Abstract
A taxonomic study was carried out on a novel aerobic bacterial strain (designated CC-LY736T) isolated from a fermentor in Taiwan. Cells of strain CC-LY736T were Gram-stain negative, spiral-shaped and motile by means of a monopolar flagellum. Strain CC-LY736T shared the greatest degree of 16S rRNA gene sequence similarity to Azospirillum irakense DSM 11586T (97.2 %), Rhodocista centenaria JCM 21060T (96.3 %) and Rhodocista pekingensis JCM 11669T (96.1 %). The major fatty acids were C16:0, C16:1 ω5c, C19:0 cyclo ω8c, C18:1 ω7c/C18:1 ω6c, C16:0 3-OH and C18:1 2-OH. The predominant polar lipids included phosphatidylcholine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylmethylethanolamine, phosphatidyldimethylethanolamine and two unidentified glycolipids. The common major respiratory quinone was ubiquinone Q-10 and predominant polyamines were sym-homospermidine and putrescine. The DNA G+C content of strain CC-LY736T was 67.6 ± 0.1 mol %. During phylogenetic analysis, strain CC-LY736T formed a unique phyletic lineage associated with Rhodocista species. However, the combination of genetic, chemotaxonomic and physiological data clearly indicated that strain CC-LY736T was a novel representative of the family Rhodospirillaceae. Based on the polyphasic comparison, the name Niveispirillum fermenti gen. nov., sp. nov. is proposed; the type strain of the type species is CC-LY736T (= BCRC 80504T = LMG 27263T). In addition, the reclassifications of Azospirillum irakense as Niveispirillum irakense comb. nov. (type strain KBC1T = ATCC 51182T = BCRC 15764T = CIP 103311T), and Azospirillum amazonense as Nitrospirillum amazonense gen. nov., sp. nov. (type strain Am14T = ATCC 35119T = BCRC 14279T = DSM 3787T) are proposed based on the polyphasic taxonomic data obtained in this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The family Rhodospirillaceae, affiliated to the subclass Alphaproteobacteria currently accommodates 34 bacterial genera that were isolated and described from various geographical locations. The members of nitrogen-fixing genus Azospirillum of the family Rhodospirillaceae are distributed in natural environments such as soils of tropical, subtropical and temperate regions, where they are frequently associated with grasses, cereals and crops (Bally et al. 1983; Döbereiner and Day 1976; Kirchhof et al. 1997; Ladha et al. 1987; Patriquin et al. 1983), discarded road tar, oil-contaminated soil and fermentative tanks (Young et al. 2008; Lin et al. 2009, 2012, 2013) and a sulfide spring (Lavrinenko et al. 2010). The other spiral-shaped phototrophic purple non-sulfur bacteria were included in the genus Rhodospirillum (Trüper and Imhoff 1989; Favinger et al. 1989; Kawasaki et al. 1993; Gest and Favinger 2001) before the proposal of the species Rhodocista centenaria (Kawasaki et al. 1992; Euzéby and Kudo 2001) and Rhodospira trueperi (Pfennig et al. 1997). The genus Rhodocista was established based on the results of 16S rRNA gene sequences, unique cyst-producing morphology and ubiquinone Q-9 as predominant respiratory quinone (Kawasaki et al. 1992). As new phototrophic bacterial strains were isolated and more information on the phenotypes and genotypes (particularly 16S rRNA gene sequences) of these and other previously described strains became available, it was evident that the original description of some of the genera within the family Rhodospirillaceae needed revision.
Materials and methods
Bacterial strains and growth conditions
While investigating bacterial diversity in a fermentative tank in central Taiwan (24°12′ N, 120°67′ E) located near a greenhouse set up at the National Chung Hsing University, Taichung. Bacteria were isolated from the fermentation medium; approximately 100 μl fermentation broth were spread on nutrient agar (NA, Hi-Media) by using the standard ten-fold dilution plating technique. After three days of aerobic incubation on nutrient agar (NA, Himedia) at 30 °C, strain CC-LY736T formed a cream-colored colony, which was isolated, purified and preserved at −80 °C as glycerol suspension.
The following type strains Azospirillum amazonense BCRC 14279T (= DSM 2787T) (Magalhães et al. 1983; Falk et al. 1985, 1986); Azospirillum brasilense BCRC 12270T (Tarrand et al. 1978); Azospirillum doebereinerae DSM 13131T (Eckert et al. 2001); Azospirillum fermentarium BCRC 80505T (Lin et al. 2013); Azospirillum formosense BCRC 80273T (Lin et al. 2012); Azospirillum irakense BCRC 15764T (= DSM 11586T) (Khammas et al. 1989); Azospirillum lipoferum BCRC 12213T (type species; Tarrand et al. 1978); Azospirillum oryzae JCM 21588T (Xie and Yokota 2005); Azospirillum picis DSM 19922T (Lin et al. 2009); Azospirillum rugosum DSM 19657T (Young et al. 2008); Azospirillum zeae LMG 23989T (Mehnaz et al. 2007b); Inquilinus limosus LMG 20952T (type species; Coenye et al. 2002); Rhodocista centenaria JCM 21060T (type species; Kawasaki et al. 1992); Rhodocista pekingensis JCM 11669T (Zhang et al. 2003) and Skermanella aerolata DSM 18479T (type species; Weon et al. 2007) were purchased for the purpose of direct comparative taxonomy. All these strains were grown on NA at 30 °C for 2 days, unless specified otherwise.
