Abstract
Two novel, Gram-negative, motile, rod-shaped, aerobic bacterial strains, MH17T and RD15, were isolated from the sterilized root and rhizosphere soil of rice, respectively. Phylogenetic analysis based on 16S rRNA gene sequences showed that the similarity between strains MH17T and RD15 was 100%. The isolates exhibit high sequence similarities to Rhizobium oryzae CGMCC 1.7048T (98.7%) and Rhizobium petrolearium SL-1T (97.0% and 97.1%), which supports that they belong to a novel species in the genus Rhizobium. Strains MH17T and RD15 exhibited growth at 15–45 °C, pH 5.0–11.0, 0–2.0% sodium chloride (w/v). Sequence analysis of housekeeping genes gyrB, recA, atpD, ropB, gltA showed that these two novel strains had less than 94% similarity with the known species, indicating the distinct position of MH17T and RD15 in the genus Rhizobium. The major cellular fatty acids were identified as summed feature 8 (C18:1 ω7c and/or C18:1 ω6c). Type strain MH17T had 87.5% DNA–DNA relatedness with RD15 by using the initial renaturation rate method. Based on draft genome sequences, strain MH17T showed 30.1% DNA–DNA hybridization values to R. oryzae CGMCC 1.7048T, the closely related strain, which supported that MH17T represents a novel species in the genus Rhizobium. Average nucleotide identity (ANI) between strains MH17T and RD15 were 97.8%, and strain MH17T showed 82.2% ANI value with R. oryzae CGMCC 1.7048T. The DNA G+C content was 60.4 mol% (Tm). Based on physiological, biochemical characteristic, genotypic data, strains MH17T and RD15 are concluded to represent a new species within the genus Rhizobium, for which the name Rhizobium rhizosphaerae sp. nov. is proposed. The type strain is MH17T (=ACCC 19963T = KCTC 52414T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Rhizobium was first proposed by Frank (1889) and revised by Young et al. (2001) and belongs to the bacterial phylum Proteobacteria. Up to now, it has more than 80 identified species, many of which have been isolated from the nodules on leguminous plants with the function of symbiotic nitrogen-fixing. However, there are a growing number of novel species that have been isolated from distinct environments, for example, Rhizobium alvei NR-22T isolated from a freshwater river (Sheu et al. 2015), Rhizobium capsici CC-SKC2T from the root tumor of a green bell pepper (Lin et al. 2015), Rhizobium flavum YW14T from soil (Gu et al. 2014), Rhizobium tarimense PL-41T isolated from Populus euphratica forest soil (Turdahon et al. 2013), Rhizobium pseudoryzae ACCC 10380T, Rhizobium rhizoryzae ACCC 05916T, Rhizobium oryzicola ACCC 05753T from the roots of rice (Zhang et al. 2011, 2014, 2015). The genus Rhizobium are usually characterized as rod-shaped, aerobic, Gram-negative bacteria, with a DNA G+C content between 57 and 66 mol% (Tighe et al. 2000; Young et al. 2001) and C18:1 ω7c as the major cellular fatty acids. In this study, we report two novel strains MH17T and RD15 of the genus Rhizobium according to their phenotypic and genotypic characteristics.
Materials and methods
Strains and culture conditions
Endophytic strain MH17T was isolated from surface sterilized rice root collected from Beijing, and RD15 was from the rhizosphere of rice sampled from Hunan Province. Surface disinfection of rice roots was performed according to the methods of Zhang et al. (2015), and the rhizosphere soil collection was as described by Zhang et al. (2011). Both of bacteria grew on yeast extract mannitol agar medium (Vincent. 1970) at 30 °C. Strains were preserved as glycerol suspension (20%, v/v) at −80 °C and at −4 °C in freeze-dried ampoules for further characterization.
