Abstract
The emergence of pathogenic bacteria resistant to antibiotics increases the need for discovery of new effective antimicrobials. Unique habitats such as marine deposits, wetlands and caves or unexplored biological communities are promising sources for the isolation of actinobacteria, which are among the major antibiotic producers. The present study aimed at examining cultivated actinobacteria strains associated with endemic Lake Baikal deepwater amphipods and estimating their antibiotic activity. We isolated 42 actinobacterial strains from crustaceans belonging to Ommatogammarus albinus and Ommatogammarus flavus. To our knowledge, this is the first report describing the isolation and initial characterization of representatives of Micromonospora and Pseudonocardia genera from Baikal deepwater invertebrates. Also, as expected, representatives of the genus Streptomyces were the dominant group among the isolated species. Some correlations could be observed between the number of actinobacterial isolates, the depth of sampling and the source of the strains. Nevertheless, >70% of isolated strains demonstrated antifungal activity. The dereplication analysis of extract of one of the isolated strains resulted in annotation of several known compounds that can help to explain the observed biological activities. The characteristics of ecological niche and lifestyle of deepwater amphipods suggests that the observed associations between crustaceans and isolated actinobacteria are not random and might represent long-term symbiotic interactions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural products remain the most important source of new pharmaceutical leads, including antibiotics (Doroghazi et al. 2014). The multiple reports on emergence of pathogenic bacteria resistant to antibiotics increases the need for effective antimicrobials with new modes of action (Haste et al. 2012; Ling et al. 2015). Despite some reduction in interest in the past, actinobacteria still remain one of the most prevalent sources of new drugs. This phenomenon is first of all associated with the diverse and extraordinary creative secondary metabolism of these bacteria. Approximately 8700 of the 16,500 known antibiotics are produced by bacteria from the order Actinomycetales (Berdy 2005). The genus Streptomyces alone accounts for 80% of the natural products produced by actinobacteria (Manivasagan et al. 2013; Jensen et al. 2005).
Since the 1940s, the main source for the isolation of antibiotics producers has been soil. The rate of discovery of novel biologically active compounds from this source has been declining since then, mostly due to re-discovery of the same compounds. Recently, a transition from the traditional to new sources, such as marine sediments (Fenical and Jensen 2006; Subramani and Aalbersberg 2012), invertebrates (Genilloud et al. 2011), caves (Maciejewska et al. 2016) has been observed. For example, new species of the genus Micromonospora were isolated from insects (Fang et al. 2015). Species of the genera Streptomyces, Nocardia, Pseudonocardia, Amycolatopsis, and Micromonospora with antimicrobial activities were isolated from termite nests (Sujada et al. 2014). The sponge-microbe associations has been a focus of many studies directed on isolation of new compounds (Taylor et al. 2007; Thomas et al. 2010). Many of them have concentrated efforts on marine animals, but several reports on freshwater sponges as a source of new actinobacterial strains and antibiotics are also published (Gernert et al. 2005; Costa et al. 2013). Actinobacteria isolated from aquatic environments are a promising source of novel biologically active compounds, such as antibiotics, enzyme inhibitors, antitumor and antiviral agents (Farnaes et al. 2014; Duncan et al. 2015). One of the unique, ancient and unstudied aquatic ecosystems in terms of actinobacterial diversity is the ecosystem of Lake Baikal and its inhabitants.
Lake Baikal, a UNESCO World Heritage Site, is located in eastern Siberia, Russia. It is one of the most ancient lakes (approximately 25–30 million years old) and the largest (by volume) in the world (Timofeyev 2016). The lake contains approximately 20% of the world’s unfrozen surface fresh water. Lake Baikal is the deepest (1642 m) lake in the world, with an average depth of 744.4 m (Kozhov 1963). The lake is inhabited by more than 2500 animal species, 80% of which are endemic (Yoshii et al. 1999; Timoshkin et al. 2001). The lake is well-oxygenated throughout the water column, and is characterized by narrow fluctuations in temperature (3.5–6 °C at depths of 30–100 m). These factors establish specific conditions for ecosystem development. Indeed, a number of endemic groups of macroinvertebrates have evolved during million years in Lake Baikal under these conditions, leading to the formation of a unique ecological system.
Amphipods are the dominant group of macroinvertebrates of Lake Baikal ecosystem (Amphipoda, Crustacea) (Timoshkin et al. 2001). Lake Baikal endemic amphipods form 45.3% of the world’s freshwater amphipod fauna. They are represented by 246 species and 78 subspecies with 100% endemicity (Takhteev 2000; Takhteev et al. 2015). They inhabit all types of ecological niches from the littoral to abyssal zones. Amphipods efficiently find and consume dead organisms and detritus. Deepwater amphipods, which are scavengers and necrophages, play a particularly important role in the self-cleaning of the lake. The life style and type of feeding of these organisms suggest constant interactions with microorganisms as competitors and possibly symbionts.
