Abstract
This study was conducted to explore fungal endophyte communities inhabiting a toxic weed (Stellera chamaejasme L.) from meadows of northwestern China. The effects of plant tissue and growth stage on endophyte assemblages were characterized. Endophytes were recovered from 50 % of the samples, with a total of 714 isolates. 41 operational taxonomical units (OTUs) were identified, consisting of 40 OTUs belonging primarily to Ascomycota and 1 OTU belonging to Basidiomycota. Pleosporales and Hypocreales were the orders contributing the most species to the endophytic assemblages. The total colonization frequency and species richness of endophytic fungi were higher in roots than in leaves and stems. In addition, for the plant tissues, the structure of fungal communities differed significantly by growth stages of leaf emergence and dormancy; for the plant growth stages, the structure of fungal communities differed significantly by plant tissues. This study demonstrates that S. chamaejasme serves as a reservoir for a wide variety of fungal endophytes that can be isolated from various plant tissues.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fungal endophytes commonly infect host plants asymptomatically and reside within internal tissues of living plants (Hyde and Soytong 2008). Recent studies of a broad range of plant species have demonstrated that endophytic fungi were ubiquitous in various geographic and climatic zones and have been found within all examined plants (Arnold 2007; Ghimire et al. 2011; Rivera-Orduña et al. 2011). It has been shown that some endophytic fungi may protect their host plants from fungal pathogens, insect pests and nematodes (Martin et al. 2012; Menjivar et al. 2012). Several studies have demonstrated that some endophytic fungi have the ability to increase the fitness of plant hosts by enhancing their resistance to abiotic (Sieber 2007; Hyde and Soytong 2008) and biotic stress (Arnold et al. 2003). Although interest in the ecological roles of endophytic fungi has increased in recent years, it has been hampered by little sampling and lack of characterization of fungal diversity (Hyde et al. 2007).
Much attention is now being paid to endophytic biodiversity, chemistry and bioactivity of endophytic metabolites, as facets of the relationship between endophytes and their host plants (Schulz et al. 2002; Tao et al. 2008). While, there is little information on whether culturable endophyte communities change seasonally, the limited evidence suggests that this might occur in both roots and leaves of plant (Quilliam and Jones 2010). Furthermore, the effect of seasonal changes on endophytic fungi remains unclear. Seasonal patterns have been detected among taxa that were not host specific, but other studies could not confirm any seasonality among endophytes (Widler and Müller 1984; Sieber and Hugentobler 1987; Joshee et al. 2009). Moreover, endophytic fungi colonize all plant parts such as roots, stems, leaves, bark and floral organs. The plant-associated habitat is a dynamic environment in which many factors affect the structure and composition of species that colonize different tissues (Rivera-Orduña et al. 2011). Surveys of endophytic communities in both above and below ground plant organs simultaneously are extremely limited so far (Saunders and Kohn 2009; Wearn et al. 2012).
Fungal endophytes have been recognized as a potential repository of novel natural products for pharmaceutical, agricultural and industrial uses (Hyde and Soytong 2008). Secondary metabolites extracted from plants are usually considered to have been produced by the plant. However, some of these metabolites may in fact have been produced by endophytic fungal colonizers in the plant (Stierle et al. 1993; Mishra et al. 2012). So, endophytic fungi are good sources of novel secondary metabolites that possess antibacterial, antifungal and anticancer properties (Ding et al. 2008; Kharwar et al. 2009; Kumara et al. 2012). In recent years, there has been particular emphasis on studying endophytes of medicinal plants in order to discover novel compounds (Zhou et al. 2007; Huang et al. 2009).
Stellera chamaejasme L., a toxic perennial weed, is a multi-stemmed herbaceous plant belonging to the family Thymelaeaceae. It has a wide geographical range from southern Russia to northern China and Mongolia, southwards as far as to the dry regions of the western Himalayas, the Tibetan Plateau and southwestern China (Zhang et al. 2011). S. chamaejasme has spread in recent decades and is now abundant in northern and western China (Sun et al. 2009; Zhang et al. 2011), and has become one of the most serious weeds damaging ecological balance of grasslands (Sun et al. 2009). It has increased the grassland degradation and the desertification process, and caused food poisoning of cattle due to eating stems and leaves. S. chamaejasme is listed as one of the most serious weeds in China (Sun et al. 2009; Zhang et al. 2011). Previous studies on this toxic weed in our lab indicated that root extracts of S. chamaejasme showed significant nematicidal activity (Guo et al. 2011), and several purified metabolites that were extracted from roots of S. chamaejasme exhibited phytotoxic activities (Yang et al. 2011). Thus, to learn if any endophytic fungi may be producing one or more of a plethora of bioactive substances known from S. chamaejasme, it would first be crucial to do a systematic study on the endophytic fungi of this plant and to know their composition and diversity in both above (leaves and stems) and below ground (roots) plant organs in the whole growing seasons. In addition, it is important to determine the species that can be cultured because this allows for the subsequent manipulation of endophytic fungi activity and natural products extraction (Wearn et al. 2012).
