Abstract
Ginsenosides are the major pharmacological components in ginseng. We isolated lactic acid bacteria from Kimchi to identify microbial modifications of ginsenosides. Phylogenetic analysis of 16S rRNA gene sequences indicated that the strain DCY65-1 belongs to the genus Lactobacillus and is most closely related to Lactobacillus brevis. On the basis of TLC and HPLC analysis, we found two metabolic pathways: F1 → 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside and C–K → 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside. These results suggest that strain DCY65-1 is capable of potent ketonic decarboxylation, ketonizing the hydroxyl group at C-3. The F1 metabolite had a more potent inhibitory effect on mushroom tyrosinase than did the substrate. Therefore, the F1 and C–K derivatives may be more pharmacologically active compounds, which should be further characterized.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ginsenosides are the major pharmacological components in ginseng. Ginsenosides can be divided into several groups, including protopanaxadiol (PPD), protopanaxatriol (PPT) and oleanane. More than 40 ginsenosides have been isolated from ginseng roots and characterized, including the major ginsenosides Rb1, Rb2, Rc, Rg1 and Re (Park 2004). The major ginsenosides found in the leaves are Rg1, Re, Rd, Rb2 and F2. Some saponins such as ginsenosides F1, F2, F3 and F5 and notoginsenoside Fe are found only in the leaves (Wang 2001).
In recent decades, many studies have reported the successful transformation of major ginsenosides into more active minor ginsenosides. Minor ginsenosides have high pharmacological activity, however much lower natural availability in ginseng.
Microbial transformation is regarded as an effective and useful technology for structural modification and metabolism, due to its significant regio- and stereoselectivities (Azerad 1999; Clark and Hufford 1991). Several microbial transformations have been used to deglycosylate ginsenosides (Kim et al. 2012a, b; Wu et al. 2012). In recent years, microbial modifications (side-chain oxidation–reduction, hydroxylation and ketonization) have been reported to form new compounds (Liu et al. 2011; Chen et al. 2013).
These reports, however, did not determine which specific enzymes were responsible for the bioconversion. Also most microorganisms used to the transform ginsenosides do not meet the generally regarded as safe (GRAS) standard. Lactic acid bacteria (LAB) are known for their probiotic properties, are GRAS and are used extensively in human nutrition and food industries.
Lactobacillus brevis has been isolated from milk, cheese, sauerkraut, the intestinal tract of humans and rats, cow manure and wine. As a member of the genus Lactobacillus and due to its long history of use in various fermented foods, L. brevis meets the GRAS standard and is regarded as a probiotic organism (O’Sullivan et al. 1992; Collins et al. 1998).
Tyrosinase inhibitors have applications in food products, medicinal uses, and as cosmetics for the browning of foods and the amelioration of skin hyperpigmentation (Parvez et al. 2007). Tyrosinase is a copper-containing enzyme which is the key step in melanin biosynthesis. It catalyzes hydroxylation of l-tyrosine to L-DOPA and further the conversion of L-DOPA to the corresponding o-quinones (Seo et al. 2003). These quinones tend to polymerize spontaneously to form brown pigments, namely melanin. Consequently, inhibiting tyrosinase activity reduces the synthesis of melanin.
This paper reports the isolation and identification of L. brevis DCY65-1 from Kimchi, a traditional Korean fermented food. In this study, we produced ketonized compounds with different structures by bioconversion with this food-grade Lactobacillus. The F1 metabolite obtained had more potent inhibitory effects against mushroom tyrosinase than did the F1 substrate.
Material and methods
Materials
Substrate ginsenosides F1 and C–K were obtained from the Ginseng Genetic Resource Bank (Suwon, Republic of Korea) and their purity was determined to be above 98 % by HPLC analysis. MRS broth was purchased from Difco (USA). Silica gel 60 F254 plates and silica gel 60 (Merck, Germany) were used for TLC and column chromatography. All chemicals and solvents were analytical or HPLC grade. HPLC-grade acetonitrile and water were purchased from SK Chemicals (Ulsan, Korea).