The uniqueness of biotic and abiotic factors makes Lake Baikal and its inhabitants a promising source of new actinobacterial species producing novel biologically active compounds with chemically and pharmaceutically interesting properties. The present study aimed to examine a cultivated actinobacterial population associated with Lake Baikal endemic deepwater amphipods for their antibiotic activity accompanied with the dereplication of secondary metabolites produced by some of them.
Materials and methods
Sampling and location
Two hundred specimens of two endemic deepwater amphipod species of the genus Ommatogammarus (O. albinus and O. flavus) were collected at different depths from the southern part of Lake Baikal near the Bolshie Koty settlement (51.9053°N, 105.0753°E) in winter 2015. No specific permissions were required for these locations and activities. The studied amphipod species are not endangered or protected species.
Samples were taken at depths of 80, 100 and 200 m. Each sample included 3–5 amphipod specimens. Five samples were collected from each depth. The amphipods were captured in traps baited with sterilized putrescent fish. Immediately after the lifting the traps, the amphipods were rinsed with 70% ethanol following by sterile water (three times) to remove transient bacteria and then homogenized in 20% sterile glycerol at an approximate ratio of 1:10. The obtained samples were stored at −20 °C until plating on the solid nutrient media. Water samples collected from the surface and the suspension of sterile putrescent fish were used as negative controls.
Isolation of actinobacteria
The actinobacterial strains were isolated by plating obtained homogenate on solid nutrient media. Six media were used: MS (soy flour 20 g/L, d-mannitol 20 g/L, agar 20 g/L, pH 7.2); ISP2 (yeast extract 4 g/L, malt extract 10 g/L, dextrose 4 g/L, agar 20 g/L, pH 7.2); starch-ammonia agar SAA ((NH4)2SO4) 2 g/L, K2HPO4 1 g/L, MgSO4 1 g/L, NaCl 1 g/L, CaCO3 3 g/L, starch 10 g/L, agar 20 g/L); Gauze’s synthetic agar (starch 20 g/L, KNO3 1 g/L, NaCl 0.5 g/L, MgSO4·7 H2O 0.5 g/L, K2HPO4 0.5 g/L, FeSO4·7 H2O 0.01 g/L, agar 15 g/L, pH 7.4); Waksman media (glycerol 3 g/L, K2HPO4 1 g/L, NaNO3 2 g/L, MgSO4·7 H2O 0.5 g/L, KCl 0.5 g/L, FeSO4·7 H2O 0.01 g/L, pH 7.0); and Czapek media (NaNO3 2 g/L, starch 30 g/L, MgSO4 7H2O 0.5 g/L, KCl 0.5 g/L, FeSO4·7H2O 0.01 g/L, K2HPO4 1 g/L, agar 20 g/L, pH 7.2). Media were supplemented with nystatin (50 μg/mL) and phosphomycin (100 μg/mL) (Kieser et al. 2000). Aliquots of collected samples were preheated for 5 min at 50 °C to activate spores germination and inactivate the vegetative cells of other bacteria.
Collected samples were diluted in a 1% sterile saline solution at 1:10, 1:100 and 1:1000 ratios. 150 μL of each dilution was plated on mentioned media in three replicates. The plates were incubated for 30 days at 28 °C and assessed every day for actinobacterial colony appearance. Actinobacteria-like strains were identified based on colony morphology (Kieser et al. 2000). The colonies were transferred from the primary plates to fresh MS plates. Pure cultures were obtained for all colonies identified as actinobacteria on the primary plates. Several isolated strains were deposited in the Russian Collection of Agricultural Microorganisms (RCAM), St. Petersburg, Russia.
16S rRNA gene sequencing and analysis
Strains were cultured in tubes for 3 days in 10 mL of TSB medium at 28 °C at a shaking rate of 180 rpm. Total DNA was isolated by the salting out procedure as described in (Kieser et al. 2000). Amplification of the 16S rRNA gene was carried out with the following primers: 8F (AGAGTTTGATYMTGGCTCAG), 1510R (TACGGYTACCTTGTTACGACTT), ACT235F (CGCGGCCTATCAGCTTGTTG) and ACT878R (CCGTACTCCCCAGGCGGGG). PCR was performed using a ScreenMix 5X PCR kit (Kat. PK041L, Evrogen, Russia). PCR was performed in a TGradient Thermocycler (Biometra, Germany) in a volume of 25 μL. The parameters of the PCR were as follows: initial denaturation at 95 °C for 5 min, followed by 25 cycles of 95 °C for 40 s, 50–55 °C for 25 s, and 72 °C for 110 s, and final elongation at 72 °C for 5 min.
