Abstract
A Gram-stain-negative, rod-shaped and aerobic bacterium, designated CL12T, was isolated from roots of Glycine max (Linn. Merr.) collected from an experimental field in the campus of South China Agricultural University, PR China (22°58′46″S, 110°51′10″E). Phylogenetic analysis based on 16S rRNA gene sequences revealed that strain CL12T belongs to the genus Rhizobium, closely related to Rhizobium wuzhouense W44T (99.3%), followed by Rhizobium rosettiformans W3T (98.0%) and Rhizobium ipomoeae Shin9-1T (97.9%). The results of analysis of sequences of four housekeeping genes (recA, atpD, rpoB and glnA) also revealed strain CL12T to be closely related to R. wuzhouense W44T with the similarities 91.0%, 95.0%, 94.2% and 90.5%, respectively. The major fatty acid of strain CL12T was Summed Feature 8 (C18:1ω7c and/or C18:1ω6c). Strain CL12T had not the nodulation genes (nodC and nodA) and nitrogenase reductase gene (nifH), and could not cause formation of nodule on soybean. The draft genome size of strain CL12T was 4.84 Mbp with a genomic DNA G + C content of 61.1 mol%. The digital DNA–DNA hybridization (dDDH) and average nucleotide identity (ANI) of strain CL12T and R. wuzhouense W44T were 27.4% and 84.7%, respectively. Based on genomic, phenotypic and phylogenetic analysis, strain CL12T is suggested to represent a new species of the genus Rhizobium, for which the name Rhizobium glycinendophyticum sp. nov. is proposed. The type strain is CL12T (=GDMCC 1.1597T = KACC 21281T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A large diversity of endophytic bacteria can help host plants cope with various biotic and abiotic stresses and support plant growth and development (Vaishnav et al. 2018; Yaish et al. 2015; Zhao et al. 2016). Rhizobium strains as endophytes have symbiotic and non-symbiotic types. Symbiotic Rhizobium strains can fix atmospheric nitrogen and reduce the need of any exogenous nitrogen fertilizer (De Lajudie et al. 2019). A large non-symbiotic Rhizobium strains co-exists with symbiotic strains represent an ill-studied reservoir of genetic diversity (Soenens and Imperial 2018). Species of the genus Rhizobium are widely distributed in various environments such as soil, legume plants and non-legume plants (Gutierrez-Zamora and Martinez-Romero 2001; Yanni et al. 2016). In Rhizobium taxonomic studies, polyphasic approaches such as phylogenetic analysis, phenotypic features, digital DDH method and genome-wide average nucleotide identity (ANI) method were used as standard criteria for the description of new bacterial species (Aserse et al. 2017b). At the time of writing, the genus Rhizobium consisted of 108 species listed in LPSN (www.bacterio.net/-allnamesac.html).
Glycine max (Linn. Merr.) naturally harbors a diverse endophytic microbial community within tissues of its interior plant compartments (Guo et al. 2016; Khan et al. 2019; Zhao et al. 2018). During an investigation on the diversity of endophytic bacteria in soybean, we characterised a bacterial isolate originated from roots of soybean and described a novel species in the genus Rhizobium.
Materials and methods
Isolation and cultivation of strain CL12T
Roots of soybean were collected from the campus of South China Agricultural University, PR China (23°9′46″S; 113°21′10″E). The surface sterilization of roots was performed according to the protocol described in previously study (Sun et al. 2008). Briefly, the entire soybean plant was washed with tap water; then the roots were cut and surface-sterilized with 70% ethanol for 1 min, 2% sodium hypochlorite for 10 min and finally rinsed five times with sterile distilled water. Tissues (1 g in 9 mL of sterile water) were ground in a sterile mortar and the extracts were further diluted with sterile water up to 10–4 and aliquots of 0.1 mL of the last three dilutions were plated onto R2A agar. Among the appeared colonies on plates, bacteria with different colony morphologies were picked up and purified by repeated streaking, and these bacteria were preserved as glycerol suspensions (20%, v/v) at -80 °C and/or as lyophilized powder in inclosed ampoules at 4 °C. We used the 16S rRNA gene sequence analysis to screen all of the isolates of endophytes, and the strain CL12T was chosen for further taxonomic analyses because of its low similarity with the defined species. The strain CL12T was routinely cultivated on R2A plates at 30 °C for 2–3 days.