Morphological tests and biochemical characterization
Cell morphology of strain CC-LY736T, including presence of flagella was determined by placing the cells (1–2 days old) on a carbon-coated copper grid followed by staining with aqueous solution of 0.2 % (w/v) uranyl acetate for 5–10 s, brief air-drying and observation under a transmission electron microscope (JEOL JEM-1400). Gram-staining was performed as described by Murray et al. (1994). Growth was tested using nutrient broth (NB, Hi-Media) at temperature 20–50 °C (at 5 °C intervals) and at pH 5–10 (at 1 unit intervals). Salt tolerance was determined by cultivating the organism in nutrient broth (Hi-Media) supplemented with 0–5 % (w/v) NaCl (at 1 % intervals). Catalase activity was determined by assessing bubble production by cells in 3 % (v/v) H2O2 and oxidase activity was determined by using 1 % (w/v) N,N,N,N,-tetramethyl-1,4-phenylenediamine reagent (bioMérieux, France). Intracellular poly-β-hydroxybutyrate (PHB) granules were detected with different methods, including Sudan Black staining (Schlegel et al. 1970) and Nile blue A staining (Ostle and Holt, 1982), which result in dark blue or fluorescent granules. DNase test was conducted by using DNase test agar (Hi-Media). Carbon source utilization pattern were determined by using Biolog GN2 MicroPlate (bioMérieux, France). Nitrate reduction, indole production, activity of β-galactosidase and urease, hydrolysis of esculin and gelatin, and assimilation of 12 substrates were tested with API 20 NE strips (bioMérieux, France). The activities of various enzymes were determined by using API ZYM system (bioMérieux, France).
Nitrogen-fixing capability
Acetylene-reduction assay described by Hardy et al. (1973) was used to assay the nitrogen-fixing capability. Vials (30 ml) containing 10 ml semisolid nitrogen-free broth (NFB) medium (Reinhold et al. 1987) were inoculated with strain CC-LY736T, A. irakense BCRC 15764T and A. brasilense BCRC 12270T, sealed with rubber septa and incubated at 30 °C in the dark incubator. After 72 h, 10 % (v/v) of the air phase was replaced with acetylene (Koch and Evans, 1966) and the vials were re-incubated. The amount of ethylene was measured for a total of 24 h by using a gas chromatograph (FID Gas Chromatograph, HITACHI, Model 163) equipped with a flame-ionization detector and a packed column (1.0 m × 2.0 mm i.d., steel column packed with Porapak-T 80–100). Conditions of analysis were: carrier gas, nitrogen; flow rate, 35 ml h−1; temperature of the flame ionization detector, 110 °C; column temperature, 65 °C.
Genomic DNA preparation and PCR amplification
Bacterial genomic DNA was isolated by using UltraClean™ Microbial Genomic DNA Isolation Kit (MO BIO, USA) by following manufacturer’s instructions. An almost full-length of 16S rRNA gene was amplified by using bacterial universal primers 1F (5′-GAG TTT GAT CAT GGC TCA GA-3′) and 9R (5′-AAG GAG GTG ATC CAA CCG CA-3′); the primers 3F (5′-CCT ACG GGA GGC AGC AG-3′), 5F (5′-AAA CTC AAA TGA ATT GAC GGG G-3′) and 4R (5′-TTA CCG CGG CTG CTG GCA C-3′) were used for sequencing reaction (Edwards et al. 1989). Gene sequencing was performed by using the Bigdye terminator kit (Heiner et al. 1998), and nucleotide sequence of PCR product was determined by an automatic DNA sequencer (ABI PRISM 310, Applied Biosystems, CA, USA) (Watts and MacBeath 2001). Obtained DNA sequences were then assembled using the Fragment Assembly System program from the Wisconsin Package (GCG 1995).
Azospirillum-specific PCR and nifH gene detection
The specific primer pair for the genus Azospirillum was designed and discussed by Lin et al. (2011). The genomic DNA of novel strain CC-LY736T as well as reference strains (including A. oryzae JCM 21588T; A. brasilense BCRC 12270T; A. canadense LMG 23617T; A. doebereinerae DSM 13131T; A. fermentarium BCRC 80505T; A. formosense BCRC 80273T; A. halopraeferens DSM 3675T; 6, A. largimobile DSM 9441T; A. lipoferum BCRC 12213T; A. melinis LMG 23364T; A. picis DSM 19922T; A. rugosum DSM 19657T; A. thiophilum DSM 21654T; A. zeae LMG 23989T) were screened by using a primer pair Azo494-F (5′-GGC CYG WTY AGT CAG RAG TG-3′) and Azo756-R (5′-AAG TGC ATG CAC CCC RRC GTC TAG C-3′) that can selectively amplify an Azospirillum-specific DNA fragment of size 263 bp. On the other hand, Gene nifH (nitrogenase reductase) was amplified to confirm the existence of sym gene.
The nifH gene was amplified by PCR using the primer set PolF (5′-TGC GAY CCS AAR GCB GAC TC-3′)/PolR (5′-ATS GCC ATC ATY NTC RCC GGA-3′) and the conditions were described previously by Poly et al. (2001). The expected size of the product was about 360 bp. Another primer set Zehrf (5′-TGY GAY CCN AAR GCN GA-3′)/Zehrr (5′-AND GCC ATC ATY TCN CC-3′) was used as described by Zehr and McReynolds (1989). The amplified patterns were screened by electrophoresis in 1.0 % (w/v) agarose gel, stained with ethidium bromide, visualized under UV radiation, and photographed.