Isolation of lactic acid bacteria from Kimchi
LAB were isolated from Kimchi by a standard dilution plating onto MRS agar. On the basis of colony shape, colour and size, single colonies were purified by transferring onto new plates.
Phylogenetic analysis
The 16S rRNA gene was sequenced using the universal bacterial primer sets 27F, 518F, 800R and 1492R (Lane 1991; Anzai et al. 2000) by Genotech (Daejeon, Republic of Korea) (Kim et al. 2005). The resulting 16S rRNA gene sequence was compared with available 16S rRNA gene sequences of related taxa by using the EzTaxon-e server (http://eztaxon-e.ezbiocloud.net/) (Kim et al. 2012a, b). A phylogenetic tree was constructed using the neighbour-joining method (Saitou and Nei 1987) with the MEGA software (Tamura et al. 2007) with bootstrap values based on 1,000 replications (Felsenstein 1985).
Biotransformation of ginsenosides
Strain DCY65-1 was grown in MRS broth at 37 °C. After sonication, crude enzyme was collected by centrifugation at 10,000×g for 15 min at 4 °C. The pellet was dissolved in 20 mM sodium phosphate buffer (pH 7.0) and mixed with 1 mM ginsenoside F1 or C–K. Subsequently, the mixture was incubated at 30 °C. The reaction mixture was extracted with water-saturated butanol and then analyzed by TLC and HPLC.
Isolation of metabolites
The extracts of transformed ginsenosides F1 and C–K were subjected to column chromatography on a silica gel column with CHCl3–CH3OH (15:1) as the solvent. The collected fraction was further purified by semi-preparative HPLC. Metabolite 1 and Metabolite 2 obtained from the ginsenosides F1 and C–K after bacterial biotransformation were dissolved in pyridine-d 5 and identified by 1H, 13C, and 2D NMR using an FT-NMR spectrometer (400 MHz; Varian Inova AS 400, Varian, Palo Alto, CA, USA).
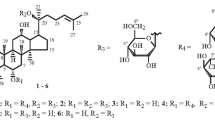
Metabolite 1: 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside
White amorphous powder; 1H-NMR (400 MHz, pyridine-d 5, δH) 5.25 (1H, dd, J = 6.8, 6.4 Hz, H-24), 5.16 (1H, d, J = 7.6 Hz, H-1′), 4.46–3.89 (sugar moieties), 1.60 (9H, s, H-28, 21, 29), 1.11 (3H, H-26), 1.03 (3H, s, H-27), 0.96 (3H, s, H-18), 0.91 (3H, s, H-19), 0.87 (3H, s, H-30); 13C-NMR (400 MHz, pyridine-d 5) spectroscopic data (Table 1).
Metabolite 2: 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside
White amorphous powder; 1H-NMR (400 MHz, pyridine-d 5, δH) 5.26 (1H, dd, J = 6.0, 6.4 Hz, H-24), 5.16 (1H, d, J = 7.6 Hz, H-1′), 4.45–3.84 (sugar moieties), 1.66 (3H, s, H-28), 1.63 (3H, s, H-21), 1.61 (3H, s, H-29), 1.59 (6H, s, H-26, 27), 1.01 (3H, s, H-18), 0.97 (3H, s, H-19), 0.82 (3H, s, H-30); 13C-NMR (400 MHz, pyridine-d 5) spectroscopic data (Table 1).
Analytical methods
TLC analysis was performed using silica gel 60 plates with CHCl3–CH3OH–H2O (65:35:10, v/v/v, lower phase) as the developing solvent. Spots on the TLC plates were detected by spraying 10 % (v/v) H2SO4 and heating at 110 °C for 10 min (Shibata et al. 1965).