Phylogenetic analysis based on 16S rRNA gene and four housekeeping genes
Genomic DNA of strain CL12T was extracted according to improved CTAB method (Chen and Ronald 1999) and 16S rRNA gene was amplified using the extracted genomic DNA as template with the universal primers 27F and 1492R (Lane 1991). PCR products were sequenced in Majorbio, China. Four housekeeping genes (recA, atpD, rpoB and glnA) were obtained from the draft genome of strain CL12T (mentioned subsequently). 16S rRNA gene and four hosekeeping genes were aligned in EzBioCloud (https://eztaxon-e.ezbiocloud.net/) and GenBank (www.ncbi.nlm.nih.gov), repectively. Maximum-likelihood (ML), neighbour-joining (NJ) and maximum evolution (ME) trees were constructed using MEGA7.0 software (Kumar et al. 2016) with bootstrap values of 1000 resamplings. Evolutionary distances were generated using Kimura’s two-parameter model (Kimura 1980).
Genome sequencing and Comparative genomic analysis
The genomic DNA of strain CL12T was sequenced with the Illumina Hiseq platform in Personalbio, Ltd. A draft genome with a mapped coverage of 206 × was assembled using A5-MiSeq v20150522 (Coil et al. 2015) and had been submitted to the National Centre for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov/genome) under the accession number VFYP00000000. The genomic features were annotated using the Prokaryotic Genome Annotation Pipeline (PGAP) at GenBank. The predicted coding sequences were translated and used as queries to search the COG database.
Based upon the close relationship with the test strain in phylogenetic analyses, the draft genome sequence of R. wuzhouense W44T was obtained from NCBI database under the accession number QJRY00000000. Digital DNA-DNA hybridization (dDDH) and average nucleotide identity (ANI) values between strain CL12T and R. wuzhouense W44T were calculated by Genome-to-Genome Distance Calculator (GGDC) (https://ggdc.dsmz.de/) and OrthANIu (www.ezbiocloud.net/tools/ani) (Yoon et al. 2017), respectively. Shared orthologous protein clusters between the genomes of strain CL12T and R. wuzhouense W44T were identified using the web-based tool InteractiVenn (Wang et al. 2015) as described previously (Aserse et al. 2017a).
Phenotypic characterisation
Three closely related type strains, R. wuzhouense W44T, R. rosettiformans W3T and R. ipomoeae Shin9-1T, were obtained from Guangdong Microbial Culture Collection Center (GDMCC), Czech Collection of Microorganism (CCM) and Korean Collection for Type Culture (KCTC) for comparison studies of phenotypic characterization and chemotaxonomic analysis. Growth on MacConkey agar, brain–heart infusion agar (BHI), trypticase soy agar (TSA), nutrient agar (NA) and 272 agar (Wang et al. 2018) was observed after incubation for 7 days at 30 °C. The following phenotypic characteristics were tested on R2A agar in parallel with three reference strains under the same conditions. Cell morphology of the strains cultured at 30 °C for 2 days was observed by both light microscopy (DM6/MC190) and transmission electron microscopy (H7650, Hitachi). The range of growth temperature was assessed at 10, 15, 25, 30, 37, 40, 42 and 45 °C for up to 7 days. pH tolerance was determined from pH 4.5 to pH 10.0 at intervals of 0.5 pH unit according to previously described method (Lv et al. 2016). Salt tolerance was evaluated at NaCl concentration range of 0–4.5% (w/v) at intervals of 0.5%. Gram-staining reaction was determined by a Gram-stain kit (bioMérieux) according to the manufacturer’s instruction. Oxidase activity was tested by oxidase test strips with 1% (w/v) tetramethyl-p-phenylenediamine (HKM). Catalase activity was determined by bubble production after mixing a loopful of cells with 3% (v/v) H2O2. Hydrolyses of starch, CM-cellulose, chitin, casein, Tween 20, 40 and 80 were tested on R2A agar with starch (1%, w/v), cellulose (1%, w/v), chitin (1%, w/v), Tween 20 (1%, v/v), Tween 40 (1%, v/v) and Tween 80 (1%, v/v), respectively. Cell motility was tested by the hanging-drop method with 0.2% agar. Some other phenotypic characteristics were determined using API 20NE, API ZYM kits (bioMérieux) and Biolog GENIII MicroPlate according to the manufacturer’s instructions.
