. Based on previous data, we hypothesised that lifelong training would preserve markers of mitochondrial content but that regardless of training history, aged muscle retains the ability to adapt to acute exercise in a manner comparable to their young counterparts.
Methods
Subjects
After providing informed written consent, 12 young (23.15 ± 3.5 years old) and 12 older (64.15 ± 4.25 years old) healthy males volunteered to take part in the study. Numbers were selected based upon a priori power calculations which suggested that n = 6 per group was needed to detect a threefold change in PGC1α mRNA with a typical SD of 1.5 (unpublished observations from our laboratory). Subjects were allocated into one of four groups according to training status: YT (n = 6); YU (n = 6), OT (n = 6) and OU (n = 6). Trained subjects were all competitive amateur cyclists that had habitually completed at least five endurance exercise sessions per week (all ≥45 min) as part of a systematic training regime. The older trained subjects were required to have adopted such an exercise regime for a minimum of 10 years. Untrained subjects did not habitually partake in endurance-based training and completed ≤3 exercise sessions per week. This study was conducted in accordance with the Declaration of Helsinki and received institutional ethical approval from Liverpool John Moore University. Baseline physical and physiological characteristics are shown in Table 1.
Assessment of physiological fitness
Maximal oxygen uptake (VO2max) and peak power output (PPO) was determined 5–7 days prior to the main experimental trial using an incremental exercise test performed until volitional exhaustion on a bicycle ergometer (Daum Electronic Ergo Bike, Daum, Germany). Oxygen uptake (Online Systems, Metamax Cortex, Germany) and heart rate (Polar S610i, Finland) were measured continuously during exercise. Following a 5 min warm-up at 50 W participants completed successive 1 min exercise bouts where wattage was increased by 30 W every minute until volitional exhaustion. Subjects were deemed to have attained VO2max if they fulfilled the following criteria: (1) heart rate within 10 beats min−1 of age-predicted maximum, (2) respiratory exchange ratio >1.1, and (3) plateau of oxygen consumption despite increased workload.
Experimental protocol
Subjects reported to the laboratory in a fasted state between 07.00 and 09.30 h after refraining from exercise, alcohol and caffeine consumption for the previous 48 h. A resting muscle biopsy was obtained using a Bard Montopy Disposable Biopsy Instrument (12 × 10 cm gauge, Bard Monotpy Systems, USA) from the left vastus lateralis muscle under local anaesthetic (0.5 % marcaine). After a 5 min warm-up at 50 % PPO, a 20 min high-intensity interval (HIT) session was completed on a bicycle ergometer (Daum Electronic Ergo Bike, Daum, Germany). The HIT session consisted of a 2 min bout at 40 % PP followed by a 2 min bout at 80 % PP. This work–rest ratio was repeated five times. We chose to use the HIT model of cycling exercise given that this form of exercise has been shown to be a tolerable, time efficient and effective method of inducting metabolic adaptations in skeletal muscle (Little et al. 2010). Oxygen uptake was recorded continuously using an online system (Metamax Cortex, Germany) whilst both heart rate (Polar S610i, Finland) and ratings of perceived exertion (RPE: Borg 6–20 scale) were recorded at 2 min intervals during exercise. Further muscle biopsies were obtained immediately post-exercise (right leg), 3 h post exercise (right leg) and 3 days post-exercise (left leg). All sites on the same leg were separated by at least 3 cm. Muscle samples (approximately 50 mg) were immediately snap frozen in liquid nitrogen and stored at −80 °C for later analysis. The 3 h time point to measure changes in mRNA was selected based upon previous titration studies from our group. Moreover, Mahoney et al. (2005) have previously reported that 3 h post cycling exercise is a suitable time-point to measure expression of exercise-responsive genes since 118 genes were significantly increased at this time-point.