The aim of the present study was to characterize endophytic fungi in leaves, stems and roots of S. chamaejasme over the course of an entire growing season. There were two specific objectives: one was to isolate and taxonomically identify leaf, stem and root-associated fungal endophytes of S. chamaejasme; the other is to evaluate these endophyte communities over the course of a growing season to provide insight into symbiont community dynamics. We used a traditional technique to isolate the endophytes, and a combination of morphological and molecular methods to identify these fungal isolates (Wang et al. 2005). The study will create a reservoir of strains that may be assayed for potential applications. To the best of our knowledge, this is the first report of the endophyte community associated with S. chamaejasme.
Materials and methods
Sampling site and procedure
The plant samples were collected at the Cuiying mountain area (35°56′N, 104°08′E) in the Yuzhong campus of Lanzhou University, province of Gansu (Northwest China). It covers 201 ha with loessial soil, and the current vegetation is dominated mainly by Melica przewalskyi, Agropyron cristatum, Artemisia frigida and S. chamaejasme. The climatic characteristics of the study area can be classified as temperate, semi-arid, continental climate with a mean annual temperature of 6.7 °C and mean annual precipitation of 381.8 mm. The average elevation of various sampling sites is 1,965.8 m above sea level (Yang et al. 2010).
The developmental stages of S. chamaejasme at Cuiying mountaintop were leafing (leaf emergence) in early May (LE), flowering (full bloom) in early June (FB), fruiting (seed maturation) in mid-July (SM) and dormancy in early November until the following March (D). To study the fluctuation of endophyte communities with different developmental stages of S. chamaejasme, plant samples were collected in the middle of May, June and July of 2011, and mid-January of 2012 according to the distribution patterns of S. chamaejasme.
Healthy, symptomless leaves (L), stems (St) and roots (R) were collected from six individual plants of S. chamaejasme at each development stage, the distance between single plants varied from 30 to 50 m depending on the distribution within the vegetation cover. One stem (length, 10–15 cm; diameter, 1–2 mm) with leaves and two root segments (length, 8–12 cm; diameter, 2–4 mm) were randomly collected from each plant in each of the four growth stages, respectively. All root samples were taken at a depth of at least 15–20 cm below ground level. These plant samples were immediately brought to the laboratory in ice boxes, stored at 4 °C and processed within 48 h.
Isolation and identification of endophytic fungi
The sampling regime was designed with the intention of isolating as many endophyte species as possible from the different tissues samples. Samples were washed thoroughly in running tap water, rinsed with distilled water, and surface-sterilized according to Zamora et al. (2008). Samples were dipped in 70 % ethanol for 1 min, immersed in aqueous solution of NaOCl (2 % available chlorine) for 5 min (leaves) and 8 min (stems and roots). The samples were then rinsed in double-distilled sterile water three times and dried under aseptic condition. Each leaf sample (leaves were removed from stem), stem and root was cut into 20 segments, measuring approximately 5 × 5 mm for leaves or 5 mm in length for stems and roots. In total, 1,440 segments (20 segments × 3 tissues × 6 individual plants × 4 growth stages) were used in this study. Sets of 20 segments were then evenly placed in a 90 mm Petri dish containing potato dextrose agar (PDA) supplemented with streptomycin (200 mg l−1) to suppress bacterial growth. The effectiveness of the surface sterilization was controlled by making imprints of disinfected sample fragments on PDA plates (Schulz et al. 1998). Petri dishes were sealed, incubated for 45–60 days at 25 ± 1 °C, and examined periodically.
When colonies developed, actively growing fungal mycelia were transferred to new PDA plates for purification and identification. Fungal isolates were selected and grouped together according to morphological characters such as spore production, spore length and morphology, aerial mycelium color, texture and form, exudates and the growth rate (Sun et al. 2011), but in addition, specimens of most isolates were also identified with internal transcribed spacer (ITS) region sequences. All isolates are maintained in Key Laboratory of Chemistry of Northwestern Plant Resources, Lanzhou Institute of Chemical Physics, Chinese Academy of Sciences.
DNA extraction, PCR amplification, and sequencing
Genomic DNA was extracted from fresh cultures following the protocol of Sim et al. (2010). The ITS (ITS1, 5.8S, ITS2) region was amplified using primer pairs ITS4 and ITS5 (White et al. 1990). PCR reactions were set-up using the following ingredients for each 50 μl reaction: PCR buffer (20 mM KCl, 10 mM (NH4)2SO4, 2 mM MgCl2, 20 mM Tris–HCl, pH8.4), 200 μM of each deoxyribonucleotide triphosphate, 15pmols of each primer, a maximum of 25 ng μl−1 of template DNA, and 2.5 units of Taq DNA polymerase (Biocolor BioScience & Technology Company, Shanghai, China). PCR amplifications were performed using the following conditions: 4 min initial denaturation at 94 °C, then 1 cycle consisting of denaturation (1 min at 94 °C), annealing (20 s at 55 °C), extension (50 s at 72 °C), and followed by 37 cycles of 20 s at 94 °C, 20 s at 55 °C, 50 s at 72 °C, and a final 4 min at 72 °C. A negative control using deionized water instead of template DNA was included in each batch of the PCRs. PCR products were checked electrophoretically in a 0.8 % (W V−1) agarose gel. The band of size approximately 550 bp in the electrophoresis pattern was excised and purified with the DNA Fragment Quick Purification/Recover Kit (Dingguo, Beijing, China) following the protocol. Purified PCR products were directly sequenced with primer pairs as mentioned above in the ABI-Prism 377 automated sequencer (Applied Biosystems, Inc. USA).