Analysis of cellular fatty acid profiles
The fatty acids of strain CL12T and its closely related reference strains, R. wuzhouense W44T, R. rosettiformans W3T and R. ipomoeae Shin9-1T, were extracted from logarithmically growing cells cultured on R2A agar at 30 °C for 2 days. Fatty acid methyl esters were prepared according to the protocol of Sherlock Microbial Identification System (MIDI), analysed via gas chromatography (modei 7890A, Hewlett Packard) and identified using the Sherlock Aerobic Bacterial Database (TSBA 6.1) (Miller 1982).
Results
Phylogenetic analysis
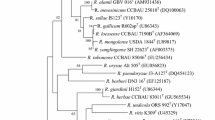
In database of EZBioCloud, the 16S rRNA gene sequence of strain CL12T shared highest identity with R. wuzhouense W44T (99.3%), followed by R. rosettiformans W3T (98.0%) and R. ipomoeae Shin9-1T (97.9%), and it exhibited less similarities than 97.4% with other members of the genus Rhizobium. In GenBank database, four housekeeping gene of strain CL12T, recA, atpD, rpoB and glnA, had similarities of 91.0%, 95.0%, 94.2% and 90.5%, respectively, with their analogues in R. wuzhouense W44T. In the ML tree, strain CL12T was clustered together with R. wuzhouense W44T and R. ipomoeae Shin9-1T with a bootstrap value of 61% (Fig. 1). Similar results were also obtained from NJ and ME trees (Fig. S1 and S2). Compared with the low resolution and confidence of trees based on 16S rRNA genes, the NJ tree based on concatenated housekeeping genes clearly showed that strain CL12T was most related to R. wuzhouense W44T, and peripherally to R. rosettiformans W3T and R. ipomoeae Shin9-1T, successively (Fig. 2). Therefore, R. wuzhouense W44T, R. rosettiformans W3T and R. ipomoeae Shin9-1T were used as references for further taxonomic studies.
Maximum-likelihood tree based on 16S rRNA gene sequences revealing the relationship between strain CL12T and other species of the genus Rhizobium. Burkholderia graminis C4D1MT was used as an out-group. GenBank accession numbers are shown in parentheses. Bootstrap values > 70% are shown. Bar, 0.02 substitutions per nucleotide position
Neighbor-joining tree based on the concatenated recA, atpD, ropB and glnA gene sequences revealing the phylogenetic relationships between strain CL12T and strains of related Rhizobium species. Bradyrhizobium japonicum USDA6T was used as an out-group. Numbers at nodes represent bootstrap values (> 70% are shown) based on 1000 resamplings. Bar, 0.02 substitutions per nucleotide position
Genomic characteristics and comparative genomics analysis
The draft genome of strain CL12T contained 16 contigs with an N50 value of 2,995,531 bp and an N90 value of 166,916 bp. The genome size was 4.84 Mbp. The genomic DNA G + C content was 61.1 mol%, which was lower than the G + C contents of R. wuzhouense W44T and R. rosettiformans W3T (61.6 and 62.3 mol%) (Kaur et al. 2011), but higher than the G + C content of R. ipomoeae Shin9-1T (58.3 mol%) (Table 1) (Sheu et al. 2016). Genes for nodulation (nodC and nodA) and nitrogen fixation (nifH) were not detected in the draft genome of strain CL12T, indicating that strain CL12T has no ability to form nodules and fix atmospheric nitrogen. The nodulation and nitrogen fixation genes were also not detected in the three reference strains except that R. rosettiformans W3T contained nifH gene (Table 1). The distribution of genes into COG functional categories revealed that the highest percentage of genes were assigned to function unknown (25.90%), transcription (7.49%), amino acid transport and metabolism (7.32%) and inorganic ion transport and metabolism (6.36%) (Table S1).
