Abstract
Myocardial ischemia and reperfusion (MIR) results in cardiomyocyte apoptosis with severe outcomes, which blocks cardiac tissue recovering from myocardial ischemia diseases. Heat shock protein 70 (HSP70) is one of protective molecule chaperones which could regulate the nucleus translocation of other proteins. In addition, eukaryotic elongation factor 2 (eEF2), which modulates protein translation process, is vital to the recovery of heart during MIR. However, the relationship between HSP70 and eEF2 and its effects on MIR are unclear. The expression and relationship between HSP70 and eEF2 is confirmed by western blot, immunoprecipitation in vitro using cardiomyocyte cell line H9c2 and in vivo rat MIR model. The further investigation was conducted in H9c2 cells with detection for cell-cycle and apoptosis. It is revealed that eEF2 interacted and be regulated by HSP70, which kept eEF2 as dephosphorylated status and preserved the function of eEF2 during MIR. In addition, HSP70 suppressed the nucleus translocation of phosphorylated eEF2, which inhibited cardiomyocyte apoptosis during myocardial reperfusion stage. Furthermore, HSP70 also interacted with C-terminal fragment of eEF2, which could reverse the nucleus translocation and cardiomyocyte apoptosis caused by N-terminal fragment of eEF2. HSP70 draw on advantage and avoid defect of MIR through regulating phosphorylation and nucleus translocation of eEF2.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myocardial ischemia and reperfusion injury (MIRI) occurs during myocardial ischemia diseases such as myocardial infarction, results in a severe decline of cardiac function which can lead to gradual heart failure development [1, 2]. Although revascularization can recover the blood flow of heart, oxidative burst and other pathological changes taking up during reperfusion stage lead to the secondary damages to the injured heart [3]. One of secondary damages during myocardial ischemia and reperfusion (MIR) is cardiomyocyte apoptosis, which can accelerate the deterioration of cardiac function [4]. On the contrary, inhibition of cardiomyocyte apoptosis can ameliorate cardiac function disorders [5]. Meanwhile, previous researches reveal that MIR triggers a complex reaction in cardiomyocytes, which has a subsequent influence on cardiac function [6]. Therefore, more basic researches need to be performed to uncover the potential mechanisms of this pathophysiological process, which can provide a novel scenario for clinical treatment [7]. More importantly, we need further researches to reveal the protective mechanism during MIR [8].
Recently, it is found that certain molecules involved in protein translation process of cardiomyocytes participate in cardiomyocyte apoptosis during MIR [9]. One of these molecules is eukaryotic elongation factor 2 (eEF2), a eukaryotic elongation factor, which has a bidirectional effects on cardiomyocyte apoptosis during MIR [10]. The function of eEF2 is regulated by its phosphorylation status, which controls the speed of ribosomal translation during nutrient deprivation to overcome the dilemma caused by energy consumption [11, 12]. eEF2 has anti-apoptosis effects during MIR, via the dephosphorylation status of eEF2 and the process was the terminal effective regulator of cellular energetic metabolism [13]. Besides the function of translational speed control, phosphorylated eEF2 is cleaved to fragments and translocate to nucleus. In detail, phosphorylated eEF2 leads to the SUMOylation of itself, and the SUMOylation of eEF2 further enhances proteolytic cleavage of eEF2, then translocates to nucleus and induces nuclear deformation and instability [14]. Finally, the instability of nucleus can induce cell apoptosis through DNA damage [15]. In addition, suppressing phosphorylation of eEF2 can inhibit cell apoptosis [16]. Although some molecules and drugs are confirmed to mediately regulate the phosphorylation of eEF2 [17, 18], more investigations are needed to reveal direct regulations for the phosphorylation of eEF2.
Heat shock protein (HSP) family inhibits the phosphorylation of some proteins and facilitates the stability and function of these proteins [19]. Heat shock protein 70 (HSP70), one major member of HSP family, binds its substance through the substrate binding domain to dephosphorylate the substrate and maintain its normal functions [20]. During MIR, HSP70 is proven to have protective effects and can suppress the detrimental effects of MIRI, which is also another potential therapeutic target for clinical research [21–23]. HSP70 specifically accelerates the dephosphorylation of JNK during MIR for cardiac protection [24]. Moreover, as a molecule chaperone, HSP70 inhibits nucleus translocation of apoptosis inducing factor (AIF) during MIR, which stabilizes nucleus and antagonizes cardiomyocyte apoptosis [25]. On the other side, deleting the domain of AIF which interacts with HSP70 enhanced AIF translocation to nucleus [26]. Thus we speculate whether HSP70 regulates the dephosphorylation and nucleus translocation of eEF2 to inhibit cardiomyocyte apoptosis during MIR.
In present study, we identified that HSP70 interacted with eEF2 to suppress the phosphorylation and nucleus translocation of eEF2 during MIR. However, HSP70 did not affect the phosphorylation and nucleus translocation of eEF2 under normal condition. Furthermore, after truncated eEF2 into three fragments, we notified that HSP70 could bind to C-terminal fragment of eEF2 which mediated the nucleus translocation. In addition, C-terminal fragment could reverse the nucleus translocation caused by N-terminal fragment, leading to attenuated cardiomyocyte apoptosis. In summary, these data indicated that HSP70 exerted an anti-apoptosis effect through suppressing phosphorylation and nucleus translocation of eEF2 during MIR.
Materials and methods
Animals care and experimental protocols
Sprague–Dawley (SD) rats, weight 220–280 g, were obtained from the Experimental Animal Center, Nantong University. All procedures were in accordance with The Guide for the Care of Use of Laboratory Animals published by the US National Institute of Health (NIH Publication No. 85–23, revised in 1996). The rats were randomly divided into the following groups (n = 6 in each group): sham group in which rats underwent operation without suture tie-down of the left anterior descending (LAD) coronary artery; experimental groups in which rats underwent 30 min ischemia with 0, 2, 4, 6, 8, 12 and 24 h reperfusion, respectively.
Animal surgery
All animals were operated under anesthesia by intraperitoneal injection with 10% chloral hydrate (300 mg/kg). After endotracheal intubation, the heart was rapidly exposed via a left thoracotomy. Then, a 6–0 polyproline ligature was placed under the left coronary artery (LCA). The ends of the tie were threaded through a small plastic tube to form a snare for reversible LCA occlusion. After ischemia, reperfusion was achieved by loosening the snare and characterized by rapid disappearance of cyanosis. Following the operation, the incisions were closed in layers. The chest and endotracheal tubes were removed. Room temperature (RT) was maintained at 37 °C.