To study the chemotaxonomic and molecular characteristics, biomass was collected from YMA medium at 30 °C for 2 days. The reference strains Rhizobium oryzae CGMCC 1.7048T, R. petrolearium SL-1T were obtained from China General Microbiological Culture Collection Center (CGMCC; China) and Agricultural Culture Collection of China (ACCC; China), respectively. The two reference strains were cultured for comparative analysis under the same conditions as strains MH17T and RD15.
Morphological, physiological and biochemical characteristics
Cell morphology was examined using light microscope (CX21; Olympus). Gram-staining was carried out using the Gram-Stain Kit (HiMedia). Growth features were tested in different temperatures at 4, 15, 25, 30, 37, 40, 45, 50 °C and pH 3.0–12.0 (at 1.0 unit intervals) on YMA medium. Salt tolerance was tested on YMA with NaCl concentration of 0, 1.0, 3.0, 5.0, 7.0, 10.0% (w/v). Oxidase and catalase activities were determined by using 1%(w/v) tetramethyl-p-phenylenediamine and 3% (v/v) H2O2, respectively. Motility was observed by semisolid culture-medium (0.4% agar added). Mean generation time was estimated based on the growth curve from YMA medium. The basic biochemical characteristics were investigated on Biolog GN2 Microplates (Hay-ward, CA) and API-20NE test strips (bioMérieux, France).
Molecular study
Genomic DNA was extracted from pure cultures using TIANamp Bacteria DNA Kit (Tiangen) based on the manufacture’s protocol. The 16S rRNA gene was amplified using the universal primers 27F and 1492R according to Lane et al. (1991). The housekeeping genes gyrB, recA, atpD, ropB, gltA were amplified using the methods of Martens et al. (2007, 2008). The 16S rRNA gene sequences similarity and multiple sequences alignment were analysed using EzTaxon-e Service (Kim et al. 2012) and CLUSTALW (Thompson et al. 1994), respectively. Similarities of housekeeping genes were performed by NCBI BLAST program (http://www.ncbi.nlm.nih.gov) and multiple sequence alignment was carried out by CLUSTALW. Phylogenetic trees were constructed using MEGA 7.0 software (Kumar et al. 2016) with neighbor-joining (Saitou and Nei 1987), minimum evolution (Rzhetsky and Nei 1992) and maximum likelihood (Felsenstein 1981) methods.
To determine the genomic DNA G+C content, Beckman DU 800 Spectrophotometer (Beckman collulter, brea, CA, USA) was used according to the thermal denaturation methods (Marmur and Doty 1962). Escherichia coli K12 was used as the reference strain. The initial renaturation rate method (De ley 1970) was used to determine DNA relatedness.
Draft genome sequences were determined using Hiseq Technology by Zeta Biosciences (Shanghai) Co., Ltd. The genomic sequences were assembled with SOAP denovo version 2.04 software and annotated with Prokaryotic Genome Annotation Pipeline (PGAP) by NCBI. The genome sequences were submitted to NCBI. The DDH (DNA-DNA hybridization) estimates were using GGDC (Genome-to-Genome Distance Calculator; http://ggdc.dsmz.de) with the BLAST+ (recommended) method (Meier-Kolthoff et al. 2012). The ANI values were estimated by using ANI Calculator in the EZBioCloud (http://www.ezbiocloud.net).
Fatty acid analysis
For cellular fatty acids analysis, strains MH17T, RD15 and R. oryzae CGMCC 1.7048T, R. petrolearium SL-1T were grown on YMA for 2 days at 28 °C. Cultures were harvested and fatty acid methyl esters were prepared and separated using methods described by Sasser (1990) and were identified with the MIDI Sherlock Microbial Identification System.