The dDDH and ANI values between strain CL12T and R. wuzhouense W44T were 27.4% and 84.7%, respectively, which were lower than the threshold values of 70% and 95–96% for species discrimination (Chun et al. 2018; Goris et al. 2007). The genomic differences of strain CL12T and R. wuzhouense W44T were shown in Table S2 and Fig. S3. Strain CL12T and R. wuzhouense W44T had 4699 and 4599 proteins, respectively, of which 917 and 906 were singletons. The orthologous clusters were shown in a Venn diagram (Fig. S3). 3700 and 3683 homologous protein clusters were identified in the genomes of strain CL12T and R. wuzhouense W44T, respectively, with 3638 clusters shared in both the two genomes. In the genomes of strain CL12T and R. wuzhouense W44T, 66 and 45 protein clusters respectively, were identified as unique clusters with no detectable homologous in each other.
Phenotypic characteristics
Strain CL12T was Gram-stain-negative, rod-shaped, aerobic, and motile bacterium. After incubation on R2A agar for 2 days at 30 °C, the colonies of strain CL12T was cream-coloured and circular; and the cells were 1.4–2.9 μm in length and 0.6–0.8 μm in width (Fig. S4). Strain CL12T was found to oxidase-positive, catalase-positive and grew well on NA, BHI and 272 agar, but didn’t grow on MacConkey and TSA agar. Phenotypic characteristics were examined and compared between strain CL12T and its closely relatives, R. wuzhouense W44T, R. rosettiformans W3T and R. ipomoeae Shin9-1T (Table 1). Strain CL12T could be distinguished from its closely relatives by the activity of arginine dihydrolase, assimilation of arabinose and GC content (mol%).
Fatty acid profiles
The predominant fatty acids of strain CL12T (> 5% of the total amounts) comprised Summed Feature 8 (C18:1ω7c and/or C18:1ω6c) (72.9%), Summed Feature 2 (iso-C16:1I and/or C14:03-OH) (6.3%) and C18:0 (5.7%). This profile was consistent with other species in the genus Rhizobium. The minor differences between strain CL12T and its closely relatives, R. wuzhouense W44T, R. rosettiformans W3T and R. ipomoeae Shin9-1T, were shown in Table 2. The presence of C20:1ω7c could be used to distinguish strain CL12T from its related type strains (Table 2).
In summary, our polyphasic taxonomy results especially the dDDH and ANI values, genomic GC content, the presence of C20:1ω7c, the activity of arginine dihydrolase and assimilation of arabinose, showed conclusively that strain CL12T represents a novel species of the genus Rhizobium, for which the name Rhizobium glycinendophyticum is proposed.
Description of Rhizobium glycinendophyticum sp. nov.
Rhizobium glycinendophyticum (gly.cin.en.do.phy'ti.cum. L. fem. n. Glycine generic name of the soy bean; Gr. pref. endo within; Gr. n. phyton plant; L. masc. suff. -icus adjectival suff. used with the sense of belonging to; N.L. masc. adj. endophyticus within plant, endophytic; N.L. neut. adj. glycinendophyticum an endophyte of soybean).