Reactants in the water-saturated n-butanol fraction were evaporated in a vacuum, and the residue was dissolved in CH3OH and subjected to HPLC analysis. Ginsenosides were resolved on a C18 column (250 × 4.6 mm, ID 5 μm) with acetonitrile (solvent A) and H2O (solvent B) at A/B ratios of 20:80, 20:80, 29:71, 30:70, 48:52, 70:30, 90:10, 90:10, 20:80 and 20:80, with run times of 0, 10, 42, 52, 65, 82, 83, 93, 94 and 100 min, respectively. The flow rate was 1.6 mL min−1 and detection wavelength was 203 nm.
1H, 13C, and 2D NMR spectra were recorded on a Varian Unity INOVA AS 400 FT-NMR instrument and chemical shifts are given in δ (ppm) based on tetramethylsilane (TMS) as an internal standard.
Inhibitory effect of ginsenosides and their metabolites on mushroom tyrosinase
The tyrosinase inhibition activity of the ginsenosides and their metabolites were measured by a modified method of Yagi et al. (1987). The reaction mixture, 20 μL of MeOH with or without ginsenosides and metabolites, was mixed with 160 μL of 0.06 mM L-DOPA solution and 20 μL of 1,500 U/mL mM mushroom tyrosinase and dissolved in 5 mM phosphate buffer (pH 6.8). The mixture was incubated at 25 °C for 30 min before measuring absorbance at 475 nm. The tyrosinase inhibition rate (%) was calculated as follows:
% Inhibition = [1−(A − B)/A] × 100
Results and discussion
Phylogenetic analysis of isolated strain
L. brevis DCY65-1 was isolated from Kimchi, a traditional Korean fermented food. The phylogenetic tree demonstrated that strain DCY65-1 should be affiliated with the genus Lactobacillus (Fig. 1). On the basis of the 16S rRNA gene sequence (GenBank accession number KM507722), the closest recognized relatives of strain DCY65-1 were identified as L. brevis ATCC 14687T (99.93 %) and L. brevis ATCC 367 (99.79 %). Strain DCY65-1 had less than 1 % difference in 16S rRNA gene sequence from the corresponding type strain and thus should be considered to belong to the same species. L. brevis has GRAS status and is regarded as a probiotic organism (O’Sullivan et al. 1992; Collins et al. 1998).
Structure of metabolites
Ginsenoside F1 was transformed by train DCY65-1 into metabolite 1. Metabolite 1 was obtained as a white powder. The 1H and 13C NMR and DEPT spectra of metabolite 1 were similar to those of ginsenoside F1 except for the appearance of a ketone (δC 218.5, C-3) and disappearance of one oxygenated methine signal from ginsenoside F1 (Liu et al. 2011). The molecular weight of metabolite 1 was 2 Da less than that of ginsenoside F1, indicating that the molecule was ketonized. The proton and carbon signals from the sugar moiety suggested the presence of a β-glucopyranosyl group (see NMR data). The correlation between δH 5.16 (H-1′) and δC 83.3 (C-20) in the HMBC spectrum and the chemical shift of the anomeric carbon signal (δC 98.2) supported the presence of a glucopyranosyl group at C-20. Therefore, the structure of metabolite 1 was determined to be 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside.
Ginsenoside C–K was transformed by train DCY65-1 into metabolite 2. Metabolite 2 was obtained as a white powder. The 13C-NMR spectrum of metabolite 2 had 36 carbon signals. By using the DEPT and HSQC spectrum and a comparison with the 13C-NMR data of the related metabolite compound K, all the signals could be assigned (Table 1). These data were similar to that for metabolite 1 with the exception of proton and carbon resonances for the lack of an oxygenated methine moiety at the C-6 position. The methylene signals from C-6 in metabolite 2 were replaced by oxygenated methine signals (δC 66.8) in metabolite 1. The molecular weight of metabolite 2 was 16 Da less than that of metabolite 1, indicating the presence of one less hydroxyl group. The proton and carbon signals from the sugar moiety suggested the presence of a β-glucopyranosyl group (Table 1). The correlation between δH 5.16 (H-1′) and δC 83.3 (C-20) in the HMBC spectrum and the chemical shift of the anomeric carbon signal (δC 98.2) supported the presence of a glucopyranosyl group at C-20. Therefore, the structure of metabolite 2 was determined to be 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside.