Results and discussion
Morphological, physiological and biochemical characteristics
Strains MH17T and RD15 were observed to be rod-shaped aerobic, motile, Gram-stain negative bacteria. Colonies were circular and pearl white on YMA medium at 30 °C. Growth occurred at 15–45 °C (optimum 30 °C) and the pH range for growth was 5.0–11.0. The tolerance of NaCl was up to 2.0% (w/v). Strains MH17T and RD15 were observed to be catalase and oxidase positive. Mean generation time of MH17T was 2.0 hours in YMA medium. Other physiological and biochemical characteristics of the novel isolates and reference strains are given in Table 1. Strains MH17T and RD15 could be distinguished from R. oryzae CGMCC 1.7048T in utilization of dextrin, i-erythritol, lactulose, α-keto glutaric acid. In contrast to R. petrolearium SL-1T, both of them could utilize adonitol, i-erythritol, lactulose, d-raffinose, succinic acid as carbon source.
Molecular results
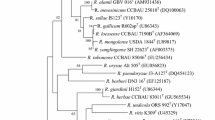
The almost complete 16S rRNA (1444 bp) gene sequences of strains MH17T and RD15 were determined and subjected to phylogenetic analysis. Similarity search in EzTaxon-e revealed that strains MH17T and RD15 are closely related to members of the genus Rhizobium, showing high 16S rRNA gene sequence similarity to R. oryzae CGMCC 1.7048T (98.7%), followed by R. petrolearium SL-1T (97.0% and 97.1%). In the phylogenetic tree based on the neighbor-joining method, and also recovered using maximum-likelihood and maximum-evolution methods, the two strains were clustered into a novel branch within the genus Rhizobium (Fig. 1).
Neighbor-joining method reconstructed from a comparative analysis of 16S rRNA gene sequences, showing the relationships between strains RD15T and MH17 and closely related species. Asterisk indicates branches that are also recovered using the ML and ME methods. The significance of each branch is indicated by a bootstrap value (%) calculated for 1000 subsets. Genbank accession numbers are given in parentheses. Bar, 0.005 substitutions per nucleotide position
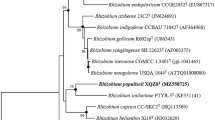
To further explore these phylogenetic relationships, five housekeeping genes, gyrB (684 bp), recA (570 bp), atpD (521 bp), ropB (935 bp), gltA (674 bp), were also sequenced. Sequence similarities of these housekeeping genes between strains MH17T and RD15 are 98–99%. Strain MH17T shared high gyrB gene sequence similarity with R. oryzae CGMCC 1.7048T (88%) and no more than 83% with other species of the genus Rhizobium. The recA and ropB gene sequences of strain MH17T showed a high degree of similarity to that of R. oryzae CGMCC 1.7048T (93%) and less than 89% sequence similarity to other members of the genus Rhizobium. The atpD gene sequence similarity between strain MH17T and R. oryzae CGMCC 1.7048T was 94%, and no more than 91% between MH17T and other species of the genus Rhizobium. Strain MH17T showed no more than 92% gltA gene sequence similarity with members of the genus Rhizobium. In addition, the phylogenetic tree reconstructed based on the concatenated housekeeping genes demonstrated that strains MH17T and RD15 formed a phylogenetic cluster with R. oryzae CGMCC 1.7048T (Fig. 2), indicating that the novel strains belonged to the genus of Rhizobium but represent a new species.
Neighbor-joining phylogenetic tree based on the comparision of the concatenated housekeeping genes (gyrB, recA, atpD, ropB, gltA) using MEGA 7.0, showing the phylogenetic relationship between novel strains and recognized species of the Rhizobium. Asterisk means branches that are also recovered using the ML and ME methods. Bootstrap values (%) based on 1000 replicates, >50%. Bar, 2% sequence divergence
The DNA G+C content of strain MH17T was determined to be 60.4 mol% (Tm), which was within the range values reported for members of the genus Rhizobium (57–66 mol%; Jordan. 1984). The DNA-DNA relatedness between strains MH17T and RD15 was 87.5% by using the initial renaturation rate method, which was higher than the species threshold of 70%. The genome sequences were submitted to NCBI and the BioProject IDs for MH17T and R. oryzae CGMCC 1.7048T are PRJNA344723 and PRJNA344729. The level of DNA-DNA relatedness between strains MH17T and R. oryzae CGMCC 1.7048T, a closely related strain, was 30.1%, according to the draft genome sequences. The ANI value between strains MH17T and RD15 was 97.8%, and strain MH17T showed 82.2% ANI value with R. oryzae CGMCC 1.7048T. These values indicate that strain MH17T represents a novel species in Rhizobium genus.