Western blot analysis
Western blot was prepared from myocardial left ventricle tissue after operation (n = 6 at each time point). Tissue was isolated from approximately 0.1 g of myocardial tissue at different time points, and tissue was minced with eye scissors on ice. Total myocardial tissue protein was then homogenized in lysis buffer and clarified by centrifugation at 13,000 rpm for 15 min in a microcentrifuge at 4 °C. The protein was separated by SDS–PAGE, transferred to PVDF membranes (Millipore; Billerica, MA) at 300 mA for different timespan according to different molecular weights. Membranes were exposed to HSP70 (mouse, 1:1000; Abcam; Cambridge, MA); eEF2 (rabbit, 1:500; Santa Cruz; Dallas, TX); glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (rabbit,1:1000; Santa Cruz); phosphorylated eEF2 at Thr-56 (rabbit, 1:1000; Abcam); cleaved caspase-3 (rabbit, 1:500; Santa Cruz); α-actinin (mouse, 1:1000; Santa Cruz); HA (mouse, 1:500; Abmart; Shanghai, China) and Flag (mouse, 1:500; Abmart) antibodies overnight at 4 °C. Then, HRP-conjugated secondary antibodies (1:1000; Southern-Biotech, Birmingham, AL) were added, and incubated for 2 h at 37 °C. Finally, membranes were analyzed by enhanced chemiluminescence system (ECL, Cell Signaling) [27].
Immunohistochemical analysis
After MIR, the left ventricle tissues of rat hearts were perfused with sterile saline and 4% formalin successively (Fisher Scientific, Fairlawn, NJ). Then, hearts were rapidly collected and immersion fixed in 4% formalin for 24 h. The sections were cut serially (5 μm). For immunohistochemical staining, the sections were boiled at 121 °C for 20 min in 10 mM citrate buffer solution (pH 6.0) for antigen retrieval. Endogenous peroxidase activity was blocked by soaking in 0.3% hydrogen peroxide. After rinsing in PBS (pH 7.2), the sections were incubated with HSP70 (mouse, 1:1000; Abcam; Cambridge, MA); eEF2 (rabbit, 1:50; Santa Cruz; Dallas, TX); phosphorylated eEF2 (rabbit, 1:50; Abcam, Cambridge, MA) for 2 h at 37 °C. After incubation, the sections were washed with PBS for three times and incubated with secondary antibody for 30 and 20 min at 37 °C separately. After three times washing by PBS, the sections were incubated with DAB in 0.05 mol/L Tris buffer (pH 7.6) containing 0.03% H2O2 for signal development. Finally, the slides were dehydrated, cleared, and cover slipped [28].
Evan’s blue-triphenyltetrazolium chloride (TTC) double staining
After MIR was established, rat hearts were perfused with 0.5% Evan’s blue through superior vena cava and when the cardiac tissue turned to blue, the hearts were harvested. Then the hearts were sliced to 2 mm-thick pieces. Then the tissue was incubated in 2% TTC solution (pH 7.2) for 30 min at 37 °C. After these procedures, the tissue was perfused with 4% formalin for 2 h at RT. The photos were taken through high resolution digital camera. All reagents were purchased from BD Bioscience Company (NJ, USA).
H9c2 cell culture
The rat cardiomyocyte-derived H9c2 cell line was obtained from the cell library of the Chinese Academy of Science (Shanghai, China) and was cultured in Dulbecco’s Modified Eagle Medium (DMEM, Invitrogen Life Technologies, Grand Island, NY, USA) with 10% fetal bovine serum (FBS, Invitrogen, USA) at 37 °C with 5% CO2. The medium was replaced every other day. When reaching 90% confluence, cells were plated at an appropriate density according to each experimental design. The cells were selected randomly to go through hypoxia (2% O2, 5% CO2, 93% N2) for 6 h with DMEM free of FBS. Then cells were treated with reoxygenation (21% O2, 5% CO2) for 0, 2, 4, 6, 8, 12 or 24 h, with DMEM containing 10% FBS. The corresponding control cells were incubated under normoxic condition for equivalent duration with DMEM containing 10% FBS.
CCK8 assay
The assay was performed using CCK8 assay kit (Biosci Biotechnology Company, Wuhan, Hubei, China) according to the instructions.
Double immunofluorescent staining in H9c2 cells
The H9c2 cells were culture in 24-wells plate (Corning Inc., Corning, NY) and after H/R stimuli, the cells were fixed. All samples were incubated overnight at 4 °C with antibodies for eEF2 (1:50; Santa Cruz), phosphorylated eEF2 (1:100; Abcam), HSP70 (1:100; Abcam), HA (1:100; Abmart) and α-actinin (cardiomyocyte marker, 1:100; Santa Cruz). After washing in PBS for three times, each time 15 min, a mixture of 4, 6-diamidino-2-phenylindole (DAPI) and FITC and Cy3-conjugated secondary antibodies were added in a dark room and incubated overnight at 4 °C. The stained sections were analyzed with a Leica fluorescence microscope (Leica DM 5000B, Germany) [29].
Nucleus and plasma separation
The nucleus and plasma separation of H9c2 cells was performed using nucleus and plasma separation kit (Sangon Biotech Cooperation, Shanghai, China) following the instructions. After separation, the samples were stored at −80 °C.
Immunoprecipitation (IP) experiments
H9c2 Cells were homogenized in lysis buffer and clarified via centrifugation at 13,000 rpm for 15 min in a microcentrifuge at 4 °C. After preclearing with 20 μl Protein G Sepharose beads for 1.5 h at 4 °C, cell lysates were incubated with 5 μl antibody and rotated overnight at 4 °C. Protein G Sepharose beads (30 μl) were added to each sample and incubated for 2 h at 4 °C. After being washed three times with lysis buffer, the immune complexes were heated in sample buffer and used for western blot analysis.
HSP70 and eEF2 full length plasmid construction
The HSP70 and eEF2 overexpression plasmid was purchased from the PPL plasmid library (Nanjing, Jiangsu, China).
Truncated plasmid construction
The fragments of eEF2 were amplified by reverse transcription polymerase chain reaction (RT-PCR) using full-length plasmid. The following primers were used: C-terminal fragment forward primer 5′-CGAATTCGGATGGTCAGCCCTGTCGTCAGGGTG-3′ and reverse primer 5′-CCTCTCGAGGTCAGTTTGTCCAGGAAGTTGTCCAGC-3′, Central fragment forward primer 5′-CGAATTCGGATGGACCGGTACTTTGACCCGGC-3′ and reverse primer 5′-CCTCTCGAGGTGCTGAACTTCATCACCCGCAT-3′, N-terminal fragment forward primer 5′-CGAATTCGGATGGTGAACTTCACAGTAGATCAGA-3′ and reverse primer 5′-CCTCTCGAGGTCCCACAGCTTCTTCATCATG-3′ (Sunny Biotechnology Company, Shanghai, China). The fragments and the vector (PPL, USA) were cut with EcoR I and Xho I. The correct insertion of fragments in pcDNA3.1-HA vector was verified by enzyme digestion and sequencing [30]. After concentration of the plasmid, the plasmids were sent to Biosune Biotechnology Company (Shanghai, China) for sequencing.