16S rRNA gene sequence analysis and phylogenetic analysis
The 16S rRNA gene sequence data were submitted to EzBioCloud server (EzTaxon-e Database, Kim et al. 2012) and NCBI GenBank for BLAST search. Subsequently, closely related 16S rRNA gene sequences were retrieved and aligned by using the CLUSTAL_X (1.83) program (Thompson et al. 1997). Unfortunately, sequence data of several type strains of Azospirillum that are available in GenBank were ambiguous due to many unidentified bases that leads to erroneous phylogenetic distinctions of some Rhodospirillaceae representatives Therefore, the 16S rRNA genes of several Azospirillum species were re-sequenced and submitted to NCBI earlier (Lin et al. 2011, 2012). The phylogenetic analysis was performed using MEGA 5 software (Molecular Evolutionary Genetics Analysis, version 5) (Tamura et al. 2011) and the topology of the resultant trees were evaluated by Neighbor-Joining (Saitou and Nei 1987), Maximum Likelihood (Felsenstein 1981) and Maximum Parsimony (Fitch 1971) methods. The bootstrap values (Felsenstein 1985) were computed after 1,000 replications.
Sequence deposition
Ambiguous 16S rRNA gene sequences of A. amazonense BCRC 14279T (X79735), A. canadense LMG 23617T (DQ393891), A. halopraeferens DSM 3675T (Z29618), A. irakense DSM 11586T (Z29583), A. lipoferum DSM 1691T (M59061) were replaced with newly obtained 16S rRNA gene sequences during current phylogenetic analysis.
DNA–DNA hybridization analysis
DNA–DNA hybridization assay was conducted between strain CC-LY736T and A. irakense BCRC 15764T using DIG DNA labeling and detection kit (Roche Diagnostics; Cat. No.11 093 657 910) according to the manufacturer’s protocol. Chromosomal DNA of strain CC-LY736T was used to construct hybridization probe by labelling with digoxigenin–11-dUTP (DIG). The experiment was carried out in triplicate for each sample.
Fatty acid methyl ester analysis
Fatty acid methyl esters (FAME) were prepared, separated and identified according to the standard protocol (Paisley 1996) of the Microbial Identification System (MIDI) (Sasser 1990) by gas chromatograph (Agilent 7890A) fitted with a flame ionization detector. The cultures were grown under the same condition on NA plates at 30 °C. After 48 h cells were harvested from the plate and subjected to saponification, methylation and extraction (Miller 1982). Identification and comparison were made by using the Aerobe (RTSBA6) database of the MIDI System (Sherlock version 6.0).
Cellular polyamines and polar lipids analyses
Polyamines were extracted as described by Scherer and Kneifel (1983), analyzed by high performance liquid chromatography (HPLC). The dansyl derivatives were separated by using a Hitachi L-2130 equipped with Hitachi L-2200 autosampler, Hitachi L-2485 fluorescence detector (excitation at 360 nm and emission at 520 nm), and a reverse-phase C18 column (Phenomenex® Synergi Fusion-RP80, 250 × 4.60 mm, 4 μm particle size). Polar lipids were extracted and analyzed by two-dimensional TLC, and isoprenoid quinones were purified by the methods according to Minnikin et al. (1984) and analyzed by HPLC as described by Collins (1985).
Determination of the predominant ubiquinone and DNA base composition
Ubiquinone is an essential component of electron transfer system in the plasma membrane of prokaryotes. The isoprenoid quinones were purified by the methods according to Minnikin et al. (1984). Extraction and assay of quinone was routinely performed according to the HPLC method described by Collins (1985). For the analysis of DNA G+C content, DNA samples were prepared and degraded enzymatically into nucleosides as described by Mesbah et al. (1989). The nucleoside mixtures obtained were then separated and analyzed via HPLC (Hitachi L-2130 chromatograph equipped with Hitachi L-2200 autosampler, Hitachi L-2455 diode array detector, and a reverse-phase C18 column (Phenomenex® Synergi 4 μ Fusion-RP80 250 × 4.60 mm)).
Results and discussion
Phenotypic and biochemical characterization
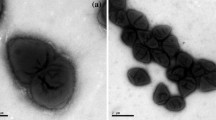
Strain CC-LY736T is Gram-stain negative, spiral shaped, 2.0–2.3 μm in length and 0.6–0.8 μm in diameter. PHB granules were observed under light microscopy and visualized by UV illumination after directly staining growing bacteria on plates containing Nile blue A. The cell morphology of strain CC-LY736T is shown in Fig. S1 and the phenotypic characteristics are given in the genus and species description. Strain CC-LY736T did not growth in nutrient broth supplemented with 3 % NaCl (w/v), which is different with strain Niveispirillum irakense BCRC 15764T. Nitrate reduction was observed in strain CC-LY736T, but Niveispirillum irakense BCRC 15764T and Nitrospirillum amazonense BCRC 14279T did not reduce this compound. Urea hydrolysis activity and indole production were negative. The new strain was able to utilize utilize α-d-lactose, d-sorbitol, acetic acid, cis-aconitic acid, γ-hydroxybutyric acid, succinic acid, succinamic acid, d-alanine, glycyl-l-aspartic acid, l-histidine, l-leucine and d-serine as carbon sources; assimilation of d-glucose, N-acetyl-glucosamine and d-maltose, which distinguished from other related type species. Distinctive phenotypic characteristics for the strains of the novel group and the type strains of the phylogenetically closest species are shown in Table 1.