Fatty acid analysis
The major cellular fatty acids of strain MH17T were summed feature 8 (C18:1 ω7c and/or C18:1 ω6c). The detailed fatty acid compositions of strains MH17T, RD15 and R. oryzae CGMCC 1.7048T, R. petrolearium SL-1T are shown in Table 2. Strains MH17T and RD15 contained the same fatty acids in similar proportions but in comparison with the close relative R. oryzae CGMCC 1.7048T, the two novel strains differed in presence of C18:1 ω9c. Strain MH17T could also be distinguished from R. petrolearium SL-1T by containing C18:1 ω7c 11-methyl, summed feature 3 (C16:1 ω7c and/or C16:1 ω6c) and lacking C19:0 cyclo ω8c.
Taxonomic conclusion
By means of biochemical, physiological and morphological characteristics, DNA-DNA hybridization and genotypic comparison of 16S rRNA and housekeeping genes, strains MH17T and RD15 are proposed to represent a novel species within the genus Rhizobium for which the name Rhizobium rhizosphaerae is proposed.
Description of Rhizobium rhizosphaerae sp. nov.
Rhizobium rhizosphaerae (rhi.zo.sphae’rae. Gr. n. rhiza, a root; L. n. sphaera, ball, sphere; N.L. fem. n. rhizosphaera, the rhizosphere; N.L. gen. n. rhizosphaerae, of the rhizosphere).
Cells are Gram-negative, motile rods and aerobic. Colonies are circular, pearl white, smooth and convex after 2 days incubation on YMA medium at 30 °C. Growth occurs from 15 to 45 °C and pH 5.0–11.0. NaCl is tolerated up to a concentration of 2% (w/v). Catalase and oxidase activities are positive. Reduces potassium nitrate. Hydrolyses esculin and gelatin. Assimilates arabinose, mannose, mannitol, N-acetyl-glucosamine, maltose, gluconate, decanoic acid, oxalic acid, malic acid, citric acid and phenylacetic acid. The following substrates are also positive: dextrin, N-acetyl-d-glucosamine, adonitol, l-arabinose, d-arabitol, cellobiose, i-erythritol, d-fructose, l-fucose, d-galactose, gentiobiose, a-d-lactose, α-d-lactose, lactulose, maltose, d-mannitol, d-mannose, d-melibiose, β-methyl-d-glucoside, d-raffinose, l-rhamnose, d-sorbitol, sucrose, d-trehalose, turanose, xylitol, methyl pyruvate, acetic acid, cis aconitic acid, formic acid, d-gluconic acid, α-keto glutaric acid, d, l-lactic acid, quinic acid, succinic acid, bromo succinic acid, l-alanyl-glycine, l-asparagine, l-glutamic, l-histidine, l-ornithine, l-proline, l-serine, γ-amino butyric acid, glycerol. The major cellular fatty acids are summed feature 8 (C18:1 ω7c and/or C18:1 ω6c).
The type strain is MH17T (= ACCC 19963T = KCTC 52414T), which was isolated from root of rice collected from Beijing, China. The DNA G+C content of the type strain is 60.4 mol%.