The amplified PCR products were visualized in 1% agarose gel, purified with Cleanup Standard PCR purification kit (Kat. BC022, Evrogen, Russia) and sequenced with primers used for amplification by SYNTOL (Moscow, Russia). The forward and reverse sequences were assembled with BioEdit software version 7.2.5 (Tom Hall) (Hall 1999).
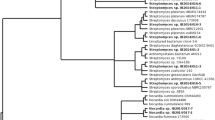
The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei 1987). The optimal tree with the sum of branch length = 0.47231526 is shown (Fig. 1). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches (Felsenstein 1985)—except for values below 50%, which were hidden for readability. The evolutionary distances were computed using the Tamura–Nei method (Tamura and Nei 1993) and are in the units of the number of base substitutions per site. The analysis involved 40 nucleotide sequences. All positions containing gaps and missing data were eliminated. There were a total of 423 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016). The sequences were deposited in the GenBank and their ID presented in Table 1 of the results section.
Cultivation and extraction of secondary metabolites
Strains were cultivated in 10 mL of TSB medium for 2 days at 28 °C at a shaking rate of 180 rpm. 2 mL of each pre-culture was used to inoculate 50 mL of production medium in a 500-mL Erlenmeyer flask with baffles. Three different liquid media were selected for metabolite production: NL19 (soy flour—20 g/L, d-mannitol—20 g/L); SG (dextrose—20 g/L, soy peptone—10 g/L, CaCO3—2 g/L, CoCl2—0.001 g/L); and ISP2 (yeast extract—4 g/L, malt extract—10 g/L, dextrose—4 g/L, pH 7.2) (Kieser et al. 2000) The strains were cultivated at 28 °C for 4 days at a shaking rate of 180 rpm. The cultural liquid and biomass were separated by centrifugation at 3000×g for 5 min. Metabolites from the liquid were extracted with an equal volume of ethyl acetate (Sigma, St. Louis, USA) (Sarker and Nahar 2012). The compounds from the biomass were extracted with 10 mL of acetone:methanol mixture (1:1). The extracts were evaporated on an IKA RV–8 rotovap (IKA, Germany) at 40 °C and dissolved in 500 µL of methanol (Sigma, St. Louis, USA).
Liquid chromatography–mass spectrometry (LC–MS) and dereplication analysis
The strain Streptomyces sp. IB 2015P113-12 was cultivated in 1.0 L of NL19 medium. The secondary metabolites from the culture liquid and biomass were extracted using ethyl acetate and the acetone-methanol mixture as described above (Sarker and Nahar 2012). The extract was dissolved in methanol and analyzed on an ultra-high resolution QTOF maXis II mass spectrometry system (Bruker, Billerica, USA) and LTQ XL Orbitrap (Thermo Fisher Scientific, USA). The sample was separated on an Ultimate 3000 HPLC system (Dionex, Sunnyvale, USA) with diode array detector using a C18 column (ACQUITY UPLC BEH C18 Column, 130Å, 1.7 µm, 2.1 mm × 100 mm) and a linear gradient of acetonitrile from 5 to 95% against a 0.1% ammonium formate solution in water at a flow rate 0.6 mL/min for 20 min. Mass detection was performed in positive mode, with the detection range set to 160–2500 Da. Data were collected and analyzed using Bruker Compass Data Analysis software, version 4.1 (Bruker, Billerica, USA) and Thermo Xcalibur, version 3.0.63 (Thermo Fisher Scientific, USA). Dereplication was performed using the Dictionary of Natural Products (DNP) database, version 6.1 (CRC Press, Boca Raton, USA) with the following search parameters: accurate molecular mass, absorption spectra and biological source. Compounds were considered to be similar when the difference in accurate mass was less than 0.005 Da, mass accuracy was less than 5 ppm, MS2 and the UV absorption spectrum were identical (Whittle and Willett 2003).
Antibiotic assay of extracts from isolated strains
The disk-diffusion method was used to determine the antimicrobial activity of obtained extracts. Seven model strains of microorganisms were chosen as the test cultures: Bacillus subtilis ATCC 6633, Pseudomonas putida KT2440, Staphylococcus carnosus ATCC 51365, Escherichia coli ATCC25922, E. coli K12, Saccharomyces cerevisiae BY4742 and Candida albicans ATCC 90027. Overnight cultures were used to inoculate fresh plates with LB (for bacteria) or YPD (for fungi) media. The paper disks (5 mm in diameter) were loaded by 25 µL of crude extracts derived either from liquid culture or biomass and dried at room temperature. The dry disks were placed on media with plated test cultures. The extracts were tested against E. coli K12 only in the case of activity against E. coli ATCC25922. Plates with disks were incubated at 37 °C (B. subtilis, E. coli, St. carnosus, P. putida) or 28 °C (S. cerevisiae, C. albicans) (Grainger et al. 2001). The zones of inhibition were manually measured with ±1 mm accuracy. Disks loaded with methanol were used as negative controls.
Bacterial test cultures were obtained from the Leibniz-Institute DSMZ-German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany).