Nitrogen-fixing capability and nifH gene detection
Strains CC-LY736T, A. irakense BCRC 15764T and A. brasilense BCRC 12270T were able to reduce acetylene to ethylene with a mean value of 0.4, 0.8 and 21.4 nmol ethylene h−1 (108 cells) at 30 °C, respectively. The free-living nitrogen fixing activity was also compared with our previously described novel bacteria namely A. formosense CC-Nfb-7T (Lin et al. 2012), A. picis IMMIB TAR-3T (Lin et al. 2009) and A. rugosum IMMIB AFH-6T (Young et al. 2008), and the free-living nitrogen fixing activities were 25, 93 and 18 nmol ethylene h−1, respectively. It demonstrates that the nitrogen-fixing ability of strains CC-LY736T and A. irakense BCRC 15764T was related lower than the other free-living nitrogen-fixing Azospirillum species. Furthermore, the PCR amplicon of nifH gene in strain CC-LY736T was not observed by using the PolF/PolR or Zehrf/Zehrr primer sets in this study.
16S rRNA gene sequence analysis
The 16S rRNA gene sequence of strain CC-LY736T showed highest pair-wise similarity to A. irakense DSM 11586T (97.2 %), R. centenaria JCM 21060T (96.3 %), R. pekingensis JCM 11669T (96.1 %), A. rugosum DSM 19657T (91.8 %), A. doebereinerae DSM 13131T (91.6 %), A. formosense BCRC 80273T (91.6 %) and A. picis DSM 19922T (91.6 %). The bacterial genera such as Skermanella and Ochrobactrum shared relatively lower (<91.1 %) sequence similarity to strain CC-LY736T. The Neighbor-Joining phylogenetic tree based on 16S rRNA gene sequences showed that the type strains of Azospirillum largely constituted one subcluster within the family Rhodospirillaceae, except for A. irakense with A. amazonense, same result was also emphasized with Maximum Likelihood and Maximum Parsimony methods (Fig. 1). The pairwise 16S rRNA gene sequence similarity data of strain CC-LY736T and other representatives of the family Rhodopsirillaceae are shown in Table 2. Low similarity data (<92 %) obtained for Azospirillum representatives and strain CC-LY736T, A. amazonense BCRC 14279T and A. irakense BCRC 15764T suggested that none of those strains belonged to the genus Azospirillum. Therefore, further detailed studies were carried out to investigate most likely taxonomic positions of strain CC-LY736T , A. amazonense DSM 2787T and A. irakense DSM 11586T.
Phylogenetic analysis of Niveispirillum, Nitrospirillum and other representatives of the family Rhodospirillaceae based on 16S rRNA gene sequences. Distances and clustering were performed by using Neighbor-Joining method with the software package MEGA 5. Filled circles indicate that the corresponding nodes were also recovered in the tree constructed based on Maximum Likelihood and Maximum Parsimony algorithm. Bootstrap values (>50 %) based on 1,000 replications are shown at the branching points. Rhizobium alkalisoli LMG 24763T and Burkholderia sabiae LMG 24235T were used as outgroups. Bar, 0.02 substitutions per nucleotide position
Azospirillum-specific PCR detection
The genus-specific primer pair (Azo494-F/Azo756-R) selectively amplified 263 bp Azospirillum-specific fragments from representative strains of large Azospirillum subcluster (including A. oryzae JCM 21588T; A. brasilense BCRC 12270T; A. canadense LMG 23617T; A. doebereinerae DSM 13131T; A. fermentarium BCRC 80505T; A. formosense BCRC 80273T; A. halopraeferens DSM 3675T; 6, A. largimobile DSM 9441T; A. lipoferum BCRC 12213T; A. melinis LMG 23364T; A. picis DSM 19922T; A. rugosum DSM 19657T; A. thiophilum DSM 21654T; A. zeae LMG 23989T). The 263 bp DNA fragment was not detected in strains CC-LY736T, A. amazonense BCRC 14279T and A. irakense BCRC 15764T. These data also clearly suggested a possible earlier misclassification of A. amazonense BCRC 14279T and A. irakense BCRC 15764T.
DNA–DNA hybridization analysis
The DNA–DNA relatedness values of strain CC-LY736T with A. irakense BCRC 15764T was 51.2 ± 2.3 %. Strain CC-LY736T showed less DNA–DNA homology with A. irakense BCRC 15764T. Using the established molecular criteria for species-level relatedness described by Wayne et al. (1987), strain CC-LY736T shows less DNA–DNA homology with other closest related species, which supports its genomic distinction as a separate species.
Cellular fatty acid analysis
The FAME profiles in strain CC-LY736T and A. irakense BCRC 15764T were similar, both strains contain C16:1ω5c, C19:0 cyclo ω8c, C16:0 3-OH, C18:1 2-OH and C18:1 ω7c/C18:1 ω6c as major (>5 % of the total) fatty acids. Strain CC-LY736T and A. irakense BCRC 15764T were distinguished from genera Azospirillum, Rhodocista and Skermanella based on the FAME features. A. amazonense BCRC 14279T also accommodated similar major fatty acids of strain CC-LY736T and A. irakense BCRC 15764T except for C16:1ω5c, which was found in minor amounts in the former. On the other hand, FAME profile of A. lipoferum BCRC 12213T was distinct when compared to strain CC-LY736T, A. amazonense BCRC 14279T and A. irakense BCRC 15764T as it accommodated C17:1ω6c, C18:1 2-OH, summed features 2, 3 and 8 in major amounts. Similarly, members of the genus Rhodocista possessed C14:0, C16:1ω5c and summed feature 8 in major amounts. There were clear qualitative and quantitative differences in terms of several fatty acids within Rhodospirillaceae members, irrespective of the presence of summed feature 8 in significant (>25 %) amounts. The details of fatty acid profiles of strain CC-LY736T and other Rhodospirillaceae representatives are given in Table 3.