Grow well on NA, BHI and 272 agar, but not on MacConkey and TSA agar. After 2 days of incubation at 30 °C on R2A agar, colonies are cream and circular; and cells are Gram-stain-negative, oxidase-positive, catalase-positive, aerobic, motile, rod-shaped and approximately 1.4–2.9 μm long and 0.6–0.8 μm wide (Fig. S4). The temperature range for growth is 10–42 °C (optimum, 30 °C). The pH range for growth is pH 5.0–9.5 (optimum, pH 7.0). Growth occurs at a NaCl concentration of 0–4.5% (optimum, 2.0%). It could not hydrolyse starch, CM-cellulose, casein, chitin, Tween 20, 40 and 80. It does not contain the nodulation genes (nodC and nodA) and nitrogenase reductase gene (nifH). The predominant fatty acids (> 5% of the total amounts) include Summed Feature 8 (C18:1ω7c and/or C18:1ω6c, 72.9%), Summed Feature 2 (iso-C16:1 I and/or C14:0 3-OH, 6.3%) and C18:0 (5.7%).
The type strain, CL12T (=GDMCC 1.1597T = KACC 21281T), was isolated from roots of G. max (Linn. Merr.). The genome size is 4.84 Mbp with a high genomic DNA G + C content of 61.1 mol%. The GenBank accession numbers of 16S rRNA, recA, atpD, rpoB, glnA gene sequences and the whole genome sequence of strain CL12T are MF383489, MN087401, MN087402, MN087403, MN087404, and VFYP00000000, respectively.
Abbreviations
- GDMCC:
-
Guangdong Microbial Culture Collection Center
- KACC:
-
Korean Agricultural Culture Collection
- KCTC:
-
Korean Collection for Type Culture
- CCM:
-
Czech Collection of Microorganism
References
Aserse AA, Woyke T, Kyrpides NC, Whitman WB, Lindstrom K (2017a) Draft genome sequence of type strain HBR26T and description of Rhizobium aethiopicum sp. nov. Stand Genomic Sci 12:14. https://doi.org/10.1186/s40793-017-0220-z
Aserse AA, Woyke T, Kyrpides NC, Whitman WB, Lindstrom K (2017b) Draft genome sequences of Bradyrhizobium shewense sp. nov. ERR11T and Bradyrhizobium yuanmingense CCBAU 10071T. Stand Genomic Sci 12:74. https://doi.org/10.1186/s40793-017-0283-x
Chen DH, Ronald PC (1999) A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol Biol Rep 17:53–57. https://doi.org/10.1023/a:1007585532036
Chun J et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. https://doi.org/10.1099/ijsem.0.002516
Coil D, Jospin G, Darling AE (2015) A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31:587–589. https://doi.org/10.1093/bioinformatics/btu661
De Lajudie PM et al (2019) Minimal standards for the description of new genera and species of rhizobia and agrobacteria. Int J Syst Evol Microbiol. https://doi.org/10.1099/ijsem.0.003426
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. https://doi.org/10.1099/ijs.0.64483-0
Guo X et al (2016) Plantactinosporasoyae sp. nov., an endophytic actinomycete isolated from soybean root [Glycine max (L.) Merr]. Int J Syst Evol Microbiol 66:2578–2584. https://doi.org/10.1099/ijsem.0.001088
Gutierrez-Zamora ML, Martinez-Romero E (2001) Natural endophytic association between Rhizobium etli and maize (Zea mays L.). J Biotechnol 91:117–126. https://doi.org/10.1016/s0168-1656(01)00332-7
Kaur J, Verma M, Lal R (2011) Rhizobium rosettiformans sp. nov., isolated from a hexachlorocyclohexane dump site, and reclassification of Blastobacter aggregatus Hirsch and Muller 1986 as Rhizobium aggregatum comb. nov. Int J Syst Evol Microbiol 61:1218–1225. https://doi.org/10.1099/ijs.0.017491-0
Khan MA, Asaf S, Khan AL, Ullah I, Ali S, Kang S-M, Lee I (2019) Alleviation of salt stress response in soybean plants with the endophytic bacterial isolate Curtobacterium sp. SAK1. Ann Microbiol 69:797–808. https://doi.org/10.1007/s13213-019-01470-x
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparatives studies of nucleotide-sequences. J Mol Evol 16:111–120. https://doi.org/10.1007/bf01731581