Biotransformation pathway
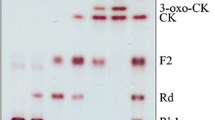
The TLC and HPLC results of ginsenosides F1 and C–K transformed by crude enzymes from strain DCY65-1 are shown in Figs. 2 and 3. The HPLC peaks with retention times of 59.04, 60.10, 74.27 and 77.03 min correspond to ginsenoside F1, 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside, ginsenoside C–K and 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside, respectively. After 48 h, ginsenoside F1 was transformed into 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside (metabolite 1), and ginsenoside C–K was transformed into 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside (metabolite 2). Based on TLC and HPLC analysis, we found two metabolic pathways: F1 → 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside and C–K → 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside (Fig. 4). These results suggested that strain DCY65-1 has potent ketonic decarboxylase activity and that ginsenosides F1 and C–K were converted into 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside and 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside, respectively, by ketonization at the C-3 position of the ginsenoside aglycone, confirming that ketonization of OH group at C-3 is the characteristic reaction of this strain. Thus a 3β-hydroxysteroid dehydrogenase from L. brevis DCY65-1 can be proposed to be an enzyme that metabolises the ginsenosides F1 and C–K by a similar pathway to progesterone biosynthesis in the adrenal gland (Cravioto et al. 1986).
TLC analysis of metabolites of ginsenosides F1 and C–K converted by strain DCY65-1. 1 ginsenoside F1 control, 2 ginsenoside F1 metabolite (6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside), 3 ginsenoside C–K control, 4 ginsenoside C–K metabolite (12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside)
HPLC profiles of metabolites of ginsenosides F1 and C–K converted by strain DCY65-1. a Ginsenoside F1 control; b ginsenoside F1 metabolite; c ginsenoside C–K control; d ginsenoside C–K metabolite. 1 F1, 2 6α,12β-dihydroxydammar-3-one-20(S)-O-β-d-glucopyranoside, 3 C–K, 4 12β-hydroxydammar-3-one-20(S)-O-β-d-glucopyranoside
Using L. brevis DCY65-1, the ginsenosides F1 and C–K were converted to new ketonized compounds. This is the first report describing ginsenosides being ketonized by L. brevis. The structures of the bioconverted compounds were analyzed, but the pharmacological effects still need to be investigated.
Inhibitory effect of ginsenosides and their metabolites on mushroom tyrosinase
Several tyrosinase inhibitors have been used in the cosmetic industry as skin whitening agents. These skin whitening agents have been screened and evaluated using DOPA oxidation by the mushroom tyrosinase because of its commercial availability. We investigated the effects of the ginsenosides and their metabolites on mushroom tyrosinase oxidation of DOPA. Neither ginsenoside C–K nor its metabolite inhibited mushroom tyrosinase activity (data not shown). Han et al. (2014) have been reported that ginsenoside F1 has a skin-whitening effect via the suppression of tyrosinase and dopachrome tautomerase. The ginsenoside F1 and metabolite concentrations that caused a 50 % loss in enzyme activity (IC50) were 130.46 and 114.12 μM, respectively. The F1 metabolite had more potent inhibitory effects against mushroom tyrosinase than did the F1 substrate. Therefore, the F1 metabolite, and possibly the C–K derivative, may be more pharmacologically active, which will be further characterized in subsequent studies.