Cellular polar lipid analysis
Strain CC-LY736T contained phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), diphosphatidylglycerol (DPG), phosphatidylmethylethanolamine (PME), and phosphatidyldimethylethanolamine (PDE), which were in line with that of A. irakense BCRC 15764T. In addition, strain CC-LY736T contained two unidentified glycolipids (GL1–2) that were absent in A. irakense BCRC 15764T. Interestingly, A. amazonense BCRC 14279T possessed DPG and three unidentified aminolipids (AL1–3), three unidentified phospholipids (PL1–3), an unidentified glycolipid (GL) and an unidentified lipid (L) besides lacking other polar lipids that were found commonly in strain CC-LY736T and A. irakense BCRC 15764T. In contrast, other Azospirillum species (n = 4) possessed PC, PE, PG, PDE and AL in common, but consistently lacked PME. Similarly, members of the genera Rhodocista and Skermanella also lacked PME, but accommodated other polar lipids in significant amounts. Taken together, polar lipid profile clearly distinguished strain CC-LY736T, A. amazonense BCRC 14279T and A. irakense BCRC 15764T from each others as well as from other Rhodospirillaceae representatives. The details of polar lipid profiles of strain CC-LY736T and other Rhodospirillaceae representatives are given in Table 4 and Fig. S2.
Cellular polyamine analysis
The distribution of polyamines within various members of the family Rhodospirillaceae is given in Fig. S3. Irrespective of their quantitative variations, putrescine and sym-homospermidine were two important polyamines detected consistently in Rhodospirillaceae representatives. Strain CC-LY736T possessed major amounts of sym-homospermidine and moderate amounts of putrescine. However, A. irakense BCRC 15764T contained both sym-homospermidine and putrescine in predominant amounts. In contrast, A. amazonense BCRC 14279T accommodated major amounts of putrescine and moderate amounts of sym-homospermidine. The polyamine data presented here revealed that sym-homospermidine is most likely to be a predominant polyamine in Azospririllum species, whereas putrescine could be a major polyamine in the members of the genera Rhodocista and Skermanella.
Cellular respiratory quinone and DNA base composition
Strain CC-LY736T and the reference strains (except for Rhodocista, which possessed ubiquinone Q-9 as a major respiratory quinone) contained ubiquinone Q-10 as the predominant respiratory quinone. The DNA G+C content of strain CC-LY736T was 67.6 ± 0.1 mol %, a value that was well within the range (64.0–70.0 mol %) reported for the representatives of the family Rhodospirillaceae.
In conclusion, based on the phylogenetic, physiological, chemotaxonomic and phenotypic characteristics, strain CC-LY736T should be classified as a novel genus of the family Rhodospirillaceae, for which we propose the name Niveispirillum fermenti gen. nov. sp. nov.; the type strain of the type species is CC-LY736T (= BCRC 80504T = LMG 27263T). We also reclassify Azospirillum irakense as Niveispirillum irakense comb. nov., and Azospirillum amazonense as Nitrospirillum amazonense gen. nov., sp. nov.
Description of Niveispirillum gen. nov
Niveispirillum (Ni.ve.i.spi.ril’lum. L. adj. niveus snow-white; L. fem. n. spirillum spiral; N.L. neut. n. Niveispirillum snow-white spiral)
Cells are Gram-stain negative and show positive reaction for catalase and oxidase. The polyamine profile consists of sym-homospermidine and putrescine. The predominant (>30 %) fatty acids are C18:1 ω7c/C18:1 ω6c. The major polar lipids are DPG, PG, PC, PE, PME, PDE and an unidentified aminolipid (AL1). The predominant quinone is ubiquinone Q-10. The type species is Niveispirillum fermenti.
Description of Niveispirillum fermenti sp. nov
Niveispirillum fermenti (fer.men’ti. L. neut. gen. n. fermenti, of a fermentation process)
Cells are spiral-shaped, which are 0.6–0.8 μm in width and 2.0–2.3 μm in length and contain a monopolar flagellum. Colonies appear cream-colored, circular and smooth when grown on R2A agar. Growth occurs at temperature 20–40 °C, pH 5.0–9.0 and <2 % (w/v) NaCl. The accumulation of intracellular granules (PHB) is observed. In the GN2 Biolog MicroPlate, cells can utilize α-d-lactose, d-melibiose, β-methyl-d-glucoside, d-sorbitol, acetic acid, cis-aconitic acid, d-galactonic acid lactone, γ-hydroxybutyric acid, itaconic acid, succinic acid, succinamic acid, d-alanine, glycyl-l-aspartic acid, l-histidine, l-leucine, L-pyroglutamic acid, d-serine, l-serine, urocanic acid, putrescine, 2-aminoethanol, 2,3-butanediol, d,l-α-glycerol phosphate and α-d-glucose-1-phosphate as sole carbon source; other carbon sources are not utilized. Nitrate and nitrite are reduced. In the API ZYM strip, positive for alkaline phosphatase, acid phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin, α-chymotrypsin, naphthol-AS-BI-phosphohydrolase and β-glucosidase; negative for other reactions. The major fatty acids are C16:0, C16:1 ω5c, C19:0 cyclo ω8c, C18:1 ω7c/C18:1 ω6c, C16:0 3-OH and C18:1 2-OH. The polar lipids are DPG, PG, PC, PE, PME, PDE, AL1, PL and GL1-2. The predominant quinone is ubiquinone Q-10 and DNA G+C content of the type strain CC-LY736T is 67.6 ± 0.1 mol %.