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Lane DJ (1991) 16S/23S rDNA sequencing. In: Stackebrandt E, Goodfellow M (eds) In Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–175
Lv YY, Wang J, Chen MH, You J, Qiu LH (2016) Dinghuibacter silviterrae gen. nov., sp. nov., isolated from forest soil. Int J Syst Evol Microbiol 66:1785–1791. https://doi.org/10.1099/ijsem.0.000940
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxy acids. J Clin Microbiol 16:584–586
Sheu SY, Chen ZH, Young CC, Chen WM (2016) Rhizobium ipomoeae sp. nov., isolated from a water convolvulus field. Int J Syst Evol Microbiol 66:1633–1640. https://doi.org/10.1099/ijsem.0.000875
Soenens A, Imperial J (2018) Novel, non-symbiotic isolates of Neorhizobium from a dryland agricultural soil. PeerJ 6:e4776. https://doi.org/10.7717/peerj.4776
Sun L, Qiu F, Zhang X, Dai X, Dong X, Song W (2008) Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA sequence analysis. Microb Ecol 55:415–424. https://doi.org/10.1007/s00248-007-9287-1
Vaishnav A, Shukla AK, Sharma A, Kumar R, Choudhary DK (2018) Endophytic bacteria in plant salt stress tolerance: current and future prospects. J Plant Growth Regul 38:650–668. https://doi.org/10.1007/s00344-018-9880-1
Wang Y, Coleman-Derr D, Chen G, Gu YQ (2015) OrthoVenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43:W78–84. https://doi.org/10.1093/nar/gkv487
Wang CL, Lv YY, Li AZ, Feng GD, Bao GG, Zhu HH, Tan ZY (2018) Chitinophaga silvisoli sp. nov., isolated from forest soil. Int J Syst Evol Microbiol 69:909–913. https://doi.org/10.1099/ijsem.0.003212
Yaish MW, Antony I, Glick BR (2015) Isolation and characterization of endophytic plant growth-promoting bacteria from date palm tree (Phoenix dactylifera L.) and their potential role in salinity tolerance. Antonie Van Leeuwenhoek 107:1519–1532. https://doi.org/10.1007/s10482-015-0445-z
Yanni YG, Dazzo FB, Squartini A, Zanardo M, Zidan MI, Elsadany AEY (2016) Assessment of the natural endophytic association between Rhizobium and wheat and its ability to increase wheat production in the Nile delta. Plant Soil 407:367–383. https://doi.org/10.1007/s11104-016-2895-0
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. https://doi.org/10.1007/s10482-017-0844-4
Zhao S, Zhou N, Zhao ZY, Zhang K, Wu GH, Tian CY (2016) Isolation of endophytic plant growth-promoting bacteria associated with the halophyte salicornia europaea and evaluation of their promoting activity under salt stress. Curr Microbiol 73:574–581. https://doi.org/10.1007/s00284-016-1096-7
Zhao L, Xu Y, Lai X (2018) Antagonistic endophytic bacteria associated with nodules of soybean (Glycine max L.) and plant growth-promoting properties. Braz J Microbiol 49:269–278. https://doi.org/10.1016/j.bjm.2017.06.007
Acknowledgements
This work was supported by the National Science Foundation of China (31800003, 31600008), the GDAS' Special Project of Science and Technology Development (2019GDASYL-0401002), Guangdong Province Science and Technology Innovation Strategy Special Fund (2018B020205003), Science and Technology Plan Project of Guangdong Province (2019B030316010).
Author information
Authors and Affiliations
Contributions
ZHH designed the study. WCL, YT and BGG performed the experiments. FGD contributed to fatty acids analysis. WCL, ZHH wrote the paper. LAZ revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The GenBank accession number for the 16S rRNA gene, recA, atpD, rpoB, glnA and the draft genome sequence of strain CL12T are MF383489, MN087401, MN087402, MN087403, MN087404 and VFYP00000000, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Li, A., Yuan, T. et al. Rhizobium glycinendophyticum sp. nov., isolated from roots of Glycine max (Linn. Merr.). Antonie van Leeuwenhoek 113, 147–154 (2020). https://doi.org/10.1007/s10482-019-01324-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-019-01324-1