Results
Isolation and phylogenetic characterization of actinobacteria from deepwater endemic amphipods of Lake Baikal
In this study, 42 individual actinobacteria-like strains were isolated (Table 1; Fig. S1). The isolates were selected by their morphological features. In total, 13 cultivated strains of actinobacteria were obtained from O. albinus. Eleven of them were obtained from the amphipods samples collected at 80 m depth. One strain was isolated from the amphipods inhabiting the 100 m, and one from 200 m depths. Twenty-nine strains were isolated from O. flavus specimens. Twenty-four of them were from the amphipods inhabiting the 200 m depth. From the representatives of O. flavus collected at the depths of 80 and 100 m, four and one strains were isolated, respectively.
The 16S rRNA gene sequence-based phylogenetic analysis indicated that 40 out of the 42 actinobacteria strains belong to the genus Streptomyces. Two other strains were identified as Micromonospora and Pseudonocardia (Table 1). Both strains were isolated from O. albinus sampled at 80 m. The 16S rDNA sequences from isolated bacteria formed a tight clade with 16S rDNA of several representatives of the respective genera (Fig. 1).
Most of the strains including the Micromonospora sp. IB 2015P61-1 were selected on MS medium. Only one strain, identified as Pseudonocardia sp. IB2015P62-1HS was obtained on starch-ammonia agar (SAA). The frequencies of strain isolation and the strains distribution in the collected samples of amphipods of the genus Ommatogammarus are shown in Table 2. In addition, four of the obtained strains were isolated from O. albinus inhabiting the 80 m depth using thermal pretreatment, including the Pseudonocardia sp. (marked as “HS”).
Analysis of the biological activities of isolated actinobacterial strains
Among 42 isolated strains, extracts of only four (Streptomyces sp. IB2015P61-1, Streptomyces sp. IB2015P113-15, Streptomyces sp. IB2015P114-1, and Streptomyces sp. IB2015P122-1) did not demonstrate antimicrobial or antifungal activity against at least one of the test organisms used (Table 3, Tables S1–S6). All other strains were found to produce compounds inhibiting growth of bacteria and/or fungi. The antimicrobial activities of the isolated strains are presented in Table 3 and Tables S1–S6. In total, the growth of Gram-positive bacteria was inhibited by extracts of 22 strains out of 42. The growth of S. carnosus and B. subtilis was inhibited by extracts obtained from five strains grown in NL19 medium, two strains incubated in SG medium and 17 ISP2 cultures (Table 3, Tables S1–S6).
The growth of Gram-negative bacteria was affected by extracts of 12 strains. P. putida and E. coli were inhibited by four isolates cultivated in NL19 medium, 1 strain grown in SG medium and eight strains cultivated in ISP2 medium. Extracts of the culture liquid of the strains Streptomyces sp. IB2015P113-12 and Streptomyces sp. IB 2015P113-2 grown in SG and NL19 media were also able to inhibit the growth E. coli ATCC25922 (tolC mutant) and E. coli K12 (Table 3, Tables S1–S6).
Thirty stains demonstrated antifungal activity. Sixteen of them were able to inhibit growth of both S. cerevisiae and C. albicans. Twelve strains demonstrated activity against S. cerevisiae only. Two strains, Streptomyces sp. IB 2015P113-13 and Streptomyces sp. IB 2015P61-3, were able to inhibit the growth of C. albicans but not S. cerevisiae. In total, 18 strains grown in NL19 medium, 25 in ISP2, and 17 in SG were found to produce fungicides.
In summary, the ISP2 medium was found to be the most efficient for production of biologically active compounds by the isolated actinobacteria, followed by NL19 and SG media. Also, it should be mentioned that activity was not always detected for both culture liquid and biomass extracts. In some cases, only one type of extract demonstrated activity against a particular test culture. For example, only culture liquid extracts of the Streptomyces sp. IB 2015P113-12 grown on NL19 or SG media were active against E. coli strains. On the other hand, the same strain accumulates compounds active against S. cerevisiae only in biomass when grown in ISP2 medium.
Dereplication of the secondary metabolites produced by Streptomyces sp. IB 2015P113-12
Strain Streptomyces sp. IB 2015P113-12 showed inhibitory activity against both bacterial and fungal test cultures. To gain deeper insights into the nature of compounds possibly contributing to observed phenomenon we conducted a dereplication analysis of the secondary metabolites produced by this strain. High-resolution mass spectrometry methods enable the general analysis of secondary metabolites and facilitate the annotation of individual compounds using information from pre-existing databases. A total of 43 major compounds were detected in the cultural liquid and biomass extracts of the Streptomyces sp. IB 2015P113-12 strain cultivated in NL19 medium (Fig. S2). Thirty-seven and twenty-nine compounds (peaks) were found in the biomass and cultural liquid extracts, respectively. Twenty-four of them were present in both extracts.