Small interfering RNA transfection
Three small interfering RNAs (siRNAs) targeting HSP70 and eEF2 gene were designed and synthesized by Shanghai Genepharma (Shanghai, China), and the most effective siRNA (siHSP70 and sieEF2) identified by western blot was applied for further experiments. The sequences of three siHSP70s are as follows: 5′-CCUGAGAAAGAGGAGUUCGTT-3′, 5′-GAAUGCGCUCGAGUCCUAUTT-3′, 5′-GCUGAGAAAGAGGAGUUCGTT-3′. The sequences of three sieEF2s are as follows: 5′-GCUUCACUGACACUCGAAATT-3′, 5′-CCGUGCUGAUGAUGAACAATT-3′, 5′-CCCAGACGGGAAGAAACUUTT-3′. Scrambled RNA oligonucleotides were used as control. Twenty-four hours prior to transfection, H9c2 cells were plated onto a 6-well plate (Corning Inc., Corning, NY) at 40–60% confluence. For each well, 33.3 nM of each of the three oligos was transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The medium was replaced with DMEM containing 10% FBS after 6 h [31].
eEF2 full length and truncated plasmid transfection
When H9c2 cells reached 85% confluence, plasmids were mixed with Lipofectamine 2000 for 20 min at RT. Then cells were incubated in DMEM for 6 h without FBS, and were replaced the medium with DMEM with 10% FBS for next 40 h. Thereafter, the culture was terminated.
LDH release assay
The assay was performed using LDH assay kit (Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China) following the instructions.
Flow cytometry analysis for H9c2 cell apoptosis
H9c2 cells were previously transfected with eEF2 plasmid for 48 h, after that were treated with H/R. After treatment, the cells were harvested at a density of 1 × 105 cells/sample. H9c2 cells without any treatments were set as negative control. The analysis was conducted following the protocol of PE-7AAD kit (BD, NZ, USA). H9c2 cell apoptosis was measured by the flow cytometer (Calibur, BD, NZ, USA).
Flow cytometry for cell cycle
H9c2 cells were harvested at a density of 1 × 105 cells/sample after transfected with eEF2 truncated plasmid for 48 h. H9c2 cells without any treatments were set as negative control. The cells were fixed in 70% cold ethanol at −20 °C for 24 h, and then the cell suspensions were centrifuged and the fixative was removed. Following 2 washes with PBS, the samples were given propidium iodide (PI) and RNase A at a final concentration of 50 µM for 30 min at 37 °C. The cell cycle distribution was measured by the flow cytometer [32].
Terminal deoxynucleotidyl transferase-mediated dUTP nickend labeling (TUNEL)
H9c2 cell apoptosis was determined by TUNEL fluorescence FITC kit according to the manufacturer’s instructions (Jiancheng Bioengineering Institute, Nanjing, Jiangsu, China). After TUNEL staining, the samples were immersed into a DAPI solution to stain nuclei. Fluorescence staining was viewed by laser scanning confocal microscopy (Leica DM 5000B; Wetzlar, Germany). Four fields were randomly selected from each sample, and at least 100 cells were counted to calculate cell apoptotic ratio.
Statistical analysis
The SPSS19.0 software was used for statistical analysis. The results were presented as means ± SD. Differences between groups were calculated using the student’s t test or one-way analysis of variance. P < 0.05 was considered statistical significant. All the experiments were repeated for at least three times with the consistent results.
Results
HSP70 expression and p-eEF2 to eEF2 ratio are up-regulated during MIR
Rat MIR model was confirmed by Evan’s blue-TTC double staining. Due to the revascularization of blood flow, the recovery of myocardial reperfusion did not completely reverse the damage caused by ischemia (Fig. 1a). In addition, the recovery of areas at risk (AAR) was significant at reperfusion stage after myocardial ischemia. However, serum LDH release increased during ischemia and dropped gradually after reperfusion, which suggested that MIR model was successfully established (Fig. 1b).
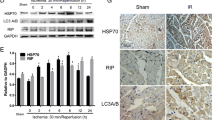
HSP70, eEF2 and p-eEF2 are associated with cardiomyocyte apoptosis in vivo. Adult SD rats were treated with ischemia for 30 min and different time points of reperfusion. a Evans blue/TTC double staining was used to detect the infarction areas (IA) and areas at risk (AAR). b LDH release in rat serum was detected. c HSP70, p-eEF2, eEF2, and c-caspase-3 expression was detected by western blot after MIR. d Statistical graphs (relative optical density) were showed for ratio of HSP70 and c-caspase-3 to GAPDH. e Statistical graphs (relative optical density) were showed for p-eEF2/eEF2 ratio. *P < 0.05 compared with sham group. (Color figure online)
Western blot indicated that after myocardial ischemia, HSP70 protein level and phosphorylated eEF2 (p-eEF2) to eEF2 ratio changed obviously. During 4–8 h reperfusion, HSP70 protein level increased dramatically. After 6 h reperfusion, p-eEF2 to eEF2 ratio dropped slightly. To detect cardiomyocyte apoptosis during MIR, cleaved caspase-3 (c-caspase-3) expression was examined, increasing significantly after reperfusion (Fig. 1c–e). These results indicated that MIR could lead to up-regulation of HSP70 expression and p-eEF2 to eEF2 ratio, which might be related to cardiomyocyte apoptosis. According to above data, 6 h of reperfusion was taken for subsequent experiments.
Subcellular localization of eEF2, HSP70 and p-eEF2 during MIR
In order to identify subcellular localization of HSP70, eEF2 and p-eEF2 during MIR, immunohistochemistry was performed. HSP70 expression increased in MIR group, localizing in nucleus and cytoplasm. Meanwhile, eEF2 expression decreased in MIR group, localizing in cytoplasm, while p-eEF2 expression increased in MIR group, translocating to nucleus (Fig. 1f–g). These results indicated that eEF2 might translocate to nucleus during MIR and interact with HSP70.
HSP70 interacts with eEF2 in vivo
To investigate the relationship between HSP70 and eEF2 during MIR, IP was performed, showing that interaction between HSP70 and eEF2 enhanced during MIR compared with sham group (Fig. 2a, b). In addition, immunofluorescent staining suggested that following MIR, HSP70 and eEF2, HSP70 and p-eEF2 co-localized in cytoplasm and nucleus separately (Fig. 2c). Therefore, we hypothesized that HSP70 interacted with eEF2 to affect the phosphorylation and nucleus translocation of eEF2.
The location of HSP70, eEF2 and p-eEF2 cardiomyocyte during ischemia and reperfusion in vivo. a HSP70, eEF2 and p-eEF2 was detected by immunohistochemical staining in sham and 6 h reperfusion groups. b Statistical graphs for HSP70, eEF2 and p-eEF2 positive areas of rat myocardial tissues.*, #, @ P < 0.05 compared with sham group
HSP70 interacts with eEF2 in vitro
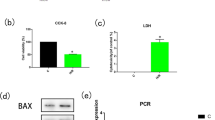
Furthermore, we utilized that H9c2 cells to mimic MIR in vitro for further investigation [33]. After hypoxia and reoxygenation (H/R) stimuli, flow cytometry was used to detect cell apoptosis, showing that secondary apoptosis after 6 h reoxygenation (Fig. 3a, b). In addition, cell viability of H9c2 cells was tested by CCK8 assay, which was in accordance with flow cytometry results (Fig. 3c).