Taxonomic assignments and phylogenetic analysis
The ITS sequences were checked for chimeras using the CHECK_CHIMERA PROGRAM (Cole et al. 2007) and Pintail (Ashelford et al. 2005) programs. Assembly and editing of good sequences were performed using BioEdit Software (Hall 1999). Operational taxonomic units (OTUs) were defined with the MOTHUR program (http://www.mothur.org/wiki/Silva_reference_alignment). The uncorrected pairwise distances between aligned sequences were calculated and the OTUs were defined using the furthest neighbour-clustering algorithm at evolutionary distances of 3 % (that is on the basis of 97 % similarity between sequences), and a representative sequence was chosen from each OTU by a majority decision (Schloss et al. 2009). A representative sequence for each OTU was used as query sequence to search for similar sequences in GenBank with BLASTN program to provide at least tentative identification for the fungus. A value of 97 % ITS region identity was used as a DNA barcoding threshold (O’Brien et al. 2005). The ITS sequence of a representative in each OTU was aligned in ClustalW for phylogeny analysis. The alignments were further used for phylogenetic trees construction using MEGA software version 5.0 (Tamura et al. 2011). The neighbor-joining (NJ) method was used to infer the evolutionary history of the fungal isolates and the bootstrapping was carried out using 1,000 replications. All of the sequences have been submitted to the GenBank. The accession numbers are the following; sequences from leaf samples: KC292821–KC292843, stem samples: KC292886–KC292914, root samples: KC292844–KC292885.
Data analysis
The colonization frequency (CF) was calculated as the total number of segments colonized by endophytic fungi divided by the total number of incubated segments. The relative species frequency (RF) was calculated as the number of isolates of one species divided by the total number of isolates (Yuan et al. 2010).
The Shannon–Weaver diversity index (H′) (Shannon and Weaver 1963) was used to show diversity of the endophytic fungal species and was calculated as: H′ =−∑p i ln p i. Shannon evenness (E) was calculated as H′/H max, where H max = ln(S), S is the total number of OTUs in the subsample. Simpson’s diversity (D = 1 − ∑p 2i , where p i is the proportion of isolates assigned to the ith OTU, that is P i = n i/N) (Simpson 1964) was calculated to compare the species richness. A Simpson diversity index is close to 1 means that the sample is highly diverse.
Difference in Shannon diversity indices of any two fungal communities were compared using a Student’s t test at a 95 % confidence level. Differences were considered statistically significant at p < 0.05 (Kutorga et al. 2012). Principal component analysis (PCA) was conducted to explore the compositions of endophytic fungal communities at different growth stages. Principal component models were built with full leverage correction and based on percentage abundance of endophytic fungal isolates at each growth stage. The statistical significance for PCA was assessed by analysis of variance of PCA scores among different growth stages (Beraldi-Campesi et al. 2012). The statistical analyses were performed using SPSS version 17.0 (SPSS Inc., Chicago, IL, USA).
Results
Isolation and identification of endophytic fungi
A total of 714 endophyte strains were isolated from 1,440 plant tissue segments of S. chamaejasme during LE, FB, SM and D growth stages. Of these strains, 224 were recovered from leaves, 236 from stems and 254 from roots. These fungal strains were sorted into 91 OTUs (22 OTUs for leaves, 29 OTUs for stems, and 40 OTUs for roots). The colonization rate of endophytic fungi was higher in roots (53 %) than in leaves (47 %) and stems (49 %). The colonization rate of endophytic fungi was significantly lower at LE stage (26 %) than at FB (56 %), SM (57 %) and D (60 %) stages.
The fungal isolates were mainly composed of Ascomycota (711, 99.6 % of the isolates), including a high relative abundance of Sordariomycetes (303, 42.6 %, 38 OTUs) and Dothideomycetes (240, 33.8 %, 30 OTUs). The unclassified Ascoymcota (53 isolates, 5 OTUs), Eurotiomycetes (45 isolates, 10 OTUs), Leotiomycetes (36 isolates, 4 OTUs) and Mucoromycotina (34 isolates, 3 OTUs) constituted only 7.5, 6.3, 5.1 and 4.8 %, respectively, of the isolated ascomycetes (Table 1). A single basidiomycete (Agaricomycetes, 3 isolates, 1 OTUs) was identified and the OTU was detected in 0.4 % of the total samples (Table 1). In the entire dataset, Alternaria (dominated in leaves and stems) and Fusarium (dominated in roots) were the most frequently isolated genera. Fusarium acuminatum was the most abundant taxon, followed by Alternaria sp., Ascomycota sp., Dothideomycetes sp. and Alternaria tenuissima.