Western blotting
Approximately 20–30 mg of frozen muscle tissue was ground to powder and homogenised in 120 μl of ice cold lysis buffer which included phosphatase inhibitors (25 mM Tris/HCl [pH 7.4], 50 mM NaF, 100,207 mM NaCl, 5 mM EGTA, 1 mM EDTA, 10 mM Na-Pyrophosphatase, 1 mM Na3VO4, 0.27 M sucrose, 1 % Triton X-100, 0.1 % 2-mercaptoethanol) and was supplemented with a protease inhibitor tablet (Complete mini, Roche Applied Science, West Sussex, UK). Homogenates were centrifuged at 14,000×g for 10 min at 4 °C and the supernatant was collected. The protein content of the supernatant was determined using a bicinchoninic acid assay (Sigma, UK). Each sample was diluted with an equal volume of 2× Laemmli buffer (National Diagnostics, USA) and boiled for 5 min at 100 °C. Samples containing 50 μg protein were separated via SDS–PAGE using self-cast gels (National Diagnostics, USA) before being transferred semi-dry onto a nitrocellulose membrane (Geneflow Ltd, UK). For each blot, samples were run alongside a pool standard and negative control. Following transfer, membranes were blocked for 1 h at room temperature in tris-buffered saline with 5 % non-fat dry milk. Membranes were washed 3 × 5 min in TBST before being incubated overnight at 4 °C with antibodies for anti-phospo p38 MAPKThr180/Tyr182, total MAPK, COXIV (Cell Signalling, UK) and PGC-1α (Calbiochem, Merck Chemicals, UK) all at concentrations of 1:1,000 in 1× TBST. Proteins of interest were normalised to the GAPDH (Cell Signalling, UK) content of the same sample to ensure equal protein loading between samples. The next morning, membranes were washed for a further 3 × 5 min in TBST and subsequently incubated with anti-species horseradish peroxidise-conjugated secondary antibody (Bio-Rad, UK or Dako, UK) for 1 h at room temperature. After a further 3 × 5 min washes in TBST, membranes were exposed in a chemiluminescence liquid (SuperSignal, Thermo Fisher Scientific, Rockford, IL, USA) for 5 min. Membranes were visualised using a Bio-Rad Chemi-doc system, and band densities were determined using Quantity One image-analysis software.
Real-time RT-PCR
Total RNA was isolated from muscle biopsies (20–30 mg) using Trizol reagent (Invitrogen), according to the manufacturer’s protocol. RNA quality and quantity were determined using Implen Nanophotometer (Implen, Munchen, Germany) and the RNA was stored at −80 °C. cDNA was synthesised using random hexamers (Applied Biosystems) and Superscript III enzyme (Invitrogen), using manufacturer’s protocol. Gene specific expression data were obtained using probes selected from Human Universal Probe Library (Roche Diagnostics) with compatible oligonucleotide primers (MWG Eurofins). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward: 5′-AGCCACATCGCTCAGACAC-3′, reverse: 5′-GCCCAATACGACCAAATCC-3′, PGC-1α forward: 5′-TGAGAGGGCCAAGCAAAG-3′, reverse: 5′-ATAATCACACGGCGCTCTT-3′, COXIV forward: 5′-CCATGTCAAGCACCTGTC-3′, reverse: 5′-CAGCAAAGCTCTCCTTGAACCTTA-3′. One microlitre of each sample was analysed in triplicate with negative controls using AB 7500 Real-Time Quantitative PCR instrument (Applied Biosystems) and Agilent Brilliant II qPCR Master Mix with Low ROX (Agilent Technologies). One microlitre of cDNA, 500 nM of primer and 200 nM of probe were used for each 20 μl reaction. The following cycling parameters were used: 50 °C for 2 min, initial denaturation at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 15 s and annealing/elongation at 60 °C for 1 min. Data were collected and analysed using AB SDS 1.43 Software (Applied Biosystems, Foster City, USA). Changes in mRNA content were calculated according to the 2−ΔΔCt method where GAPDH was used as the reference gene. Several reference genes, including GAPDH, Tubulin and beta-2-microglobulin were tested prior to starting the qPCR analysis on the same samples as presented in the present manuscript. GADPH was selected as the reference gene since it was the most stable of the reference genes tested across the control conditions and post-exercise as previously reported from our group (Bartlett et al. 2012) and others (Mahoney et al. 2004) following endurance based exercise.