Type strain is CC-LY736T (= BCRC 80504T = LMG 27263T) isolated from a fermentor in Taiwan.
Description of Niveispirillum irakense comb. nov
Niveispirillum irakense (i.ra.ken’se. N.L. neut. adj. irakense, pertaining to Iraq, where the organism was first isolated)
Gram-stain negative, aerobic, curved rods or S-shaped, motile with winding or snake-like movements. Catalase- and oxidase-positive. The growth temperature ranges from 20 to 35 °C, pH 5.5–8.5 (optimum pH is 6.5) and tolerates up to 3 % (w/v) NaCl. A monopolar flagellum is observed when the cells are grown in a liquid medium. Intracellular granules (poly-β-hydroxybutyrate) are observed. The predominant quinone is Q-10. In the GN2 Biolog MicroPlate, cells can utilize dextrin, adonitol, l-arabinose, d-arabitol, d-cellobiose, d-fructose, l-fucose, d-galactose, α-d-glucose, maltose, d-mannitol, d-mannose, l-rhamnose, sucrose, D-trehalose, acetic acid, citric acid, l-histidine, l-leucine, l-ornithine, l-proline, and d-serine as sole carbon source; other substrates are not utilized. Nitrate is reduced to nitrite. In the API 20NE system, cells assimilate d-glucose, l-arabinose, d-mannose, d-mannitol, N-acetyl-glucosamine, d-maltose, potassium gluconate, capric acid and malic acid. In the API ZYM system, positive for alkaline phosphatase, acid phosphatase, esterase (C4), esterase lipase (C8), leucine arylamidase, valine arylamidase, cystine arylamidase and trypsin. The DNA G+C content of the type strain KBC1T is 64–67 mol %.
Type strain is KBC1T (= ATCC 51182T = BCRC 15764T = CIP 103311T) isolated from rice rhizosphere in Iraq.
Description of Nitrospirillum gen. nov
Nitrospirillum (Ni.tro.spi.ril’lum. Gr. n. nitron soda; L. neut. n. spirillum spiral; N.L. neut. n. Nitrospirillum soda spiral)
Cells are Gram-stain negative, diazotrophic and show positive reaction for catalase and oxidase. The polyamine patterns include major amounts of putrescine and moderate amounts of sym-homospermidine. The predominant (>27 %) fatty acids are C18:1 ω7c/C18:1 ω6c. The polar lipid profile constitutes major amounts of DPG, three unidentified phospholipids (PL1–3) and two unidentified aminolipids (AL2–3). The predominant quinone is ubiquinone Q-10. The type species is Nitrospirillum amazonense.
Description of Nitrospirillum amazonense sp. nov
Nitrospirillum amazonense (a.ma.zon.en’se N.L. neut. adj. amazonense, pertaining to the Amazon region of Brazil)
Cells are spiral-shaped, which are 0.9–1.0 μm in width and 3.5 μm in length. Colonies appear cream-colored, circular and smooth when grown on R2A agar. Optimal growth occurs at temperature 35 °C, pH 5.7–7.8 and <3 % (w/v) NaCl. In the GN2 Biolog MicroPlate, cells are able to utilize α-cyclodextrin, dextrin, d-cellobiose, α-d-glucose, maltose and d-melibiose as sole carbon source; other carbon sources are not utilized. Nitrate and nitrite are not reduced. Assimilate d-glucose, l-arabinose and d-mannose. In the API ZYM strip, positive for esterase (C4), esterase lipase (C8), α-chymotrypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase and α-mannosidase; other reactions are negative. The major fatty acids are C16:0, C19:0 cyclo ω8c, C16:0 3-OH, C18:1 2-OH and C18:1 ω7c/C18:1 ω6c. The polar lipid profile constitutes DPG, three unidentified phospholipids (PL1–3) and three unidentified aminolipids (AL1–3), an unidentified lipid (L) and glucolipid (GL) each. The predominant quinone is ubiquinone Q-10 and DNA G+C content of the type strain Am14T is 67–68 mol %.
Type strain is Am14T (= ATCC 35119T = BCRC 14279T = DSM 3787T) was isolated from grass rhizosphere in Brazil.
Emended description of the genus Rhodocista Kawasaki et al. 1992
The genus description of Rhodocista is given by Kawasaki et al. (1992) with following amendments: both nitrate and nitrite are reduced. The predominant (>51 %) fatty acids are C18:1 ω7c/C18:1 ω6c. The major polar lipids are PC, PG, DPG, PE, PDE, PL and two unidentified aminolipids (AL2 and AL4). Polyamine profile contains major amounts of putrescine and moderate amounts of sym-homospermidine. The range of DNA G+C content is 68.8–70 mol %.