Endophyte community in leaf, stem and root tissues
The leaf endophyte community was composed of 224 fungal isolates that belong to 22 OTUs (Table 2; Fig. 1a). The representation of individual species in the community was variable, as some species were recovered frequently in two or three collections (e.g. Alternaria tenuissima, Alternaria alternata, Ascomycota sp., Dothideomycetes sp., Fusarium acuminatum) and others were recovered infrequently and/or from a single collection (e.g. Alternaria brassicae, Aporospora terricola, Fusarium sporotrichioides, Sordaria lappae). The number of isolates recovered from different growth stages varied, with 49, 34, 67 and 74 isolates obtained at LE, FB, SM and D stage, respectively. At LE stage, the fungal isolates were grouped into 4 OTUs, the majority of the isolates were Alternaria tenuissima (65.3 %), followed by Alternaria alternata (22.4 %). At FB stage, the fungal isolates were also grouped into 4 OTUs, the majority of the isolates were Dothideomycetes sp. (35.3 %) and Sporormiella sp. (35.3 %), followed by Alternaria solani (17.6 %) and A. tenuissima (11.8 %). At SM stage, the fungal isolates were grouped into 7 OTUs, the majority of the isolates were Fusarium acuminatum (35.8 %), followed by Acremonium exuviarum (20.9 %) and Ascomycota sp. (14.9 %). The genus of Fusarium was predominant in the leaf tissues at SM stage. At D stage, the fungal isolates were grouped into 7 OTUs, the majority of the isolates were Fusarium torulosum (36.5 %), followed by Fusarium acuminatum (16.2 %) and Dothideomycetes sp. (14.9 %). As for SM stage, the genus of Fusarium was predominant in the leaf tissues.
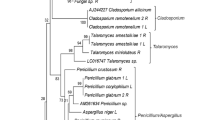
Phylogenetic tree constructed using the neighbor-joining method based on sequences of the ITS region showing closest relatives of endophytic filamentous fungi isolated from the leaves (a), stems (b) and roots (c) of S. chamaejasme at different growth stages. Bootstrap values (≥50 %) from 1,000 replicates are shown. The culture collection accession numbers of all reference strains are shown. The number in parentheses of each isolate indicates accession number of the gene sequences deposited in the GenBank sequence library, and the relative proportions of the fungal isolates are also presented (LE leaf emergence; FB full bloom; SM seed maturation; D dormancy; l leaves; st stems; r roots.)
The stem endophyte community was composed of 236 fungal isolates that belong to 29 OTUs (Table 2; Fig. 1b). As for the leaf tissues, a number of fungal isolates obtained from stem tissues varied among growth stages. The highest number of isolates were obtained from D stage (n = 82) and the least from LE stage (n = 18). A total of 67 and 69 isolates were obtained from FB and SM stages, respectively. As observed for leaf, the representation of individual species in the stem endophyte community was variable. At LE stage, the fungal isolates were grouped into 4 OTUs, the majority of the isolates were Botryotinia fuckeliana (61.1 %), followed by Aporospora terricola (16.7 %). The genus of Botryotinia was predominant in the stem tissues at LE stage. At FB stage, the fungal isolates were also grouped into 4 OTUs, the majority of the isolates were Alternaria sp. (82.1 %), followed by Dothideomycetes sp. (9.0 %). The genus of Alternaria was predominant in the stem tissues at FB stage. At SM stage, the fungal isolates were grouped into 17 OTUs, the majority of the isolates were Ascomycota sp. (20.3 %), followed by Fusarium cf. acuminatum (14.5 %) and Fusarium avenaceum (7.2 %). The genus of Fusarium was predominant in the leaf tissues at SM stage. At D stage, the fungal isolates were grouped into 4 OTUs, the majority of the isolates were Fusarium acuminatum (51.2 %), followed by Ascomycota sp. (24.4 %) and Dothideomycetes sp. (22.0 %). As for SM stage, the genus of Fusarium was predominant in the stem tissues.
The root endophyte community was composed of 254 fungal isolates that belong to 40 OTUs (Table 2; Fig. 1c). As for the leaf and stem tissues, a number of fungal isolates were obtained varied among growth stages. The highest number of isoltes were obtained from FB stage (n = 102) and the least from LE stage (n = 25). A total of 68 and 59 isolates were obtained from SM and D stages, respectively. As observed for leaf and stem, the representation of individual species in the root endophyte community was variable. Bionectria ochroleuca, Fusarium solani, Penicillium chrysogenum, etc. were recovered frequently in two or four collections, and Eucasphaeria capensis, Penicillium lanosum, Scytalidium circinatum, etc. were recovered infrequently and/or from a single collection. At LE stage, the fungal isolates were grouped into 11 OTUs, the majority of the isolates were Eupenicillium sp. (24.0 %) and Fusarium oxysporum (24.0 %). The genus of Fusarium was predominant in the root tissues at LE stage. At FB stage, the fungal isolates were also grouped into 10 OTUs, the majority of the isolates were Fusarium oxysporum (27.5 %), followed by Geomyces sp. (16.7 %). As for LE stage, the genus of Fusarium was predominant at FB stage. At SM stage, the fungal isolates were grouped into 14 OTUs, the majority of the isolates were Neonectria sp. (14.7 %) and Penicillium commune (14.7 %), followed by Penicillium chrysogenum (11.8 %) Cadophora sp. (10.3 %). The genus of Penicillium was predominant in the root tissues at SM stage. At D stage, the fungal isolates were grouped into 5 OTUs, the majority of the isolates were Mucor hiemalis (37.3 %), followed by Fusarium solani (25.4 %) and Fusarium acuminatum (18.6 %). The genera of Mucor and Fusarium were predominant in the root tissues at D stage.