Mitochondrial DNA abundance (mtDNA)
Mitochondrial number was estimated using a real-time PCR based method. Briefly, total DNA was isolated from biopsy samples of approximately 2–5 mg using a commercially available kit (DNeasy, Qiagen, USA). Nuclear DNA (nDNA) was quantified using primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward: 5′-CGGGGCTCTCCAGAACATC-3′, reverse: 5′-ATGACCTTGCCCACAGCCT-3′. Mitochondrial DNA (mtDNA) was quantified using primers for the mitochondrial encoded ATP synthase 6 (ATP-6) forward: 5′-ACCAATAGCCCTGGCCGTAC-3′ and reverse: 5′-GGTGGCGCTTCCAATTAGTG-3′. Separate reactions for GAPDH and ATP6 reactions were set up as follows: 25 ng of total DNA, 12.5 μl 2× SensiMix Sybr Green master mix (Bioline Ltd, UK), 2 μl each forward and reverse primer (0.8 μM final concentration) and 6 μl DNase free water. Real-time PCR was performed on a Bio-Rad iCycler with an iCycler iQ Multicolour real-time PCR detection system (Bio-Rad, Hercules, USA) with the following conditions: 95 °C for 3 min, 26 cycles of: 95 °C for 20 s, 62 °C for 20 s and 72 °C 20 s. The ratio of nDNA:mtDNA was calculated from the Ct values for GAPDH and ATP-6.
Statistical analysis
A one-way ANOVA was employed to analyse baseline differences between groups. If any F values were observed, least-significant difference tests were performed to determine where any significant differences occurred. A three-way ANOVA was utilised to explore the influence of exercise on the outcome variable (e.g. pre vs. post exercise) and the influence of age (e.g. young vs. old) and training status (e.g. trained vs. untrained). Significant differences were further analysed with least-significant differences tests. An alpha value of P ≤ 0.05 was used for all tests. All statistical analysis were performed with the statistical package for social sciences version 17.0 (SPSS, England). All data in text, tables and figures are presented as mean ± SD.
Results
Physiological responses to the exercise protocol
The physiological responses to the exercise protocol are summarised in Table 2. There was a significant difference in workload between the four groups at both 40 % (P = 0.01) and 80 % (P = 0.01) of peak power. Workload was significantly higher in YT compared with all other groups. There were no significant differences between groups for percentage of VO2max, percentage of maximum heart rate and RPE at both exercise intensities (P > 0.05).
Baseline muscle characteristics
mtDNA
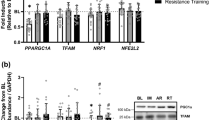
mtDNA was significantly different between groups (P = 0.002; Fig. 1a). mtDNA was significantly lower in YU (P = 0.002) and OU (P = 0.001) compared with YT subjects. mtDNA was significantly lower in OU (P = 0.020) compared with OT subjects. No differences in mtDNA between young and OU (P = 0.465) and trained (P = 0.090) were observed.
Baseline muscle characteristics measured in young trained (YT), young untrained (YU), old trained (OT) and old untrained (OU) participants. Mitochondrial DNA (mtDNA) was significantly reduced in untrained compared with trained subjects (a). PGC-1α protein content was significantly greater in old trained subjects compared with both old untrained and young untrained subjects (b). COXIV protein content was greater in trained compared with untrained subjects and was greater in old trained compared with young trained subjects (c). *Denotes statistical significance (P < 0.05)
P38MAPK phosphorylation measured in young trained (YT), young untrained (YU), old trained (OT) and old untrained (OU) participants immediately pre-exercise (Pre) and immediately post-exercise (Post). It was observed that P38MAPK phosphorylation was significantly increased post-exercise irrespective of age or training status. *Denotes significant main effect of exercise (P < 0.05)
Protein content
Baseline PGC-1α protein content was significantly lower in OU (P = 0.001) and YU (P = 0.002) compared with OT subjects (Fig. 1b). PGC-1α was also significantly lower in OU (P = 0.026) compared with YT subjects (P = 0.026). COXIV protein content was significantly different between groups at baseline (P = 0.001; Fig. 1c). COXIV protein content was significantly lower in YU (P = 0.001) and OU (P = 0.003) compared with YT subjects. Moreover, COXIV protein content was also significantly lower in YU (P = 0.001) and OU (P = 0.001) compared with OT subjects.
p38MAPK signalling
p38MAPK phosphorylation (Fig. 2) significantly increased immediately post-exercise (P = 0.001). Increased p38 MAPK phosphorylation was not influenced by group (P = 0.292), age (P = 0.393) or training status (P = 0.435).