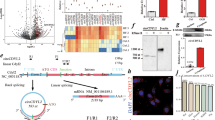
HSP70 interacts with eEF2 in vivo. Myocardial tissue from rats of sham and MIR groups was extracted. a The interaction of HSP70 and eEF2 was detected by IP in sham group. b The interaction of HSP70 and eEF2 was detected by IP in MIR group (IB immunoblotting, IP immunoprecipitation). c Double immunofluorescence staining was performed for HSP70 with eEF2 and p-eEF2 in sham and IR groups. Scale bars 25 μm
Western blot indicated that after H/R stimuli, the changes of HSP70 expression, p-eEF2 to eEF2 ratio and c-caspase-3 expression were in line with the results in vivo (Fig. 3d–f). Additionally, IP and immunofluorescent staining data using H9c2 cells were consistent with in vivo results (Fig. 4). These results further demonstrated the interaction between HSP70 and eEF2.
HSP70, eEF2 and p-eEF2 are associated with cardiomyocyte apoptosis in vitro. H9c2 cells were treated with 6 h hypoxia and different time of reoxygenation. a Flow cytometry was performed to detect H9c2 cell apoptosis. b Statistical graphs for ratio of apoptotic index. c CCK-8 assay was used to detect H9c2 cell viability. d HSP70, eEF2, p-eEF2 and c-caspase-3 expression was detected by western blot after H/R. e Statistical graphs (relative optical density) for HSP70 and c-caspase-3 to GAPDH. f Statistical graphs (relative optical density) for p-eEF2/eEF2 ratio. *P < 0.05 compared with normal group
HSP70 interacts with the C-terminal fragment of eEF2
In order to identify the specific domain of eEF2 which interacted with HSP70, we truncated full-length eEF2 into three fragments based on previous researches and domain analysis, and named them as N-terminal fragment, Central fragment and C-terminal fragment (Fig. 5a). After labeled by HA, the plasmids were transfected into H9c2 cells with Flag-HSP70 for 48 h and IP was used to clarify the interactive region, showing that HSP70 interacted with C-terminal fragment of eEF2 (Fig. 5b).
HSP70 interacts with eEF2 in vitro. H9c2 cells of normal and H/R (hypoxia: 6 h, reoxygenation: 6 h) stimuli groups were used. a The interaction of HSP70 and eEF2 was detected by IP in normal group. b The interaction of HSP70 and eEF2 was detected by IP in H/R stimuli group (IB immunoblotting, IP immunoprecipitation). c Double immunofluorescence staining was performed for HSP70 with eEF2 and p-eEF2 in normal and H/R stimuli groups. Scale bars 50 μm
HSP70 knockdown promotes phosphorylation and nucleus translocation of eEF2
To explore the effects of HSP70 on eEF2 after H/R stimuli, we utilized siHSP70 to knock down HSP70 expression, finding that HSP70 siRNA3 was the most efficient (Fig. 6a). After HSP70 knockdown, p-eEF2 to eEF2 ratio increased significantly after H/R stimuli, while normal group did not change remarkably (Fig. 6b–d). However, eEF2 knockdown did not change HSP70 expression in H/R stimuli and sham groups (Fig. 6e–g).
HSP70 interacts with C-terminal fragment of eEF2 in HEK293T cells. HEK293T cells were transfected with Flag-HSP70 and HA-labeled wild-type and three fragments of eEF2. a The graphic demonstration for truncated fragments of eEF2. b The interaction of Flag-HSP70 and HA-labeled wild-type and three fragments of eEF2 was detected by IP in H/R stimuli group (IB immunoblotting, IP immunoprecipitation)
In addition, we used immunofluorescence to detect nucleus translocation of eEF2 with siHSP70 transfection, finding that HSP70 knockdown promoted phosphorylation and nucleus translocation of eEF2 (Fig. 7). These results indicated that HSP70 might inhibit phosphorylation and nucleus translocation of eEF2, while eEF2 had no effects on HSP70 expression.
HSP70 knockdown promotes phosphorylation of eEF2. H/R: hypoxia for 6 h and reoxygenation for 6 h. a HSP70 protein expression after HSP70 knockdown with different siRNA. b HSP70, eEF2 and p-eEF2 expression was detected by western blot. c Statistical graph (relative optical density) for p-eEF2/eEF2 ratio. d Statistical graph (relative optical density) for HSP70 to GAPDH. e eEF2 protein expression after eEF2 knockdown with different siRNA. f Statistical graph (relative optical density) for the ratio of HSP70 to GAPDH. *P < 0.05 compared with control group
HSP70 overexpression inhibits phosphorylation and nucleus translocation of eEF2
To further explore the effects of HSP70 on phosphorylation and nucleus translocation of eEF2, Flag-HSP70 was transfected in H9c2 cells to up-regulate HSP70 expression. After Flag-HSP70 transfection, p-eEF2 to eEF2 ratio increased after H/R stimuli, while in normal group it did not change significantly (Fig. 8a–c). Furthermore, immunofluorescence confirmed that HSP70 overexpression suppressed phosphorylated and nucleus translocation of eEF2 after H/R stimuli (Fig. 8d). These results indicated that HSP70 overexpression inhibited phosphorylation and nucleus translocation of eEF2.
HSP70 knockdown promotes p-eEF2 nucleus translocation. Distribution of p-eEF2 and eEF2 in H9c2 cells after HSP70 knockdown. H9c2 cells fixed on the small plates were detected for eEF2, p-eEF2 with α-actinin. DAPI was used to label nuclei (H/R: hypoxia for 6 h and reoxygenation for 6 h). Scale bars 25 μm
Next, we utilized IP to detect SUMOylation of eEF2 after HSP70 overexpression. After Flag-HSP70 transfection, SUMOylation of eEF2 was inhibited after H/R stimuli (Fig. 9a). The results indicated that HSP70 might reduce SUMOylation of eEF2 during MIR.
HSP70 overexpression inhibits phosphorylation and nucleus translocation of eEF2. H9c2 cells transfected with Flag-HSP70 were treated with 6 h hypoxia and 6 h reoxygenation. a HSP70, eEF2 and p-eEF2 expression after HSP70 overexpression. b Statistical graphs (relative optical density) for HSP70 to GAPDH. c Statistical graphs (relative optical density) for p-eEF2/eEF2 ratio. d Distribution of p-eEF2 and eEF2 in H9c2 cells after HSP70 overexpression. (H/R: hypoxia for 6 h and reoxygenation for 6 h). Scale bars 25 μm. *P < 0.05 compared with normal group
HSP70 reverses detrimental effect of N-terminal fragment of eEF2 through binding to C-terminal fragment of eEF2
In addition, three fragments of eEF2 were transfected into H9c2 cells respectively and western blot was used to detect the efficiency of transfection. After transfection, N-terminal fragment led to more c-caspase-3 expression than C-terminal and central fragment. C-caspase-3 expression was reduced after C-terminal fragment and Flag-HSP70 transfection, compared with only Flag-HSP70 transfection (Fig. 9b, c). Furthermore, H9c2 cell apoptosis was further confirmed by TUNEL staining (Fig. 9d, e). These results showed that HSP70 blocked the detrimental effect of N-terminal fragment of eEF2 through binding to C-terminal fragment of eEF2.