References
Bally R, Thomas-Bauzon D, Heulin T, Balandreau J, Richard C, De Ley J (1983) Determination of the most frequent N2-fixing bacteria in a rice rhizosphere. Can J Microbiol 29:881–887
Coenye T, Goris J, Spilker T (2002) Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol 40:2062–2069
Collins MD (1985) Isoprenoid quinone analysis in classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Döbereiner J, Day JM (1976) Associative symbioses in tropical grasses: characterization of microorganisms and dinitrogen-fixing sites. In: Newton WE, Nyman CJ (eds) Proceedings of the first international symposium on N2 fixation. Washington State University Press, Pullman, pp 518–538
Eckert B, Weber OB, Kirchhof G, Halbritter A, Stoffels M, Hartmann A (2001) Azospirillum doebereinerae sp. nov., a nitrogen-fixing bacterium associated with the C(4)-grass Miscanthus. Int J Sys Evol Microbiol 51:17–26
Edwards U, Rogall T, Blocker H, Emde M, Bottger EC (1989) Isolation and direct complete nucleotide determination of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res 17:7843–7853
Euzéby JP, Kudo T (2001) Corrigenda to the validation lists. Int J Sys Evol Microbiol 51:1933–1938
Falk EC, Döbereiner J, Johnson JL, Krieg NR et al (1985) Deoxyribonucleic acid homology of Azospirillum amazonense Magalhães, 1984 and emendation of the description of the genus Azospirillum. Int J Sys Bacteriol 35:117–118
Falk EC, Johnson JL, Baldani VLD, Döbereiner J, Krieg NR (1986) Deoxyribonucleic and ribonucleic acid homology studies of the genera Azospirillum and Conglomeromonas. Int J Sys Bacteriol 36:80–85
Favinger J, Stadtwald R, Gest H (1989) Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie Van Leeuwenhoek 55:291–296
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Sys Zool 20:406–416
GCG (1995). Wisconsin package version 8.1 program manual. Genetics Computer Group, Madison
Gest H, Favinger J (2001) Taxonomic ambiguities: a case history. Int J Sys Evol Microbiol 51:707–710
Hardy R, Burns RC, Holsten RD (1973) Application of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol Biochem 5:47–81
Heiner CR, Hunkapiller LK, Chen SM, Glass JI, Chen EY (1998) Sequencing multimegabase-template DNA using BigDye terminator chemistry. Genome Res 8:557–561
Kawasaki H, Hoshino Y, Kuraiski Y, Yamasato K (1992) Rhodocista centenaria gen. nov., sp. nov., a cyst-forming anoxygenic photosynthetic bacterium and its phylogenetic position in the Proteobacteria alpha group. J Gen Appl Microbiol 38:541–551
Kawasaki H, Hoshino Y, Yamasato K (1993) Phylogenetic diversity of phototrophic purple non-sulfur bacteria in the Proteobacteria group. FEMS Microbiol Lett 112:61–66
Khammas KM, Ageron E, Grimont PAD, Kaiser P (1989) Azospirillum irakense sp. nov., a new nitrogen-fixing bacterium associated with rice roots and rhizosphere soil. Res Microbiol 140:679–693
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Sys Evol Microbiol 62:716–721
Kirchhof G, Reis VM, Baldan JI, Eckert B, Döbereiner J, Hartmann A (1997) Occurrence, physiological and molecular analysis of endophytic diazotrophic bacteria in gramineous energy plants. Plant Soil 194:45–55
Koch B, Evans HJ (1966) Reduction of acetylene to ethylene by soybean root nodules. Plant Physiol 41:1748–1750
Ladha JK, So RB, Watanabe I (1987) Composition of Azospirillum species associated with wetland rice plants grown in different soils. Plant Soil 102:127–129
Lavrinenko K, Chernousova E, Gridneva E, Dubinina G, Akimov V, Kuever J, Lysenko A, Grabovich M (2010) Azospirillum thiophilum sp. nov., a novel diazotrophic bacterium isolated from a sulfide spring. Int J Sys Evol Microbiol 60:2832–2837
Lin S-Y, Young C-C, Hupfer H, Siering C, Arun AB, Chen W-M, Lai W-A, Shen F-T, Rekha PD, Yassin AF (2009) Azospirillum picis sp. nov., isolated from discarded tar. Int J Sys Evol Microbiol 59:761–765
Lin S-Y, Shen F-T, Young C-C (2011) Rapid detection and identification of the free-living nitrogen fixing genus Azospirillum by 16S rRNA-gene-targeted genus-specific primers. Antonie Van Leeuwenhoek 99:837–844
Lin S-Y, Shen F-T, Young L-S, Zhu Z-L, Chen W-M, Young C-C (2012) Azospirillum formosense sp. nov., a novel diazotrophic bacterium isolated from agricultural soil. Int J Sys Evol Microbiol 62:1185–1190
Lin S-Y, Liu Y-C, Hameed A, Hsu Y-H, Lai W-A, Shen F-T, Young C–C (2013) Azospirillum fermentarium sp. nov., a nitrogen-fixing species isolated from a fermenter. Int J Sys Evol Microbiol 63:3762–3768
Magalhães FM, Baldani JL, Souto SM, Kuykendall JR, Döbereiner J (1983) A new acid-tolerant Azospirillum species. An Acad Bras Cien 55:417–430
Mehnaz S, Weselowski B, Lazarovits G (2007a) Azospirillum canadense sp. nov., a nitrogen-fixing bacterium isolated from corn rhizosphere. Int J Sys Evol Microbiol 57:620–624
Mehnaz S, Weselowski B, Lazarovits G (2007b) Azospirillum zeae sp. nov., a diazotrophic bacterium isolated from rhizosphere soil of Zea mays. Int J Sys Evol Microbiol 57:2805–2809