Gene expression
The expression of PGC-1α mRNA increased significantly (P = 0.002) 3 h post-exercise, although this increase was not influenced by age (P = 0.604) or training status (P = 0.992; Fig. 3a). COXIV mRNA increased significantly 3 h post-exercise (P = 0.001; Fig. 3b). This increase was not influenced by subject group (P = 0.795) and thus occurred independently of age (P = 0.864) and training (P = 0.411).
mRNA and protein responses to the exercise challenge in young trained (YT), young untrained (YU), old trained (OT) and old untrained (OU) participants measured pre exercise and 3 h post-exercise. PGC-1α (a) and COXI (b) mRNA increased 3 h post-exercise irrespective of age or training status. No significant increase in PGC-1α protein content at 3 days post-exercise was observed in any group (c). COXIV protein content (d) increased 3 days post-exercise in all groups (main effect of exercise) although this increase was greater in both trained groups compared with untrained groups (main effect of training status). *Denotes significant main effect of exercise (P < 0.05). #Denotes significant main effect of training status (P < 0.05)
Protein content
Despite increased mRNA expression 3 h post-exercise PGC-1α protein content did not increase at 3 days following the acute exercise session (P = 0.137; Fig. 3c). A trend towards increased PGC-1α protein content in YT subjects was observed (P = 0.069). The increase in COXIV mRNA at 3 h post exercise was followed by a significant increase in COXIV protein content at 3 days following the acute exercise session (P = 0.002; Fig. 3d). The increase in COXIV protein content was dependent upon training status (P = 0.046) and not age (P = 0.401), in that, trained persons were able to increase COXIV protein content to a greater extent than untrained persons irrespective of age.
Discussion
The aim of the present study was to examine the effects of ageing and training status on markers of skeletal muscle mitochondrial content at rest and also the ability to activate the acute signalling pathways associated with regulating exercise-induced mitochondrial biogenesis. To the author’s knowledge, this is the first study to incorporate a four-group design whereby both young and old and trained and untrained individuals were simultaneously compared in both the rested and exercised state. We confirm recent observations from human studies (Lanza et al. 2008; Safdar et al. 2010) in that lifelong physical activity can apparently preserve mitochondrial content (as evidenced by resting mtDNA, PGC-1α and COXIV protein content) so much so that muscles from old trained individuals were comparable to young untrained individuals. Additionally, we provide novel data by demonstrating that despite a lifetime of sedentary behaviour, muscles from untrained elderly individuals retain the ability to respond to an acute exercise stress (as evidenced by increased p38MAPK phosphorylation and its downstream effects on PGC-1α mRNA and COXIV mRNA and protein content changes) in a manner comparable to young individuals. As such, we consider our data to have immediate translational potential as they demonstrate that the potential therapeutic effects of exercise to induce skeletal muscle mitochondrial biogenesis persist late in adulthood, even after a lifetime of physical inactivity.
Sedentary ageing is associated with reduced abundance and activity of ETC subunits, impaired mitochondrial function, reduced mtDNA content and lower abundance of nuclear and mitochondrial transcription factors in both human and rodent skeletal muscle (Joseph et al. 2012; Lanza et al. 2008; Ljubicic and Hood 2009; Ljubicic et al. 2010; Reznick et al. 2007; Safdar et al. 2010; Ljubicic et al. 2009). In agreement, the present study demonstrates that sedentary ageing is associated with decreased mtDNA abundance, reduced PGC-1α and COXIV protein content. Conversely, life-long training maintained mtDNA abundance, PGC-1α and COXIV protein content to a level comparable to YU individuals (see Fig. 1). The PGC-1α protein is now well recognised for its role as a transcriptional co-activator of downstream targets such as mitochondrial transcription factor A (TFAM) and nuclear respiratory factor 1 (NRF1) and therefore has regulatory influences on both nuclear and mitochondrial encoded genes (Ljubicic et al. 2010). The preservation of PGC-1α content in OT muscles therefore provides a likely contributory mechanism to the comparable mtDNA and COXIV protein levels observed between OT and YU individuals.