To further investigate nucleus translocation of eEF2, we used three fragments of eEF2 and Flag-HSP70 transfection. N-terminal fragment translocated to the nucleus while the other two fragments did not. However, N-terminal fragment nucleus translocation could not be reversed by HSP70 overexpression, but could be blocked by C-terminal fragment transfection (Fig. 10a). Besides, cell cycle flow cytometry analysis revealed that cell cycle arrest effect of N-terminal fragment could be blocked by Flag-HSP70 and C-terminal fragment transfection (Fig. 10a, b). These results indicated that HSP70 reduced pro-apoptotic effect of N-terminal fragment with assistance of C-terminal fragment via breaking cell cycle arrest (Fig. 11a, b).
SUMOylation modification of eEF2 and pro-apoptotic function of N-terminal fragment of eEF2. a IP for SUMO2/3 of eEF2 in normal and H/R (hypoxia for 6 h and reoxygenation for 6 h) groups. b C-caspase-3 expression of H9c2 cells transfected with Flag-HSP70, HA-labeled wild-type and three fragments of eEF2. c Statistical graphs (relative optical density) for c-caspase-3 to GAPDH. d TUNEL staining for H9c2 cells transfected with Flag-HSP70, HA-labeled wild-type and three fragments of eEF2. e Statistical graphs for TUNEL positive cell count. Scale bars 25 μm. *P < 0.05 compared with normal group
N-terminal fragment of eEF2 translocates to nucleus and leads to cell cycle arrest of H9c2 cells. a Double immunofluorescence staining and cell cycle flow cytometry for H9c2 cells transfected with Flag-HSP70, HA-labeled wild-type and three fragments of eEF2. b Statistical graphs for different cell cycle. *P < 0.05 compared with normal group
Discussion
The study reveals that HSP70 inhibits eEF2 nucleus translocation during MIR via promoting dephosphorylation of eEF2 and inhibiting SUMOylation of eEF2. After full length eEF2 is truncated, N-terminal fragment of eEF2 promotes eEF2 nucleus translocation, leads to more H9c2 cell apoptosis through cell cycle arrest, which is in line with previous study [34]. The nucleus translocation and pro-apoptotic effects of N-terminal fragment can be blocked by HSP70 overexpression and C-terminal fragment of eEF2 transfection.
HSP70 is up-regulated during MIR due to reactive oxygen species (ROS), which results from oxygen supply rescue during reperfusion stage [35, 36]. During MIR, HSP70 exerts cardioprotection roles, including maintaining chromatin stability, inhibiting cardiomyocyte apoptosis and promoting recovery of cardiac function [5, 22]. However, HSP70, with the assistance of other molecule chaperones, promotes virus protein translation in eukaryotic cells [37]. In addition, HSP70 blocks AIF nucleus translocation during MIR and deletion the domain of AIF interacting with HSP70 results in enhancement of nucleus translocation [25, 26]. In our study, HSP70 interacts with eEF2 and this interaction is more intensive during MIR than normal condition, indicating that HSP70 might affect nucleus translocation of eEF2 during MIR.
During MIR, HSP70 inhibits the phosphorylation of proteins such as protein kinase C (PKC), to stabilize function and structure of them [38]. In our study, we also find HSP70 overexpression reduces the phosphorylation of eEF2. Recent research reveals that HSP70 regulates the phosphorylation process of other protein not through combining with the phosphorylation sites [39]. In our study, HSP70 interacts with C-terminal fragment of eEF2 rather than the fragment containing phosphorylation site.
The functions of eEF2 during MIR are of great significance, and have different roles during ischemia and reperfusion stages [13]. In addition, phosphorylation of eEF2 after nutrient deprivation leads to less apoptosis and more resistance to the environment [11]. In the circumstance of stress, inhibiting eEF2 activity results in the rapid arrest of protein synthesis and cell cycle arrest, apoptosis and cell death displaying consecutively [40, 41], which cause irreversible damage to cardiac function [42]. Additionally, previous studies suggest that activity of eEF2 is regulated by phosphorylation through its kinase, eEF2K, which is regulated by multiple signaling pathways including adenosine 5′-monophosphate (AMP)-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) during MIR [13, 43]. These studies focus on the energetic regulatory function of eEF2, illustrating that inhibiting the activity of eEF2 can reduce apoptosis through enhancing the resistance to hypoxia or ischemia damage [10, 44]. Therefore, in our study, we focused on phosphorylation and nucleus translocation of eEF2 during MIR, identifying that HSP70 reduced phosphorylation and nucleus translocation of eEF2 during reperfusion stage.
As reported previously that eEF2 has small ubiquitin-related modifier (SUMO) after phosphorylated and modification type is 1 or 2/3 [14]. In lung adenocarcinoma cells, eEF2 has SUMO1ylation which leads to protein stability and anti-apoptotic effects [45]. We utilize the software GPS-SUMO 1.0 to predict potential SUMOylation type of eEF2, finding that eEF2 has both SUMO1-ylation and SUMO2/3-ylation [46]. Moreover, unlike SUMO1-ylation, SUMO2/3-ylation has effects on nucleus translocation of some proteins [47]. In our study, after H/R stimuli, HSP70 overexpression leads to less SUMO2/3-ylation of eEF2. All of SUMO2/3-yaltion regions are in N-terminal fragment of eEF2.
eEF2 contains ribosomal protein S5 domain, which is presented in various kinds of DNA/RNA binding proteins [48]. According to domain analysis, this domain forms a hexamer and is oriented towards the surface of ring formed by ATPase domain [49]. In addition, ribosomal protein S5 domain can function as GHMP kinases contains galacto-, homoserine, mevalonate and phosphomevalonate kinases, GHMP kinase, which transfers phosphoryl group from ATP to an acceptor [50]. Phosphoryl groups are used by kinase to catalyze phosphorylation reaction [51], suggesting that this domain might assist eEF2K to promote phosphorylation of eEF2. HSP70 has an ATPase domain which hydrolyses ATP to ADP and produces phosphate radical, leading to apoptosis after inhibiting by some drugs [52]. Thus, it is reasonable to predict the HSP70 might interact with C-terminal fragment of eEF2 to maintain the function of eEF2 and inhibit phosphorylation of eEF2.
On the basis of software analysis, eEF2 also has two more SUMO2/3-ylation interaction motifs (SIM). SUMOylation occurs in the unstable region or domain of peptide chain, and needs one another functional region which is known as SIM to assist [53]. SIM promotes SUMOylation of proteins and truncating without SIM induces dysfunction of proteins [54]. In addition, SIM has effects on the distribution of protein within cells through regulating SUMOylation [55]. HSP70 might be one of targets of SUMO using stable isotope labeling with amino acids (SILAC) quantitative proteomic technique [56], while the software shows that no SUMO sites are predicted. Thus, we predict that HSP70 might competitively bind to SIM, causing less SUMO2/3-ylation of eEF2. Besides, HSP70 competitively binds to the functional domain of estrogen receptor, resulting in functional suppression [57], indicating that as a molecule chaperone, HSP70 functions as competitive binding. In a system analysis, both HSP70 and eEF2 combine with AKT2 [58], suggesting that HSP70 and eEF2 have certain resemblance in domain structure and might have competitive binding effect. Our results indicate that HSP70 only interacts with N-terminal fragment of eEF2. However, the unknown exact mechanism needs further investigation.