References
De Ley J (1970) Reexamination of the association between melting point, buoyant density, and chemical base composition of deoxyribonucleic acid. J Bacteriol 101:738–754
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Frank B (1889) Ueber die Pilzsymbiose der Leguminosen. Ber DtschBot Ges 7:332–346 (in German)
Gu T, Sun LN, Zhang J, Sui XH, Li SP (2014) Rhizobium flavum sp. nov., a triazophos-degrading bacterium isolated from soil under the long-term application of triazophos. Int J Syst Evol Microbiol 64:2017–2022
Jordan DC (1984) Genus I. Rhizobium Frank 1889, 338AL. In: Krieg NR, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 1. Williams & Wilkins, Baltimore, pp 235–242
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1–6
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Lin SY, Hung MH, Hameed A, Liu YC, Hsu YH, Wen CZ, Arun AB, Busse HJ, Glaeser SP, Kämpfer P, Young CC (2015) Rhizobium capsici sp. nov., isolated from root tumor of a green bell pepper (Capsicum annuum var. grossum) plant. Antonie Van Leeuwenhoek 107:773–784
Marmur J, Doty P (1962) Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol 5:109–118
Martens M, Delaere M, Coopman R, De Vos P, Gillis M, Willems A (2007) Multilocus sequence analysis of Ensifer and related taxa. Int J Syst Evol Microbiol 57:489–503
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2012) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:1–14
Rzhetsky A, Nei M (1992) A simple method for estimating and testing minimum evolution trees. Mol Biol Evol 9(5):945–967
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI Technical Note 101. MIDI Inc, Newark, DE
Sheu S, Huang H, Young C, Chen W (2015) Rhizobium alvei sp. nov., isolated from a freshwater river. Int J Syst Evol Microbiol 65:472–478
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tighe SW, de Lajudie P, Dipietro K, Lindström K, Nick G, Jarvis BD (2000) Analysis of cellular fatty acids and phenotypic relationships of Agrobacterium, Bradyrhizobium, Mesorhizobium, Rhizobium and Sinorhizobium species using the Sherlock microbial identification system. Int J Syst Evol Microbiol 50:787–801
Turdahon M, Osman G, Hamdun M, Yusuf K, Abdurehim Z, Abaydulla G, Abdukerim M, Fang C, Rahman E (2013) Rhizobium tarimense sp. nov., isolated from soil in the ancient Khiyik River. Int J Syst Evol Microbiol 63:2424–2429
Vincent JM (1970) The cultivation, isolation and maintenance of rhizobia. In: Vincent JM (ed) A manual for the practical study of the root-nodule bacteria. Blackwell Scientific, Oxford, pp 1–13
Young JM, Kuykendall LD, Martínez-Romero E, Kerr A, Sawada H et al (2001) A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie, 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int J Syst Evol Microbiol 51:89–103
Zhang X, Sun L, Ma X, Sui XH, Jiang R (2011) Rhizobium pseudoryzae sp. nov., isolated from the rhizosphere of rice. Int J Syst Evol Microbiol 61:2425–2429
Zhang X, Tang X, Ali Sheirdil R, Sun L, Ma X (2014) Rhizobium rhizoryzae sp. nov., isolated from rice roots. Int J Syst Evol Microbiol 64:1373–1377
Zhang X, Gao J, Cao Y, Sheirdil R, Wang X, Zhang L (2015) Isolation and proposal novel rice promoting endophytic bacteria, Rhizobium oryzicola sp. nov. Int J Syst Evol Microbiol 65:2931–2936
Acknowledgements
This work was supported by the Grants from the Chinese Ministry of Science and Technology (2013AA102802) and National Natural Science Foundation of China (316700005; 41271273).
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession numbers for the 16S rRNA, gyrB, recA, atpD, ropB, gltA gene sequences of strain MH17T are KX129902, KX434874, KX434876, KY321909, KY321911, KY321915, respectively.
Rights and permissions
About this article
Cite this article
Zhao, JJ., Zhang, J., Zhang, RJ. et al. Rhizobium rhizosphaerae sp. nov., a novel species isolated from rice rhizosphere. Antonie van Leeuwenhoek 110, 651–656 (2017). https://doi.org/10.1007/s10482-017-0831-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0831-9