Although our data are comparable to other authors in that we show lifelong exercise has an apparent protective effect on mitochondrial number and content (Lanza et al. 2008; Safdar et al. 2010), it is noteworthy that we show that muscles from our OT group were also comparable to our YT group and not just the YU males per se (see Fig. 1). In fact, in the case of COXIV protein content, we observed higher baseline protein levels in OT subjects compared with YT subjects. These data are somewhat different to Lanza et al. (2008) who observed in their chosen cohort of subjects (samples consisted of mixed sex), that endurance training attenuates but not prevents age-related loss of mtDNA, PGC-1α and TFAM protein content. Disparate findings between studies could relate to subject group differences between studies such as mixed sex versus single sex designs and importantly, training history of the subjects. Indeed, Lanza et al. (2008) classified subjects as trained if they presented with >4 years of training history whereas our OT subjects had >10 years of competitive race cycling experience. When taken together, such data support the notion that the extent of training history (likely related to habitual training intensities, volume and frequencies) may be a predominant factor in determining the magnitude of loss of mitochondrial mass and function which accompanies healthy ageing. In this regard, future studies should ascertain the optimal dose of physical exercise that is required to maintain mitochondrial mass and function during healthy ageing.
The emergence of PGC-1α as a potential target to combat age-related disorders (Wenz 2011), has arisen from transgenic models demonstrating that overexpression of PGC-1α in skeletal muscle protects against sarcopenia, improves insulin sensitivity and exercise capacity (Wenz et al. 2010). The potential of exercise to induce PGC-1α-induced protection appears limited in rodent muscle, however, given that aged animals do not increase PGC-1α content in response to endurance training (Derbre et al. 2012), which may be due to a failure to activate intracellular signalling cascades involving AMPK and p38MAPK in response to muscle contraction (Ljubicic and Hood 2009). In contrast, data from human studies recently demonstrated that muscles from elderly individuals retain the ability for exercise-induced phosphorylation of p38MAPK and AMPK as well as PGC-1α mRNA in the hours following exercise, when compared with trained elderly individuals (Iversen et al. 2011). We confirm and extend these data by observing herein that OU males adapt to an acute exercise stress with similar upstream signalling responses (e.g. p38MAPK phosphorylation) and downstream adaptations in the hours (e.g. PGC-1α and COXIV mRNA) and days (COXIV protein content) following exercise in a manner that is comparable to YU individuals. This highlights the interspecies differences in molecular plasticity of skeletal muscle between human and animal models of ageing (Rennie et al. 2010).
Although we observed similar exercise-induced changes in PGC-1α mRNA at 3 h post-exercise independent of age or training status, it is noteworthy that we observed no concomitant increases in PGC-1α protein at 3 days post-exercise. Time-course studies in young recreationally active males demonstrate that acute exercise induces measurable changes PGC-1α protein content at 24 h post-exercise (Perry et al. 2010) and thus it is possible that in relation to our chosen sampling point of 3 days post-exercise, PGC-1α had returned towards basal levels. In contrast, COXIV protein content exhibited significant increases at 3 days post-exercise in all groups though trained individuals responded with greater increases than untrained individuals regardless of training status. This finding may seem somewhat paradoxical at first given that trained individuals typically display blunted transcriptional responses to habitual exercise (Nordsborg et al. 2010). However, it is worth noting that our trained subjects performed the exercise protocol at higher absolute workloads than untrained individuals (see Table 2) and hence, the enhanced COXIV response may simply be a consequence of higher work done. Alternatively, trained individuals may exhibit post-transcriptional regulation such that a more efficient translation of mRNA to protein is evident with training (Benziane et al. 2008) though further studies are required to test this hypothesis.
In summary, we provide novel data by demonstrating that lifelong endurance training maintains markers of mitochondrial mass (mtDNA, PGC-1α and COXIV protein content) so much so that muscles from old trained males were comparable to muscles from young untrained and young trained individuals. Furthermore, despite a lifetime of physical inactivity that is typically associated with mitochondrial dysfunction and aberrant mitochondrial homeostasis, muscles from OU males retain the ability to respond to an acute exercise challenge via upstream signalling through p38MAPK and its downstream influence on PGC-1α. When taken together, these data demonstrate that the potential therapeutic effects of exercise to induce skeletal muscle mitochondrial biogenesis persist late in adulthood, even after a lifetime of physical inactivity. Future studies are now required (longitudinal in design) to further characterise the time-course of muscle adaptations to sustained endurance training of previously sedentary elderly individuals.