There are several limitations in our study. Firstly, we only focus on the investigation of mechanism and the outcome of fragments we constructed. Thus in vivo experiments are the keystone of our future study. Secondly, due to the large amount of in vitro experiments and considering siRNA and plasmid transfection efficiency, we utilized H9c2 cell lines to partially represent the features of neonatal cardiomyocytes. Although previous report confirms the cell line H9c2 cells can mimic the pathological changes in vivo [33], we still hope to use primary cardiomyocytes in our future study. Thirdly, we could not exclude the probabilities that other molecules have effects on the interaction of HSP70 and eEF2 during MIR.
References
Wassenaar JW, Gaetani R, Garcia JJ, Braden RL, Luo CG, Huang D, DeMaria AN, Omens JH, Christman KL (2016) Evidence for mechanisms underlying the functional benefits of a myocardial matrix hydrogel for post-MI treatment. J Am Coll Cardiol 67(9):1074–1086. doi:10.1016/j.jacc.2015.12.035
Dharmakumar R (2015) Building a unified mechanistic insight into the bimodal pattern of edema in reperfused acute myocardial infarctions: observations, interpretations, and outlook. J Am Coll Cardiol 66(7):829–831. doi:10.1016/j.jacc.2015.05.074
Eisenhardt SU, Weiss JB, Smolka C, Maxeiner J, Pankratz F, Bemtgen X, Kustermann M, Thiele JR, Schmidt Y, Bjoern Stark G, Moser M, Bode C, Grundmann S (2015) MicroRNA-155 aggravates ischemia-reperfusion injury by modulation of inflammatory cell recruitment and the respiratory oxidative burst. Basic Res Cardiol 110(3):32. doi:10.1007/s00395-015-0490-9
Joiner ML, Koval OM, Li J, He BJ, Allamargot C, Gao Z, Luczak ED, Hall DD, Fink BD, Chen B, Yang J, Moore SA, Scholz TD, Strack S, Mohler PJ, Sivitz WI, Song LS, Anderson ME (2012) CaMKII determines mitochondrial stress responses in heart. Nature 491(7423):269–273. doi:10.1038/nature11444
Gottlieb RA, Burleson KO, Kloner RA, Babior BM, Engler RL (1994) Reperfusion injury induces apoptosis in rabbit cardiomyocytes. J Clin Invest 94(4):1621–1628. doi:10.1172/JCI117504
Whelan RS, Kaplinskiy V, Kitsis RN (2010) Cell death in the pathogenesis of heart disease: mechanisms and significance. Annu Rev Physiol 72:19–44. doi:10.1146/annurev.physiol.010908.163111
Moludi J, Keshavarz S, Tabaee AS, Safiri S, Pakzad R (2016) Q10 supplementation effects on cardiac enzyme CK-MB and troponin in patients undergoing coronary artery bypass graft: a randomized, double-blinded, placebo-controlled clinical trial. J Cardiovasc Thorac Res 8(1):1–7. doi:10.15171/jcvtr.2016.01
Gottlieb RA, Finley KD, Mentzer RM Jr (2009) Cardioprotection requires taking out the trash. Basic Res Cardiol 104(2):169–180. doi:10.1007/s00395-009-0011-9
Salloum FN (2015) Hydrogen sulfide and cardioprotection–Mechanistic insights and clinical translatability. Pharmacol Ther 152:11–17. doi:10.1016/j.pharmthera.2015.04.004
Kim AS, Miller EJ, Wright TM, Li J, Qi D, Atsina K, Zaha V, Sakamoto K, Young LH (2011) A small molecule AMPK activator protects the heart against ischemia-reperfusion injury. J Mol Cell Cardiol 51(1):24–32. doi:10.1016/j.yjmcc.2011.03.003
Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Kwok B, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins VP, Jones RG, Pollak M, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PH (2013) The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153(5):1064–1079. doi:10.1016/j.cell.2013.04.055
Kaul G, Pattan G, Rafeequi T (2011) Eukaryotic elongation factor-2 (eEF2): its regulation and peptide chain elongation. Cell Biochem Funct 29(3):227–234. doi:10.1002/cbf.1740
Crozier SJ, Vary TC, Kimball SR, Jefferson LS (2005) Cellular energy status modulates translational control mechanisms in ischemic-reperfused rat hearts. Am J Physiol Heart Circ Physiol 289(3):H1242–H1250. doi:10.1152/ajpheart.00859.2004
Yao Q, Liu BQ, Li H, McGarrigle D, Xing BW, Zhou MT, Wang Z, Zhang JJ, Huang XY, Guo L (2014) C-terminal Src kinase (Csk)-mediated phosphorylation of eukaryotic elongation factor 2 (eEF2) promotes proteolytic cleavage and nuclear translocation of eEF2. J Biol Chem 289(18):12666–12678. doi:10.1074/jbc.M113.546481
Bayram F, Bitgen N, Donmez-Altuntas H, Cakir I, Hamurcu Z, Sahin F, Simsek Y, Baskol G (2014) Increased genome instability and oxidative DNA damage and their association with IGF-1 levels in patients with active acromegaly. growth hormone & IGF research : official journal of the growth hormone research society and the International IGF Research. Society 24(1):29–34. doi:10.1016/j.ghir.2013.12.002
Faller WJ, Jackson TJ, Knight JR, Ridgway RA, Jamieson T, Karim SA, Jones C, Radulescu S, Huels DJ, Myant KB, Dudek KM, Casey HA, Scopelliti A, Cordero JB, Vidal M, Pende M, Ryazanov AG, Sonenberg N, Meyuhas O, Hall MN, Bushell M, Willis AE, Sansom OJ (2015) mTORC1-mediated translational elongation limits intestinal tumour initiation and growth. Nature 517(7535):497–500. doi:10.1038/nature13896
Chen Z, Gopalakrishnan SM, Bui MH, Soni NB, Warrior U, Johnson EF, Donnelly JB, Glaser KB (2011) 1-Benzyl-3-cetyl-2-methylimidazolium iodide (NH125) induces phosphorylation of eukaryotic elongation factor-2 (eEF2): a cautionary note on the anticancer mechanism of an eEF2 kinase inhibitor. J Biol Chem 286(51):43951–43958. doi:10.1074/jbc.M111.301291
Iizuka A, Sengoku K, Iketani M, Nakamura F, Sato Y, Matsushita M, Nairn AC, Takamatsu K, Goshima Y, Takei K (2007) Calcium-induced synergistic inhibition of a translational factor eEF2 in nerve growth cones. Biochem Biophys Res Commun 353(2):244–250. doi:10.1016/j.bbrc.2006.11.150