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Sys Bacteriol 39:159–167
Miller LT (1982) Single derivatization method for routine analysis of bacterial whole-cell fatty acid methyl esters, including hydroxyl acids. J Clin Microbiol 16:584–586
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Murray RGE, Doetsch RN, Robinow CF (1994) Methods for general and molecular bacteriology. In: Gerhardt P, Murray RGE, Wood WA, Krieg NR (eds) Determination and cytological light microscopy. American Society for Microbiology, Washington, DC, pp 31–32
Ostle AG, Holt JG (1982) Nile blue A as a fluorescent stain for poly-β-hydroxybutyrate. Appl Environ Microbiol 44:238–241
Paisley R (1996) MIS whole cell fatty acid analysis by gas chromatography training manual. MIDI, Newark
Patriquin DG, Döbereiner J, Jain DK (1983) Sites and processes of association between diazotrophs and grasses. Can J Microbiol 29:900–915
Pfennig N, Lünsdorf H, Süling J, Imhoff JF (1997) Rhodospira trueperi, gen. nov., sp. nov., a new phototrophic Proteobacterium of the alpha group. Arch Microbiol 168:39–45
Poly F, Monrozier LJ, Bally R (2001) Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res Microbiol 152:95–103
Reinhold B, Hurek T, Fendrik I, Pot B, Gillis M, Kersters K, Thielemans S, Ley JD (1987) Azospirillum halopraeferens sp. nov., a nitrogen-fixing organism associated with roots of Kallar grass (Leptochloa fusca (L.) Kunth). Int J Sys Bacteriol 37:43–51
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical note 101, MIDI Inc, Newark
Scherer P, Kneifel H (1983) Distribution of polyamines in methanogenic bacteria. J Bacteriol 154:1315–1322
Schlegel HG, Lafferty R, Krauss I (1970) The isolation of mutants not accumulating poly-β-hydroxybutyric acid. Arch Microbiol 71:283–294
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tarrand JJ, Krieg NR, Döbereiner J (1978) A taxonomic study of the Spirillum lipoferum group, with descriptions of a new genus, Azospirillum gen. nov., and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol 24:967–980
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Trüper HG, Imhoff JF (1989) Bergey’s manual of systematic bacteriology. In: Staley JT, Bryant MP, Pfennig N, Holt JG (eds) Genus Rhodospirillum. Williams and Wilkins, Baltimore, pp 1662–1666
Watts D, MacBeath JR (2001) Automated fluorescent DNA sequencing on the ABI PRISM 310 genetic analyzer. Meth Mol Biol 167:153–170
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE et al (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Sys Bacteriol 37:463–464
Weon H-Y, Kim B-Y, Hong S-B, Joa J-H, Nam S-S, Lee K-H, Kwon S-W (2007) Skermanella aerolata sp. nov., isolated from air, and emended description of the genus Skermanella. Int J Sys Evol Microbiol 57:1539–1542
Xie CH, Yokota A (2005) Azospirillum oryzae sp. nov., a nitrogen-fixing bacterium isolated from the roots of the rice plant Oryza sativa. Int J Sys Evol Microbiol 55:1435–1438
Young C-C, Hupfer H, Siering C, Ho M-J, Arun AB, Lai W-A, Rekha PD, Shen F-T, Hung M-H, Chen W-M, Yassin AF (2008) Azospirillum rugosum sp. nov., isolated from oil-contaminated soil. Int J Sys Evol Microbiol 58:959–963
Zehr JP, McReynolds LA (1989) Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol 55:2522–2526
Zhang D, Yang H, Zhang W, Huang Z, Liu S-J (2003) Rhodocista pekingensis sp. nov., a cyst-forming phototrophic bacterium from a municipal wastewater treatment plant. Int J Sys Evol Microbiol 53:1111–1114
Acknowledgments
Authors would like to thank Mr. Wen-Shao Yen for the isolation of this novel bacterium. This research work was kindly supported by grants from the National Science Council, the Council of Agriculture, Executive Yuan and in part by the Ministry of Education, Taiwan, ROC under the ATU plan.
Author information
Authors and Affiliations
Corresponding author
Additional information
The GenBank/EMBL/DDBJ accession number for the 16S rRNA sequence of strain CC-LY736T is JX843283. In this study, the GenBank/EMBL/DDBJ accession number for the 16S rRNA sequence of strains Azospirillum amazonense BCRC 14279T, A. halopraeferens DSM 3675T, A. irakense DSM 11586T, A. lipoferum DSM 1691T, A. canadense LMG 23617T were GU256437, GU256439, GU256440, GU256441 and HM636056, respectively. A transmission electron micrograph of cells of strain CC-LY736T, polar lipid profile and polyamine compositions of all strains used in this study are available as supplementary material with the online version of this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lin, SY., Hameed, A., Shen, FT. et al. Description of Niveispirillum fermenti gen. nov., sp. nov., isolated from a fermentor in Taiwan, transfer of Azospirillum irakense (1989) as Niveispirillum irakense comb. nov., and reclassification of Azospirillum amazonense (1983) as Nitrospirillum amazonense gen. nov. Antonie van Leeuwenhoek 105, 1149–1162 (2014). https://doi.org/10.1007/s10482-014-0176-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0176-6