References
Bartlett JD, Hwa Joo C, Jeong TS, Louhelainen J, Cochran AJ, Gibala MJ, Gregson W, Close GL, Drust B, Morton JP (2012) Matched work high-intensity interval and continuous running induce similar increases in PGC-1alpha mRNA, AMPK, p38, and p53 phosphorylation in human skeletal muscle. J Appl Physiol 112(7):1135–1143. doi:10.1152/japplphysiol.01040.2011
Benziane B, Burton TJ, Scanlan B, Galuska D, Canny BJ, Chibalin AV, Zierath JR, Stepto NK (2008) Divergent cell signaling after short-term intensified endurance training in human skeletal muscle. Am J Physiol Endocrinol Metab 295(6):E1427–E1438. doi:10.1152/ajpendo.90428.2008
Carter CS, Hofer T, Seo AY, Leeuwenburgh C (2007) Molecular mechanisms of life- and health-span extension: role of calorie restriction and exercise intervention. Appl Physiol Nutr Metab 32(5):954–966. doi:10.1139/H07-085
Close GL, Kayani A, Vasilaki A, McArdle A (2005) Skeletal muscle damage with exercise and aging. Sports Med 35(5):413–427
Derbre F, Gomez-Cabrera MC, Nascimento AL, Sanchis-Gomar F, Martinez-Bello VE, Tresguerres JA, Fuentes T, Gratas-Delamarche A, Monsalve M, Vina J (2012) Age associated low mitochondrial biogenesis may be explained by lack of response of PGC-1alpha to exercise training. Age (Dordr) 34(3):669–679. doi:10.1007/s11357-011-9264-y
Gianni P, Jan KJ, Douglas MJ, Stuart PM, Tarnopolsky MA (2004) Oxidative stress and the mitochondrial theory of aging in human skeletal muscle. Exp Gerontol 39(9):1391–1400. doi:10.1016/j.exger.2004.06.002
Iversen N, Krustrup P, Rasmussen HN, Rasmussen UF, Saltin B, Pilegaard H (2011) Mitochondrial biogenesis and angiogenesis in skeletal muscle of the elderly. Exp Gerontol 46(8):670–678. doi:10.1016/j.exger.2011.03.004
Janssen I, Shepard DS, Katzmarzyk PT, Roubenoff R (2004) The healthcare costs of sarcopenia in the United States. J Am Geriatr Soc 52(1):80–85
Joseph AM, Adhihetty PJ, Buford TW, Wohlgemuth SE, Lees HA, Nguyen LM, Aranda JM, Sandesara BD, Pahor M, Manini TM, Marzetti E, Leeuwenburgh C (2012) The impact of aging on mitochondrial function and biogenesis pathways in skeletal muscle of sedentary high- and low-functioning elderly individuals. Aging Cell. doi:10.1111/j.1474-9726.2012.00844.x
Kowald A, Kirkwood TB (1996) A network theory of ageing: the interactions of defective mitochondria, aberrant proteins, free radicals and scavengers in the ageing process. Mutat Res 316(5–6):209–236
Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS (2008) Endurance exercise as a countermeasure for aging. Diabetes 57(11):2933–2942. doi:10.2337/db08-0349
Lezza AM, Boffoli D, Scacco S, Cantatore P, Gadaleta MN (1994) Correlation between mitochondrial DNA 4977-bp deletion and respiratory chain enzyme activities in aging human skeletal muscles. Biochem Biophys Res Commun 205(1):772–779. doi:10.1006/bbrc.1994.2732
Little JP, Safdar A, Wilkin GP, Tarnopolsky MA, Gibala MJ (2010) A practical model of low-volume high-intensity interval training induces mitochondrial biogenesis in human skeletal muscle: potential mechanisms. J Physiol 588(Pt 6):1011–1022. doi:10.1113/jphysiol.2009.181743
Ljubicic V, Hood DA (2009) Diminished contraction-induced intracellular signaling towards mitochondrial biogenesis in aged skeletal muscle. Aging Cell 8(4):394–404. doi:10.1111/j.1474-9726.2009.00483.x
Ljubicic V, Joseph AM, Adhihetty PJ, Huang JH, Saleem A, Uguccioni G, Hood DA (2009) Molecular basis for an attenuated mitochondrial adaptive plasticity in aged skeletal muscle. Aging (Albany NY) 1(9):818–830