Diversity of endophytic fungi
The species diversity indices for different fungal communities were presented in Table 1. There was a higher species richness of endophytic fungi in roots than in leaves and stems. The Shannon diversity indices and Shannon evenness indices were the highest in roots and lowest in leaves. The Simpson’s diversity index was the highest in roots and lowest in stems.
The leaf fungal community from D stage had the highest species diversity whereas the least diversity was found in LE stage community. Species diversity of leaf-associated fungi was rather similar between SM and D stages (P > 0.500), but all the other communities from leaf differed in species diversity (P < 0.001) (Table 2). The stem fungal community from SM stage had the highest species diversity whereas the least diversity was found in FB stage community. All pairwise comparisons from stem fungal community differed in species diversity except between LE and D stage communities (Table 2). As for root fungal community, SM stage had the highest species diversity and D stage had the least. Species diversity were similar between LE and FB stages, and between LE and SM stages (P > 0.100), the remaining pairwise comparisons from root fungal community differed in species diversity (Table 2).
All fungal communities from leaf, stem and root tissues at different growth stages differed in species diversity except between stem and root communities at SM stage (P > 0.100) (Table 2). Species diversity was rather similar between leaf-associated fungi and stem-associated fungal community (P > 0.500), but both differed from that found in root-associated fungal community (Table 2).
PCA analysis
Results from PCA of different growth stages based on fungal community composition of leaves, stems and roots of S. chamaejasme were presented in Fig. 2. The first two principal components (PCs) represent 79.43 % of the variance. A two-dimensional scores plot was performed, from the X axis defined by PC1 (47.05 %), the scores for each of the three tissue samples from the same growth stage were different. The score for stem sample at LE stage was lower (0.11) than the similar scores for leaf sample (0.74) and root sample (0.76), while at D stage, the score for leaf sample was higher (0.94) than the similar scores for stem sample (0.56) and root sample (0.36). The scores for leaf, stem and root samples from FB and SM stages were similar. Leaf samples and root samples were easily distinguished from the Y axis, defined by PC2 (32.38 %), since the scores of the root samples at four different growth stages were all more than zero, whereas those of leaf samples were all less than zero. The score for stem samples were divided into two groups, one group included LE and FB stages, the scores were less than zero; the other group included SM and D stages, the scores were more than zero (Fig. 2). PCA showed that, for the plant organs, the structure of endophytic fungal communities differed significantly by growth stages of LE and D; for the plant growth stages, the structure of endophytic fungal communities differed significantly by plant organs (Fig. 2).
Principal component analysis (PCA) of all tissue samples of S. chamaejasme at different growth stages. X axis was defined by PC1 which represented 47.05 % of the total variance and Y axis was defined by PC2 which represented 32.38 % of the total variance (LE leaf emergence; FB full bloom; SM seed maturation; D dormancy; l leaves; st stems; r roots.)
Discussion
This study constitutes the first research on the endophytic fungal community of S. chamaejasme. The study’s analysis of endophytic diversity was performed at four growth stages characterized by different developmental stages and seasonal variation. The fungal communities were studied with a cultivation-based approach on samples from three different plant tissues, the leaves, stems and roots. The isolated fungal species were identified using molecular methods. The culture method was chosen in order to be in a position to conduct experiments on the ecological role of these fungi (especially for their ability to produce natural secondary metabolites which can be applied as biocontrol agents) (Wearn et al. 2012).
The total colonization frequency and species richness of endophytic fungi were higher in the below ground part (roots) than in the aboveground parts (leaves and stems) in our study. One likely reason is soil-borne fungi are typically much more prevalent and diversified than those that infect aerial plant tissues (Ghimire et al. 2011). The other reason is the main sources of easily accessible substrate are roots, and roots might be considered as a relatively stable environment adequate for many fungal species (Garbeva et al. 2004). It may be that the roots of S. chamaejasme release a wide variety of compounds/exudates into the surrounding soil and create a nutrient rich environment favoring fungal survival and coexistence in roots organs. Similar results have been reported in previous endophytes studies (Ghimire et al. 2011; Angelini et al. 2012). However, this point has disagreement with some other studies. For example, Mishra et al. (2012) reported that frequency of colonization and species richness of fungal communities associated with Tinospora cordifolia were highest in the leaves and lowest in the roots. In Holcus leaves and roots, the number of species identified and the Shannon index were very similar for both organs (Sánchez Márquez et al. 2010). In our study, the total colonization frequency and species richness of endophytic fungi were slightly higher in the stems than that in the leaves. This is in agreement with other previous studies (Guo et al. 2008; Sun et al. 2012). This may be due to the structure and substrates are different between stem and leaf tissues, which influence the infection of endophytic fungi (Rodrigues 1994), or that the stems are more permanent than leaves, so that the stems accumulate more fungal species (Guo et al. 2008; Sun et al. 2012). In addition, the relative abundance of each endophytic species identified in S. chamaejasme reflects a very unequal distribution of isolate richness among species. Such unequal distributions of endophytic fungal species have been reported in other plants (Sánchez Márquez et al. 2010; Ghimire et al. 2011).