Twenty-three compounds were preliminarily annotated based on exact mass, source, and absorption spectrum parameters search in the Dictionary of Natural Products (DNP) database (Whittle and Willett 2003). Another 20 did not give positive hits in the DNP and could not be annotated based on the available mass-spec data and might represent new findings. Results of dereplication analysis with a list of the detected adducts and possible database hits are shown in Figs. S2–S5 and Table 4.
The following compounds could be annotated in both culture liquid and biomass extracts of Streptomyces sp. IB2015P113-12: N6-Deoxy-nocardimine (terragine E) (m/z 585.312 [M + H]+, CRC code—MCG61; 4.6 ppm) and three derivatives of bafilomycin - 2-O-desmethyl-Leucanicidin (m/z 769.4738 [M + H]+, CRC code—CJW04; 1.6 ppm), Leucanicidin (m/z 783.4859 [M + H]+, CRC code—DON49; 4.5 ppm) and Antibiotic NK 155141 (m/z 797.5005 [M + H]+, CRC code—CKN32; 5.8 ppm). Terragine E, that seems to act as a siderophore, originally was obtained by expression of metagenomics DNA in Streptomyces lividans (Wang et al. 2000). Finding of terragine E in the producing strain isolated from a deepwater endemic organism confirms the specificity of Lake Baikal ecosystem. Bafilomycins are macrolides, produced by Streptomyces griseus and act as inhibitors of membrane ATPases. These compounds have antibacterial, antifungal, antineoplastic and immunosuppressive activities (Bowman et al. 1988). In addition, another polyketide antibiotic BE 67251 (m/z 454.2957 [M + H]+, CRC code—KFM90, 0.7 ppm.) was found in the biomass extract of Streptomyces sp. IB2015P113-12 (Wu et al. 2009; Conti et al. 2016) (Fig. S2; Table 4).
In the crude extract of the Streptomyces sp. IB2015P113-12 strain several ionophore antibiotics of the macrotetrolide family could be annotated. We were able to identify nonactin (m/z 737.4498 [M + H]+, CRC-code HDN12), monactin (751.4655 m/z, CRC code CKH81) and dinactin/isodinactin (m/z 765.4819 [M + H]+, CRC code CKH81). These compounds (Figs. S2–S4) are known for their broad spectrum antibacterial activity, as well as for insecticidal, anthelmintic, larvicidal and coccidiostatic activities (Zhan and Zheng 2016). In addition, several less characterized representatives of the macrotetrolide group can be annotated in the extract of the strain, including macrotetrolide G (m/z 779.5028 [M + H]+, CRC code CKW16), macrotetrolide B (m/z 821.5379 [M + H]+, CRC code CKW13), macrotetrolide C (m/z 807.5231 [M + H]+, CRC code CKW14), macrotetrolide D (m/z 793.5123 [M + H]+, CRC code CKW15) (Figs. S2, S5). These compounds are derivatives of nactins with partially defined structures and possess antibacterial activity against Gram-positive bacteria (Jizba et al. 1991; Botti et al. 2010). We also managed to identify several precursors of macrotetrolides: homononactic acid, nonactyl nonactoate, homononactyl nonactoate (Table 4; Fig. S2). Based on these findings, we can assume that the activity of Streptomyces sp. IB2015P113-12 against Gram-positive bacteria can be explained at least partially by the accumulation of macrotetrolide antibiotics. Also, nonactins could be responsible for the activity against Gram-negative bacteria and fungi (Becerril Espinosa et al. 2012).
Discussion
Previous studies have demonstrated a high abundance of actinobacteria in Lake Baikal water, sediments, and sponge microbial communities (Kaluzhnaya et al. 2012). These observations suggested that the lake endemic animals can be also a source of novel actinobacterial species. Herein, we report isolation and initial characterization of 42 strains of actinobacteria from Lake Baikal endemic deepwater amphipod species O. albinus and O. flavus. Among them, 40 were identified as representatives of the genus Streptomyces and two belongs to the genera Micromonospora and Pseudonocardia. This finding correlates with the previous observations of phylogenetic diversity of microbial communities of Lake Baikal water and sediments (Terkina et al. 2002). It is generally accepted that the minor genera of actinobacteria are potentially more interesting in terms of secondary metabolites novelty. Thus, the relatively high frequency of isolation of strains belonging to Micromonospora and Pseudonocardia (appr. 5% of all isolated specimens) from the sources used is promising and will further stimulate the research in this direction.