Ljubicic V, Joseph AM, Saleem A, Uguccioni G, Collu-Marchese M, Lai RY, Nguyen LM, Hood DA (2010) Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: effects of exercise and aging. Biochim Biophys Acta 1800(3):223–234. doi:10.1016/j.bbagen.2009.07.031
Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, Tarnopolsky MA (2004) Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics 18(2):226–231. doi:10.1152/physiolgenomics.00067.2004
Mahoney DJ, Parise G, Melov S, Safdar A, Tarnopolsky MA (2005) Analysis of global mRNA expression in human skeletal muscle during recovery from endurance exercise. FASEB J 19(11):1498–1500. doi:10.1096/fj.04-3149fje
McArdle A, Dillmann WH, Mestril R, Faulkner JA, Jackson MJ (2004) Overexpression of HSP70 in mouse skeletal muscle protects against muscle damage and age-related muscle dysfunction. FASEB J 18(2):355–357. doi:10.1096/fj.03-0395fje
McKenzie D, Bua E, McKiernan S, Cao Z, Aiken JM (2002) Mitochondrial DNA deletion mutations: a causal role in sarcopenia. Eur J Biochem 269(8):2010–2015
Nordsborg NB, Lundby C, Leick L, Pilegaard H (2010) Relative workload determines exercise-induced increases in PGC-1alpha mRNA. Med Sci Sports Exerc 42(8):1477–1484. doi:10.1249/MSS.0b013e3181d2d21c
Perry CG, Lally J, Holloway GP, Heigenhauser GJ, Bonen A, Spriet LL (2010) Repeated transient mRNA bursts precede increases in transcriptional and mitochondrial proteins during training in human skeletal muscle. J Physiol 588(Pt 23):4795–4810. doi:10.1113/jphysiol.2010.199448
Rennie MJ, Selby A, Atherton P, Smith K, Kumar V, Glover EL, Philips SM (2010) Facts, noise and wishful thinking: muscle protein turnover in aging and human disuse atrophy. Scand J Med Sci Sports 20(1):5–9. doi:10.1111/j.1600-0838.2009.00967.x
Reznick RM, Zong H, Li J, Morino K, Moore IK, Yu HJ, Liu ZX, Dong J, Mustard KJ, Hawley SA, Befroy D, Pypaert M, Hardie DG, Young LH, Shulman GI (2007) Aging-associated reductions in AMP-activated protein kinase activity and mitochondrial biogenesis. Cell Metab 5(2):151–156. doi:10.1016/j.cmet.2007.01.008
Safdar A, Hamadeh MJ, Kaczor JJ, Raha S, Debeer J, Tarnopolsky MA (2010) Aberrant mitochondrial homeostasis in the skeletal muscle of sedentary older adults. PLoS ONE 5(5):e10778. doi:10.1371/journal.pone.0010778
Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM (2006) PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci USA 103(44):16260–16265. doi:10.1073/pnas.0607795103
Wenz T (2011) Mitochondria and PGC-1alpha in aging and age-associated diseases. J Aging Res 2011:810619. doi:10.4061/2011/810619
Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT (2009) Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci USA 106(48):20405–20410. doi:10.1073/pnas.0911570106
Wenz T, Williams SL, Bacman SR, Moraes CT (2010) Emerging therapeutic approaches to mitochondrial diseases. Dev Disabil Res Rev 16(2):219–229. doi:10.1002/ddrr.109
Acknowledgments
Age UK are thanked for providing financial support. We also acknowledge the efforts of all the subjects for their outstanding efforts during a demanding exercise protocol. Finally, we thank Rob Allan and Chang Haw-Joo for providing technical support during data collection.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cobley, J.N., Bartlett, J.D., Kayani, A. et al. PGC-1α transcriptional response and mitochondrial adaptation to acute exercise is maintained in skeletal muscle of sedentary elderly males. Biogerontology 13, 621–631 (2012). https://doi.org/10.1007/s10522-012-9408-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10522-012-9408-1