References
Anzai Y, Kim H, Park JY, Wakabayashi H, Oyaizu H (2000) Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol 50:1563–1589
Azerad R (1999) Microbial models for drug metabolism. Adv Biochem Eng Biotechnol 63:169–218
Chen G, Yang M, Nong S, Yang X, Ling Y, Wang D, Wang X, Zhang W (2013) Microbial transformation of 20(S)-protopanaxadiol by Absidia corymbifera. Cytotoxic activity of the metabolites against human prostate cancer cells. Fitoterapia 84:6–10
Clark AM, Hufford CD (1991) Use of microorganisms for the study of drug metabolism: an update. Med Res Rev 11:473–501
Collins JK, Thornton G, Sullivan GO (1998) Selection of probiotic strains for human applications. Int Dairy J 8:487–490
Cravioto MD, Ulloa-Aguirre A, Bermudez JA, Herrera J, Lisker R, Mendez JP, Perez-Palacios G (1986) A new inherited variant of the 3β-hydroxysteroid dehydrogenase-isomerase deficiency syndrome: evidence for the existence of two isoenzymes. J Clin Endocrinol Metab 63(2):360–367
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Han J, Lee E, Kim E, Yeom M, Kwon OS, Yoon TH, Lee TR, Kim K (2014) Role of epidermal γδ T cell-derived interleukin 13 in the skin-whitening effect of Ginsenoside F1. Exp Dermatol. doi:10.1111/exd12531 [Epub ahead of print]
Kim MK, Im WT, Ohta H, Lee M, Lee ST (2005) Sphingopyxis granuli sp. nov., a ß-glucosidase-producing bacterium in the family Sphingomonadaceae in α-4 subclass of the Proteobacteria. J Microbiol 43:152–157
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012a) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kim SH, Min JW, Quan LH, Lee S, Yang DU, Yang DC (2012b) Enzymatic Transformation of Ginsenoside Rb1 by L. pentosus Strain 6105 from Kimchi. J Ginseng Res 36:291–297
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, Chichester, pp 115–176
Liu X, Qiao LR, Xie D, Dai JG (2011) Microbial deglycosylation and ketonization of ginsenosides Rg1 and Rb1 by Fusarium oxysporum. J Asian Nat Prod Res 13:652–658
O’Sullivan MG, Thornton G, O’Sullivan GC, Collins JK (1992) Probiotic bacteria: myth or reality? Trends Food Sci Technol 3:309–314
Park JH (2004) Sun ginseng-a new processed ginseng with fortified activity. Food Nutr 9:23–27
Parvez S, Kang M, Chung HS, Bae H (2007) Naturally occurring tyrosinase inhibitors: mechanism and applications in skin health, cosmetics and agriculture industries. Phytother Res 21(9):805–816
Saitou N, Nei M (1987) The neighbour - joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Seo SY, Sharma VK, Sharma N (2003) Mushroom tyrosinase: recent prospects. J Agric Food Chem 51(10):2837–2853
Shibata S, Tanaka O, Soma K, Ando T, Iida Y, Nakamura H (1965) Studies on saponins and sapogenins of ginseng: the structure of panaxatriol. Tetrahedron Lett 42:207–213
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wang FS (2001) China Ginseng. Liaoning Science and Technology Press, Shenyang, p 692
Wu LP, Jin Y, Yin CR, Bai LV (2012) Co-transformation of Panax major ginsenosides Rb1 and Rg1 to minor ginsenosides C-K and F1 by Cladosporium cladosporioides. J Ind Microbiol Biotechnol 39:521–527
Yagi A, Kanbara T, Morinobu N (1987) Inhibition of mushroom-tyrosinase by Aloe extract. Planta Med 53(6):515–517
Acknowledgments
This research was supported by Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry & Fisheries (KIPET NO: 313038-03-1-SB010) and Next-Generation BioGreen 21 Program (SSAC, Grant #: PJ009529032014), Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yan Jin and Sun Young Jung have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Jin, Y., Jung, S.Y., Kim, YJ. et al. Microbial ketonization of ginsenosides F1 and C–K by Lactobacillus brevis . Antonie van Leeuwenhoek 106, 1215–1221 (2014). https://doi.org/10.1007/s10482-014-0291-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0291-4