The isolation-based study and molecular identification showed that the total fungal diversity was relatively high, consisting of 40 OTUs belonging primarily to the Ascomycota (99.6 % of the total number of isolates). Only a few basidiomycota isolates were obtained, and represented by a single OTU. This finding is consistent with the results of previous studies in which a predominance of ascomycetes were reported as a characteristic of endophytic mycobiota (Rakotoniriana et al. 2008; Angelini et al. 2012). Basidiomycetes are generally recorded only rarely as stem endophytes. The single basidiomycete species (Schizophyllum commune, 0.4 % of the total number of isolates) in our study was found only associated with the stem tissues at D stage.
Pleosporales (25.6 % of the total number of isolates) and Hypocreales (42.4 % of the total number of isolates) were the orders contributing the most species to the endophytic assemblages of S. chamaejasme. This result was consistent with the previous studies in many semiarid plants (Porras-Alfaro et al. 2007; Herrera et al. 2010). It seems that these fungi form important, intimate and long lasting relationships with S. chamaejasme.
In the present study, Alternaria was a dominant genus associated the leaf and stem tissues of S. chamaejasme. Although species of Alternaria are mainly saprophytes that are commonly found in soil or on decaying plant tissues, many recent studies have demonstrated that species belonging to the Alternaria genus are true endophytes associated with temperate and tropical plants (Morakotkarn et al. 2006) and an Antarctic grass (Deschampsia antarctica) (Rosa et al. 2009). The genus Fusarium represented the most abundant endophytic fungi recovered from the roots of S. chamaejasme at almost all growth stages, but was also present in leaf and stem tissues at FB and SM stages. The species of Fusarium often are found among the most frequently isolated endophytes in tropical plants (Vega et al. 2010). The plant tissue becomes highly resistant to subsequent invasions by other fungi when occupied by species of Fusarium. This was be because it is a protected niche in which the primary colonizer has an increased capability to resist replacement by other endophytic fungi (Lockwood 1981).
The aboveground parts (leaf and stem) and belowground part (root) are the interfaces which interact most dynamically with the environment due to the very different ecological niches (different water content, organic matter, light and UV radiation levels) they represent. It is reasonable to expect that different plant organs are inhabited by distinctive endophytic communities (Arnold 2007; Angelini et al. 2012). As expected, tissue specificity of endophytes was demonstrated in this present study. For example, Sporormiella sp. and Sordaria lappae were only isolated from leaves; Botryotinia fuckeliana, Schizophyllum commune, Dothiorella iberica and Cadophora malorum were only isolated from stems; and Bionectria ochroleuca, Ilyonectria sp., and Mucor sp. were only isolated from roots. Some previous studies in other plants have also indicated that endophytes exhibit tissue specificity (Rodrigues 1994; Kumar and Hyde 2004; Sun et al. 2012). Particular endophytes found only in one tissue suggest that certain species survive within a specific chemistry or texture of different tissues (Rodrigues 1994).
Endophyte assemblages were also influenced by growth stages of S. chamaejasme in our study. There were not only differences in colonization frequency and species richness (Table 1), but also differences in fungi in certain orders (Table 2) throughout the growing season. The variations observed indicate that for most endophytes, individual infections are not persistent. Ongoing turnover of endophyte populations within a single plant organ may occur (Joshee et al. 2009). One possibility for this variation may be that there are very few endophytes in newly developed leaves and stems of S. chamaejasme, but as the leaves and stems mature, the density and diversity of fungi increase. Over a period of several months, fungal diversity declines, but density continues to increase. In addition, the root tissues of S. chamaejasme remain more persistent throughout the growing season, while leaf and stem tissues of senescing or pre-senescents have lower nutrient content, which could be less suitable for endophytic growth. The other reason for this variation may be environmental factors, such as temperature, humidity, and ecological niches. These factors may determine spread and germination success of endophytic fungal spores (Schulthess and Faeth 1998).
Endophytic fungi are considered potential sources of novel antimicrobials metabolites after the discovery of taxol from Tinospora andreanae, an endophyte of Taxus brevifolia (Stierle et al. 1993; Mishra et al. 2012). Species of the genera Chaetomium, Penicillium and Streptomyces which were isolated from Oryza sativa exhibited antifungal activity against various phytopathogens (Shankar et al. 2007). Alternaria sp., Nigrospora oryzae and Papulospora sp. which were isolated from medicinal plants showed inhibitory activity against both Gram positive and Gram negative bacteria and also against Candida albicans (Raviraja et al. 2006). Endophytic Penicilllium spp. are well known to produce a plethora of secondary metabolites with antibiotic, anticancer and/or cytotoxic activity (Wang et al. 2010; Mishra et al. 2012). Some endophytic Penicillium species also produce plant growth hormones such as gibberellins (Hamayun et al. 2010). At present, more than 150 bioactive compounds were isolated during a screening of 6,500 endophytic fungi, and more than 50 % of these compounds were significantly active against various pathogenic microbes (Hyde and Soytong 2008). The endophytes isolated from S. chamaejasme in this study offer an excellent platform for the discovery of novel bioactive (antibacterial, antifungal, anticancer and cytotoxic) compounds.