Yu N, Kakunda M, Pham V, Lill JR, Du P, Wongchenko M, Yan Y, Firestein R, Huang X (2015) HSP105 recruits protein phosphatase 2 A to dephosphorylate beta-catenin. Mol Cell Biol 35(8):1390–1400. doi:10.1128/MCB.01307-14
Connarn JN, Assimon VA, Reed RA, Tse E, Southworth DR, Zuiderweg ER, Gestwicki JE, Sun D (2014) The molecular chaperone Hsp70 activates protein phosphatase 5 (PP5) by binding the tetratricopeptide repeat (TPR) domain. J Biol Chem 289(5):2908–2917. doi:10.1074/jbc.M113.519421
Vicencio JM, Yellon DM, Sivaraman V, Das D, Boi-Doku C, Arjun S, Zheng Y, Riquelme JA, Kearney J, Sharma V, Multhoff G, Hall AR, Davidson SM (2015) Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J Am Coll Cardiol 65(15):1525–1536. doi:10.1016/j.jacc.2015.02.026
Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G (2006) Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell cycle 5(22):2592–2601
Feng Y, Huang W, Meng W, Jegga AG, Wang Y, Cai W, Kim HW, Pasha Z, Wen Z, Rao F, Modi RM, Yu X, Ashraf M (2014) Heat shock improves Sca-1 + stem cell survival and directs ischemic cardiomyocytes toward a prosurvival phenotype via exosomal transfer: a critical role for HSF1/miR-34a/HSP70 pathway. Stem cells 32(2):462–472. doi:10.1002/stem.1571
Gabai VL, Meriin AB, Yaglom JA, Wei JY, Mosser DD, Sherman MY (2000) Suppression of stress kinase JNK is involved in HSP72-mediated protection of myogenic cells from transient energy deprivation. HSP72 alleviates the stewss-induced inhibition of JNK dephosphorylation. J Biol Chem 275(48):38088–38094. doi:10.1074/jbc.M006632200
Choudhury S, Bae S, Ke Q, Lee JY, Kim J, Kang PM (2011) Mitochondria to nucleus translocation of AIF in mice lacking Hsp70 during ischemia/reperfusion. Basic Res Cardiol 106(3):397–407. doi:10.1007/s00395-011-0164-1
Gurbuxani S, Schmitt E, Cande C, Parcellier A, Hammann A, Daugas E, Kouranti I, Spahr C, Pance A, Kroemer G, Garrido C (2003) Heat shock protein 70 binding inhibits the nuclear import of apoptosis-inducing factor. Oncogene 22(43):6669–6678. doi:10.1038/sj.onc.1206794
Tao T, Cheng C, Ji Y, Xu G, Zhang J, Zhang L, Shen A (2012) Numbl inhibits glioma cell migration and invasion by suppressing TRAF5-mediated NF-kappaB activation. Mol Biol Cell 23(14):2635–2644. doi:10.1091/mbc.E11-09-0805
Wang Y, Liu F, Mao F, Hang Q, Huang X, He S, Wang Y, Cheng C, Wang H, Xu G, Zhang T, Shen A (2013) Interaction with cyclin H/cyclin-dependent kinase 7 (CCNH/CDK7) stabilizes C-terminal binding protein 2 (CtBP2) and promotes cancer cell migration. J Biol Chem 288(13):9028–9034. doi:10.1074/jbc.M112.432005
Zhang D, Sun L, Zhu H, Wang L, Wu W, Xie J, Gu J (2012) Microglial LOX-1 reacts with extracellular HSP60 to bridge neuroinflammation and neurotoxicity. Neurochem Int 61(7):1021–1035. doi:10.1016/j.neuint.2012.07.019
Wang J, Li Y, Liu Y, Li Y, Gong S, Fang F, Wang Z (2015) Overexpression of truncated AIF regulated by Egr1 promoter radiation-induced apoptosis on MCF-7 cells. Radiat Environ Biophys 54(4):413–421. doi:10.1007/s00411-015-0619-0
Wan C, Hou S, Ni R, Lv L, Ding Z, Huang X, Hang Q, He S, Wang Y, Cheng C, Gu XX, Xu G, Shen A (2015) MIF4G domain containing protein regulates cell cycle and hepatic carcinogenesis by antagonizing CDK2-dependent p27 stability. Oncogene 34(2):237–245. doi:10.1038/onc.2013.536
Lu D, Qian J, Li W, Feng Q, Pan S, Zhang S (2015) beta-hydroxyisovaleryl-shikonin induces human cervical cancer cell apoptosis via PI3K/AKT/mTOR signaling. Oncol Lett 10(6):3434–3442. doi:10.3892/ol.2015.3769
Kuznetsov AV, Javadov S, Sickinger S, Frotschnig S, Grimm M (2015) H9c2 and HL-1 cells demonstrate distinct features of energy metabolism, mitochondrial function and sensitivity to hypoxia-reoxygenation. Biochim Biophys Acta 1853(2):276–284. doi:10.1016/j.bbamcr.2014.11.015
Ranjani J, Pushpanathan M, Mahesh A, Niraimathi M, Gunasekaran P, Rajendhran J (2015) Pseudomonas aeruginosa PAO1 induces distinct cell death mechanisms in H9C2 cells and its differentiated form. J Basic Microbiol 55(10):1191–1202. doi:10.1002/jobm.201500037
Suzuki K, Murtuza B, Sammut IA, Latif N, Jayakumar J, Smolenski RT, Kaneda Y, Sawa Y, Matsuda H, Yacoub MH (2002) Heat shock protein 72 enhances manganese superoxide dismutase activity during myocardial ischemia-reperfusion injury, associated with mitochondrial protection and apoptosis reduction. Circulation 106 (12 Suppl 1):I270–1276
Sun L, Fan H, Yang L, Shi L, Liu Y (2015) Tyrosol prevents ischemia/reperfusion-induced cardiac injury in H9c2 cells: involvement of ROS, Hsp70, JNK and ERK, and apoptosis. Molecules 20(3):3758–3775. doi:10.3390/molecules20033758 pii]
Hafren A, Hofius D, Ronnholm G, Sonnewald U, Makinen K (2010) HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22(2):523–535. doi:10.1105/tpc.109.072413
Gao T, Newton AC (2002) The turn motif is a phosphorylation switch that regulates the binding of Hsp70 to protein kinase C. J Biol Chem 277(35):31585–31592. doi:10.1074/jbc.M204335200
Gao T, Newton AC (2006) Invariant Leu preceding turn motif phosphorylation site controls the interaction of protein kinase C with Hsp70. J Biol Chem 281(43):32461–32468. doi:10.1074/jbc.M604076200
Wullner U, Neef I, Eller A, Kleines M, Tur MK, Barth S (2008) Cell-specific induction of apoptosis by rationally designed bivalent aptamer-siRNA transcripts silencing eukaryotic elongation factor 2. Curr Cancer Drug Targets 8(7):554–565