The materials obtained in this study are summarized in Table 5. Some correlations could be observed between the number of actinobacterial isolates, the depth of sampling and the species of amphipod used as a source. For example, the number of actinobacteria decreased in the case of O. albinus along the depth gradient from 80 (n = 11) to 200 m (n = 1). In the case of O. flavus the opposite trend was observed: the number of actinobacteria strains increased with the depth. A deeper understanding of ecology of the studied amphipods can help to explain the observed phenomena. Both species are known for their seasonal migration activities (from depth of 2.5–1313 m in the case of O. flavus and from depth of 100–1600 m in the case of O. albinus) (Timoshkin et al. 2001). These migrations are caused by factors related to feeding and probably can influence the microbial associations of amphipods depending on the substrates exploited.
We also found that a large proportion (>70%) of strains isolated from amphipods have antifungal activity. This makes us to believe that these associations are not random and could represent symbiotic interactions between bacteria and amphipods. The amphipods consume the organic matter and detritus which are naturally inhabited by fungi as decomposers (Su et al. 2015). Fungi enter the gastrointestinal tract of amphipods together with food and may have undesirable effects on the metabolism and physiology. Actinobacterial produced compounds may therefore be involved in the fungal resistance of amphipods. It is known that fungi are one of the main causes of infections in aquatic crustaceans (Armstrong et al. 1976; Hatai 2012). In some cases, the crustaceans are remarkably resistant to fungal infection. Gil-Turnes et al. described the resistance of embryos of the shrimp Palaemon macrodactylus to the action of the parasitic fungus Lagenidium callinestes (Gil-Turnes et al. 1989). The cause of the tolerance was the bacteria Alteromonas sp. isolated from the surfaces of the embryos. It produces the highly effective antifungal compound 2,3-indolinedione or tribulin. Similar studies led to the discovery of another antifungal metabolite, 4-hydroxyphenethyl alcohol or tyrosol, isolated from the embryos of the American lobster Homarus americanus (Gil-Turnes and Fenical 1992). Both compounds have a narrow activity against L. callinestes, supporting the idea of the adaptive nature of bacteria-crustaceans’ interaction.
Among the animals of Lake Baikal, the microbial communities inhabiting sponges are the most extensively studied. Actinobacteria of the genera Streptomyces and Micromonospora were found to be permanent components of microbial communities of sponges belonging to the genera Swartschewskia, Baicalospongia, and Lubomirskia (Parfenova et al. 2008; Kaluzhnaya et al. 2011). However, other ecological niches and endemic animals of the lake are less studied. We attempted for the first time to explore the cultivable actinobacterial populations from deepwater amphipods of Lake Baikal. These macroinvertebrates are a key element of the food chains of the Baikal ecosystem. Due to their lifestyle, they are facing a number of challenges, including permanent competition with fungi and bacteria. However, little is known about the protection mechanisms that amphipods are using to fight these competitors and potential pathogens. A significant role in this could be played by the associations with actinobacteria producing biologically active metabolites. Also, this is the first report of isolation of actinobacteria belonging to the genera Micromonospora and Pseudonocardia from Lake Baikal amphipods. In our previous study, ten strains of the genera Aeromicrobium, Nocardia, and Streptomyces were isolated from the amphipod species Brandtia sp. (Axenov-Gribanov et al. 2015). One strain identified as Nocardia sp. was obtained from the amphipod Pallasea cancellus.
To conclude, in the present study we isolated 42 actinobacteria strains from Lake Baikal endemic deepwater amphipods O. albinus and O. flavus. The high frequency of isolation of strains producing biologically active and possibly new compounds supports the idea that actinobacteria from unique and extreme ecosystems might be a prominent source of novel metabolites for pharmaceutical and biotechnological needs. This is especially relevant in the light of global problems caused by development and spread of antibiotic resistance among pathogens (Stadler and Dersch 2016).