Although the ITS region is typically variable and rich in informative sites and has the highest probability of successful identification for the broadest range of fungi, it is unlikely that a single-marker barcode system will be capable of identifying every specimen or culture to species level. Furthermore, the limitations of ITS sequences for identifying species in some groups and the failure of the universal ITS primers to work in a minority of other groups will have to be carefully documented (Oechsler et al. 2009; Schoch et al. 2012). Taking these factors into account, the species level identification of Fusarium isolates from ITS sequences were tentative, and phylogenetic species were recognized if they received ≥50 % bootstrap values from 1,000 replicates in this study.
This study shows that S. chamaejasme harbours a wide diversity of fungal endophytes, and provides a reservoir of strains for the search of bioactive secondary metabolites which can be screened against different drugable targets. Based on this purpose, the fungal communities were determined using a culture-based technique. However, this approach may have overlooked the presence of uncluturable or slow-growing species (Hyde and Soytong 2008). Culture-independent (direct molecular analysis) approaches can be used to more comprehensively and systematically explore the fungal endophytes associated with S. chamaejasme.
References
Angelini P, Rubini A, Gigante D, Reale L, Pagiotti R, Venanzoni R (2012) The endophytic fungi communities associated with the leaves and roots of the common reed (Phragmites australis) in lake Trasimeno (Perugia, Italy) in declining and healthy stands. Fungal Ecol 5:683–693
Arnold AE (2007) Understanding the diversity of foliar endophytic fungi: progress, challenges, and frontiers. Fungal Biol Rev 21:51–66
Arnold AE, Mejia LC, Kyllo D, Rojas EI, Maynard Z, Robbins N, Herre EA (2003) Fungal endophytes limit pathogen damage in a tropical tree. Pros Natl Acad Sci USA 100:15649–15654
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microb 71:7724–7736
Beraldi-Campesi H, Arenas-Abad C, Garcia-Pichel F, Arellano-Aguilar O, Auqué L, Vázquez-Urbez M, Sancho C, Osácar C, Ruiz-Velasco S (2012) Benthic bacterial diversity from freshwater tufas of the Iberian Range (Spain). FEMS Microb Ecol 80:363–379
Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35 (database issue):D169–D172
Ding G, Liu S, Guo L, Zhou Y, Che Y (2008) Antifungal metabolites from the plant endophytic fungus Pestalotiopsis foedan. J Nat Prod 71:615–618
Garbeva P, Van Veen JA, Van Elsas JD (2004) Microbial diversity in soil: selection of microbial population by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270
Ghimire SR, Charlton ND, Bell JD, Krishnamurthy YL, Craven KD (2011) Biodiversity of fungal endophyte communities inhabiting switchgrass (Panicum virgatumL.) growing in the native tallgrass prairie of northern Oklahoma. Fungal Divers 47:19–27
Guo LD, Huang GR, Wang Y (2008) Seasonal and tissue age influences on endophytic fungi of Pinus tabulaeformis (Pinaceae) in the Dongling Mountains, Beijing. J Integr Plant Biol 50:997–1003
Guo X, Ding WJ, Yang JY, Xu R, Yan ZQ, Qin B (2011) Studies on the nematicidal effective fraction of Stellera chamaejasme root extracts to Ditylenchus destructor. Plant Protection 31:128–131 (in Chinese with English abstract)
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98
Hamayun M, Khan SA, Iqbal I, Ahmad B, Lee I-J (2010) Isolation of a gibberellin-producing fungus (Penicillium sp. MH7) and growth promotion of crown caisy (Chrysanthemum coronarium). J Microbiol Biotechnol 20:202–207
Herrera J, Khidir HH, Eudy DM, Porras-Alfaro A, Natvig DO, Sinsabaugh RL (2010) Shifting fungal endophyte communities colonize Bouteloua gracilis: effect of host tissue and geographical distribution. Mycologia 102:1012–1026
Huang WY, Cai YZ, Surveswaran S, Hyde KD, Corke H, Sun M (2009) Molecular phylogenetic identification of endophytic fungi isolated from three Artemisia species. Fungal Divers 36:69–88
Hyde KD, Soytong K (2008) The fungal endophyte dilemma. Fungal Divers 33:163–173
Hyde KD, Bussaban B, Paulus B, Crous PW, Lee S, Mckenzie EHC, Photita W, Lumyong S (2007) Diversity of saprobic microfungi. Biodivers Conserv 16:17–35
Joshee S, Paulus BC, Park D, Johnston PR (2009) Diversity and distribution of fungal foliar endophytes in New Zealand Podocarpaceae. Mycol Res 113:1003–1015
Kharwar RN, Verma VC, Kumar A, Gond SK, Strobel G (2009) Javanicin, an antibacterial naphthaquinone from an endophytic fungus of Neem, Chloridium sp. Curr Microbiol 58:233–238
Kumar DSS, Hyde KD (2004) Biodiversity and tissue-recurrence of endophytic fungi in Tripterygium wilfordii. Fungal Divers 17:69–90