Gismondi A, Caldarola S, Lisi G, Juli G, Chellini L, Iadevaia V, Proud CG, Loreni F (2014) Ribosomal stress activates eEF2K-eEF2 pathway causing translation elongation inhibition and recruitment of terminal oligopyrimidine (TOP) mRNAs on polysomes. Nucleic Acids Res 42(20):12668–12680. doi:10.1093/nar/gku996
Mazelin L, Panthu B, Nicot AS, Belotti E, Tintignac L, Teixeira G, Zhang Q, Risson V, Baas D, Delaune E, Derumeaux G, Taillandier D, Ohlmann T, Ovize M, Gangloff YG, Schaeffer L (2016) mTOR inactivation in myocardium from infant mice rapidly leads to dilated cardiomyopathy due to translation defects and p53/JNK-mediated apoptosis. J Mol Cell Cardiol. doi:10.1016/j.yjmcc.2016.04.011
Heise C, Taha E, Murru L, Ponzoni L, Cattaneo A, Guarnieri FC, Montani C, Mossa A, Vezzoli E, Ippolito G, Zapata J, Barrera I, Ryazanov AG, Cook J, Poe M, Stephen MR, Kopanitsa M, Benfante R, Rusconi F, Braida D, Francolini M, Proud CG, Valtorta F, Passafaro M, Sala M, Bachi A, Verpelli C, Rosenblum K, Sala C (2016) eEF2K/eEF2 pathway controls the excitation/inhibition balance and susceptibility to epileptic seizures. Cereb Cortex. doi:10.1093/cercor/bhw075
Terai K, Hiramoto Y, Masaki M, Sugiyama S, Kuroda T, Hori M, Kawase I, Hirota H (2005) AMP-activated protein kinase protects cardiomyocytes against hypoxic injury through attenuation of endoplasmic reticulum stress. Mol Cell Biol 25(21):9554–9575. doi:10.1128/MCB.25.21.9554-9575.2005
Chen CY, Fang HY, Chiou SH, Yi SE, Huang CY, Chiang SF, Chang HW, Lin TY, Chiang IP, Chow KC (2011) Sumoylation of eukaryotic elongation factor 2 is vital for protein stability and anti-apoptotic activity in lung adenocarcinoma cells. Cancer Sci 102(8):1582–1589. doi:10.1111/j.1349-7006.2011.01975.x
Zhao Q, Xie Y, Zheng Y, Jiang S, Liu W, Mu W, Liu Z, Zhao Y, Xue Y, Ren J (2014) GPS-SUMO: a tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucl Acids Res 42 (Web Server issue):W325–330. doi:10.1093/nar/gku383
Sedek M, Strous GJ (2013) SUMOylation is a regulator of the translocation of Jak2 between nucleus and cytosol. Biochem J 453(2):231–239. doi:10.1042/BJ20121375
Ramakrishnan V, Davies C, Gerchman SE, Golden BL, Hoffmann DW, Jaishree TN, Kyila JH, Porter S, White SW (1995) Structures of prokaryotic ribosomal proteins: implications for RNA binding and evolution. Biochemistry Cell Biol 73 (11–12):979–986
Botos I, Melnikov EE, Cherry S, Tropea JE, Khalatova AG, Rasulova F, Dauter Z, Maurizi MR, Rotanova TV, Wlodawer A, Gustchina A (2004) The catalytic domain of Escherichia coli Lon protease has a unique fold and a Ser-Lys dyad in the active site. J Biol Chem 279(9):8140–8148. doi:10.1074/jbc.M312243200
Andreassi JL 2nd, Leyh TS (2004) Molecular functions of conserved aspects of the GHMP kinase family. BioChemistry 43(46):14594–14601. doi:10.1021/bi048963o
Ul-Haq Z, Gul S, Usmani S, Wadood A, Khan W (2015) Binding site identification and role of permanent water molecule of PIM-3 kinase: a molecular dynamics study. J Mol Gr Model 62:276–282. doi:10.1016/j.jmgm.2015.07.004
Ko SK, Kim J, Na DC, Park S, Park SH, Hyun JY, Baek KH, Kim ND, Kim NK, Park YN, Song K, Shin I (2015) A small molecule inhibitor of ATPase activity of HSP70 induces apoptosis and has antitumor activities. Chem Biol 22 (3):391–403. doi:10.1016/j.chembiol.2015.02.004
Lin D, Tatham MH, Yu B, Kim S, Hay RT, Chen Y (2002) Identification of a substrate recognition site on Ubc9. J Biol Chem 277(24):21740–21748. doi:10.1074/jbc.M108418200
Kolesar P, Altmannova V, Silva S, Lisby M, Krejci L (2016) Pro-recombination role of Srs2 protein requires SUMO (Small Ubiquitin-like Modifier) but Is independent of PCNA (Proliferating Cell Nuclear Antigen) Interaction. J Biol Chem 291(14):7594–7607. doi:10.1074/jbc.M115.685891
Yang SH, Sharrocks AD (2010) The SUMO E3 ligase activity of Pc2 is coordinated through a SUMO interaction motif. Mol Cell Biol 30(9):2193–2205. doi:10.1128/MCB.01510-09
Wen D, Xu Z, Xia L, Liu X, Tu Y, Lei H, Wang W, Wang T, Song L, Ma C, Xu H, Zhu W, Chen G, Wu Y (2014) Important role of SUMOylation of spliceosome factors in prostate cancer cells. J Proteome Res 13(8):3571–3582. doi:10.1021/pr4012848
Ogawa S, Oishi H, Mezaki Y, Kouzu-Fujita M, Matsuyama R, Nakagomi M, Mori E, Murayama E, Nagasawa H, Kitagawa H, Yanagisawa J, Yano T, Kato S (2005) Repressive domain of unliganded human estrogen receptor alpha associates with Hsc70. Genes Cells 10 (12):1095–1102. doi:10.1111/j.1365-2443.2005.00904.x
Bottermann K, Reinartz M, Barsoum M, Kotter S, Godecke A (2013) Systematic analysis reveals elongation factor 2 and alpha-enolase as novel interaction partners of AKT2. PloS ONE 8(6):e66045. doi:10.1371/journal.pone.0066045
Acknowledgements
This work was supported by the National Basic Research Program of China (973 Program, No. 2012CB822104); National Natural Science Foundation of China (Nos. 81401365, 31500647, 81171140, 81471258, 31440037 and 31270802) The Natural Science Foundation of the Jiangsu Higher Education Institutions of China (14KJB180018); Nantong science and technology project (MS12015056, MS12015067); a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No competing financial interests exist.
Additional information
Chao Zhang, Xiaojuan Liu, Xiang Wu and Aiguo Shen have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhang, C., Liu, X., Miao, J. et al. Heat shock protein 70 protects cardiomyocytes through suppressing SUMOylation and nucleus translocation of phosphorylated eukaryotic elongation factor 2 during myocardial ischemia and reperfusion. Apoptosis 22, 608–625 (2017). https://doi.org/10.1007/s10495-017-1355-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-017-1355-5