References
Armstrong DA, Buchanan DV, Caldwell RS (1976) A mycosis caused by Lagenidium sp. in laboratory-reared larvae of the Dungeness crab, Cancer magister, and possible chemical treatments. J Invertebr Pathol 28:329–336. doi:10.1016/0022-2011(76)90007-0
Axenov-Gribanov D, Rebets Y, Tokovenko B et al (2015) The isolation and characterization of actinobacteria from dominant benthic macroinvertebrates endemic to Lake Baikal. Folia Microbiol. doi:10.1007/s12223-015-0421-z
Barazi HO, Zhou L, Templeton NS et al (2002) Identification of heat shock protein 60 as a molecular mediator of α3β1 integrin activation. Cancer Res 62(5):1541–1548
Becerril Espinosa A, Guerra-Rivas G, Ayala-Sánchez NE, Soria Mercado IE (2012) Antitumor activity of actinobacteria isolated in marine sediment from Todos Santos Bay, Baja California, Mexico. Rev Biol Mar y Oceanogr 47(2):317–325. ISSN-e 0717-3326
Berdy J (2005) Bioactive microbial metabolites. J Antibiot 58:1–26. doi:10.1038/ja.2005.1
Botti P, Tchertchian S, Theurillat D (2010) Orthoester derivatives of crown ethers as carriers for pharmaceutical and diagnostic compositions
Bowman EJ, Siebers A, Altendorf K (1988) Bafilomycins: a class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc Natl Acad Sci USA 85:7972–7976
Conti R, Chagas FO, Caraballo-Rodriguez AM et al (2016) Endophytic actinobacteria from the Brazilian medicinal plant Lychnophora ericoides and the biological potential of their secondary metabolites. Chem Biodivers 13:727–736. doi:10.1002/cbdv.201500225
Costa R, Keller-Costa T, Gomes NCM et al (2013) Evidence for selective bacterial community structuring in the freshwater sponge Ephydatia fluviatilis. Microb Ecol 65:232–244. doi:10.1007/s00248-012-0102-2
Doroghazi J, Albright J, Goering A et al (2014) A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol 10:963–968
Duncan KR, Crüsemann M, Lechner A et al (2015) Molecular networking and pattern-based genome mining improves discovery of biosynthetic gene clusters and their products from Salinispora species. Chem Biol 22:460–471. doi:10.1016/j.chembiol.2015.03.010
Fang B, Liu C, Guan X et al (2015) Two new species of the genus Micromonospora: Micromonospora palomenae sp. nov. and Micromonospora. Antonie Van Leeuwenhoek 108:141–150. doi:10.1007/s10482-015-0472-9
Farnaes L, Coufal NG, Kauffman CA et al (2014) Napyradiomycin derivatives, produced by a marine-derived actinomycete, illustrate cytotoxicity by induction of apoptosis. J Nat Prod 77:15–21. doi:10.1021/np400466j
Felsenstein J (1985) Phylogenies and the comparative method. Am Nat 125:1–15. doi:10.1086/284325
Fenical W, Jensen PR (2006) Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol 2:666–673. doi:10.1038/nchembio841
Genilloud O, González I, Salazar O et al (2011) Current approaches to exploit actinomycetes as a source of novel natural products. J Ind Microbiol Biotechnol 38:375–389. doi:10.1007/s10295-010-0882-7
Gernert C, Glöckner FO, Krohne G, Hentschel U (2005) Microbial diversity of the freshwater sponge Spongilla lacustris. Microb Ecol 50:206–212. doi:10.1007/s00248-004-0172-x
Gil-Turnes MS, Fenical W (1992) Embryos of Homarus americanus are protected by epibiotic bacteria. Biol Bull 182:105–108. doi:10.2307/1542184
Gil-Turnes MS, Hay ME, Fenical W (1989) Symbiotic marine bacteria chemically defend crustacean embryos from a pathogenic fungus. Science 246:116–118
Grainger J, Hurst J, Burdass D (2001) Basic practical microbiology Compiled by. Soc Gen Microbiol 1–30
Hall T (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Haste NM, Thienphrapa W, Tran DN et al (2012) Activity of the thiopeptide antibiotic nosiheptide against contemporary strains of methicillin-resistant Staphylococcus aureus. J Antibiot 65:593–598. doi:10.1038/ja.2012.77
Hatai K (2012) Diseases of fish and shellfish caused by marine fungi. Springer, Berlin, pp 15–52
Jensen PR, Mincer TJ, Williams PG, Fenical W (2005) Marine actinomycete diversity and natural product discovery. Antonie Van Leeuwenhoek 87:43–48. doi:10.1007/s10482-004-6540-1
Jizba J, Sedmera P, Zima J et al (1991) Macrotetrolide antibiotics produced by Streptomyces globisporus. Folia Microbiol 36:437–443. doi:10.1007/BF02884062
Kaluzhnaya O, Itskovich V, McCormack G (2011) Phylogenetic diversity of bacteria associated with the endemic freshwater sponge Lubomirskia baicalensis. World J Microbiol Biotechnol 27:1955–1959. doi:10.1007/s11274-011-0654-1
Kaluzhnaya O, Krivich A, Itskovich V (2012) Diversity of 16S rRNA genes in metagenomic community of the freshwater sponge Lubomirskia baicalensis. Russ J Genet 48:855–858. doi:10.1134/S1022795412070058
Keller-Schierlein W, Gerlach H (1968) Makrotetrolide. Springer, Vienna
Kieser B, Buttner M, Charter K, Hopwood B (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kozhov M (1963) Lake Baikal and its life: Monographiae Biologicae. W. Junk, The Hague
Kumar S, Stecher G, Tamura K et al (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi:10.1093/molbev/msw054
Ling LL, Schneider T, Peoples AJ et al (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459. doi:10.1038/nature14098
Maciejewska M, Adam D, Martinet L et al (2016) A phenotypic and genotypic analysis of the antimicrobial potential of cultivable Streptomyces isolated from cave moonmilk deposits. Front Microbiol 7:1455. doi:10.3389/fmicb.2016.01455
Manivasagan P, Venkatesan J, Sivakumar K, Kim SK (2013) Marine actinobacterial metabolites: current status and future perspectives. Microbiol Res 168:311–332. doi:10.1016/j.micres.2013.02.002
Parfenova VV, Terkina IA, Kostornova TI et al (2008) Microbial community of freshwater sponges in Lake Baĭkal. Izv Akad Nauk Ser Biol 35:435–441. doi:10.1134/S1062359008040079
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sarker SD, Nahar L (2012) An introduction to natural products isolation. Methods Mol Biol 864:1–25
Smith LL (1975) An additional source of macrotetrolide antibiotics. J Antibiot 28(12):1000–1003. doi:10.7164/antibiotics.28.1000
Stadler M, Dersch P (eds) (2016) How to overcome the antibiotic crisis. Springer, Cham
Su R, Kuehn KA, Phipps SW (2015) Fungal contributions to carbon flow and nutrient cycling during decomposition of standing Typha domingensis leaves in a subtropical freshwater marsh. Freshw Biol 60:2100–2112. doi:10.1111/fwb.12635
Subramani R, Aalbersberg W (2012) Marine actinomycetes: an ongoing source of novel bioactive metabolites. Microbiol Res 167:571–580. doi:10.1016/j.micres.2012.06.005
Sujada N, Sungthong R, Lumyong S (2014) Termite nests as an abundant source of cultivable actinobacteria for biotechnological purposes. Microbes Environ 29:211–219. doi:10.1264/jsme2.ME13183
Tahteev V (2000) Essayes on the amphipods of Lake Baikal: systematics, comparative ecology, evolution. Irkutsk State University Press, Irkutsk
Takhteev VV, Berezina NA, Sidorov DA (2015) Checklist of the Amphipoda (Crustacea) from continental waters of Russia, with data on alien species. Arthropoda Sel 24:335–370
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Taylor MW, Radax R, Steger D et al (2007) Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev 71:295–347. doi:10.1128/MMBR.00040-06
Terkina IA, Drukker VV, Parfenova VV, Kostornova TY (2002) The biodiversity of actinomycetes in Lake Baikal. Microbiology 71:346–349. doi:10.1023/A:1015871115187
Thomas TRA, Kavlekar DP, Lokabharathi PA (2010) Marine drugs from sponge-microbe association—a review. Mar Drugs 8:1417–1468. doi:10.3390/md8041417
Timofeyev MA (2016) Monitoring: safeguarding the world’s largest lake. Nature. doi:10.1038/538041a
Timoshkin OA, Sitnikova TY, Rusinek OT et al (2001) Index of animal species inhabiting Lake Baikal and its catchment area, vol 1. Nauka, Novosibirsk
Wang G-Y-S, Graziani E, Waters B et al (2000) Novel natural products from soil DNA libraries in a Streptomycete host. Org Lett 2:2401–2404. doi:10.1021/ol005860z
Whittle M, Willett P (2003) Evaluation of similarity measures for searching the dictionary of natural products database. J Chem Inf Comput Sci 43:449–457
Wu YC, Wu WKK, Li Y, Yu L, Li ZJ, Wong CCM, Cho CH (2009) Inhibition of macroautophagy by bafilomycin A1 lowers proliferation and induces apoptosis in colon cancer cells. Biochem Biophys Res Commun 382(2):451–456
Yoshii K, Melnik NG, Timoshkin OA, Bondarenko NA, Anoshko PN, Yoshioka T, Wada E (1999) Stable isotope analyses of the pelagic food web in Lake Baikal. Limnol Oceanogr 44(3):502–511. doi:10.4319/lo.1999.44.3.0502
Zhan Y, Zheng S (2016) Efficient production of nonactin by Streptomyces griseus subsp. griseus. Can J Microbiol 62:711–714. doi:10.1139/cjm-2016-0248
Acknowledgements
We acknowledge the Irkutsk regional veterinary laboratory where the antibiotic assay of crude extracts against C. albicans was conducted.
Funding
This study was supported by the Ministry of education and science of Russian Federation as a part of Goszadanie projects (6.9654.2017/8.9), Russian science foundation (17-14-01063), Russian foundation for basic research (projects N 16-34-00686, 16-34-60060), Grants of Irkutsk State University for researchers and Deutscher Akademischer Austauschdienst.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants and/or animals
Statement of human rights: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Statement on the welfare of animals: This article does not contain any studies with human participants or vertebrate animals performed by any of the authors. As stated in the article, Baikalian macroinvertebrate species are not endangered or protected species. No specific permissions were required for sampling of invertebrates species.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Protasov, E.S., Axenov-Gribanov, D.V., Rebets, Y.V. et al. The diversity and antibiotic properties of actinobacteria associated with endemic deepwater amphipods of Lake Baikal. Antonie van Leeuwenhoek 110, 1593–1611 (2017). https://doi.org/10.1007/s10482-017-0910-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0910-y