Kumara PM, Zuehlke S, Priti V, Ramesha BT, Shweta S, Ravikanth G, Vasudeva R, Santhoshkumar TR, Spiteller M, Shaanker RU (2012) Fusarium proliferatum, an endophytic fungus from Dysoxylum binectariferum Hook.f, produces rohitukine, a chromane alkaloid possessing anti-cancer activity. Antonie van Leeuwenhoek 101:323–329
Kutorga E, Adamonytė G, Iršėnaitė R, Juzėnas S, Kasparavičius J, Markovskaja S, Motiejūnalitė J, Treigienė A (2012) Wildfire and post-fire management effects on early fungal succession in pinus mugo plantations, located in Curonian Spit (Lithuania). Geoderma 191:70–79
Lockwood JL (1981) Exploitation competion. In: Wicklow DT, Carrol GC (eds) The fungal community: its organization and role in ecosystem. Marcel Dekker, New York, pp 319–349
Martin LL, Ross Friedman CM, Phillips LA (2012) Fungal endophytes of the obligate parasitic dwarf mistletoe Arceuthobium americanum (Santalaceae) act antagonistically in vitro against the native fungal pathogen Cladosporium (Davidiellaceae) of their host. Am J Bot 99:2027–2034
Menjivar RD, Cabrera JA, Kranz J, Sikora RA (2012) Induction of metabolite organic compounds by mutualistic endophytic fungi to reduce the greenhouse whitefly Trialeurodes vaporariorum (Westwood) infection on tomato. Plant Soil 352:233–241
Mishra A, Gond SK, Kumar A, Sharma VK, Verma SK, Kharwar RN, Sieber TN (2012) Season and tissue type affect fungal endophyte communities of the Indian medicinal plant Tinospora cordifolia more strongly than geographic location. Microb Ecol 64:388–398
Morakotkarn D, Kawasaki H, Seki T (2006) Molecular diversity of bamboo-associated fungi isolated from Japan. FEMS Microbiol Lett 266:10–19
O’Brien HE, Parrent JL, Jackson JA, Moncalvo J-M, Vi lgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Appl Environ Microbiol 71:5544–5550
Oechsler RA, Feilmeier MR, Ledee DR, Miller D, Diaz MR, Elizabeth Fini M, Fell JW, Alfonso EC (2009) Utility of molecular sequence analysis of the ITS rRNA region for identification of Fusarium spp. from ocular sources. Invest Ophth Vis Sci 50:2230–2236
Porras-Alfaro A, Herrera J, Natvig DO, Sinsabaugh RL (2007) Effect of long term nitrogen fertilization on mycorrhizal fungi associated with a dominant grass in a semiarid grassland. Plant Soil 296:65–75
Quilliam RS, Jones DL (2010) Fungal root endophytes of the carnivorous plant Drosera rotundifolia. Mycorrhiza 20:341–348
Rakotoniriana EF, Munaut F, Decock C, Randriamampionona D, Andriambololoniaina M, Rakotomalala T, Rakotonirina EJ, Rabemanantsoa C, Cheuk K, Ratsimamanga SU, Mahillon J, EI-Jaziri M, Quetin-Leclercq J, Corbisier AM (2008) Endophytic fungi from leaves of Centella asiatica: occurrence and potential interactions within leaves. Antonie van Leeuwenhoek 93:27–36
Raviraja NS, Maria GL, Sridhar KR (2006) Antimicrobial evaluation of endophytic fungi inhabiting medicinal plants of the Western Ghats of India. Eng Life Sci 6:515–520
Rivera-Orduña FN, Suarez-Sanchez RA, Flores-Bustamante ZR, Gracida-Rodriguez JN, Flores-Cotera LB (2011) Diversity of endophytic fungi of Taxus globosa (Mexican yew). Fungal Divers 47:65–74
Rodrigues KF (1994) The foliar fungal endophytes of the Amazonian Palm Euterpe oleracea. Mycologia 86:376–385
Rosa LH, Vaz ABM, Caligiorne RB, Campolina S, Rosa CA (2009) Endophytic fungi associated with the Antarctic grass Deschampsia antarctica Desv. (Poaceae). Polar Biol 32:161–167
Sánchez Márquez S, Bills GF, Domínguez Acuña L, Zabalgogeazcoa I (2010) Endophytic mycobiota of leaves and roots of the grass Holcus lanatus. Fungal Divers 41:115–123
Saunders M, Kohn LM (2009) Evidence for alteration of fungal endophyte community assembly by host defense compounds. New Phytol 182:229–238
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: opensource, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, André levesque C, Chen W, Fungal Barcoding Consortium (2012) Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Pros Natl Acad Sci USA 109:6241–6246
Schulthess FM, Faeth SH (1998) Distribution, abundances and association of the endophytic fungal community of Arizona fescue (Festuca arizonica). Mycologia 90:569–578
Schulz B, Guske S, Dammann U, Boyle C (1998) Endophyte-host interactions II. Defining symbiosis of the endophyte-host interaction. Symbiosis 25:213–227
Schulz B, Boyle C, Draeger S, Römmert AK, Krohn K (2002) Endophytic fungi: a source of novel biologically active secondary metabolites. Mycol Res 106:996–1004
Shankar NB, Shashikala J, Krishnamurthy YL (2007) Study on diversity of endophytic communities from rice (Oryza sativa L.) and their antagonistic activities in vitro. Microbiol Res 164:290–296
Shannon CE, Weaver W (1963) The mathematical theory of communication. University of Illinois Press, Urbana
Sieber TN, Hugentobler C (1987) Endophytische Pilze in Blättern und Ästen gesunder und geschädigter Buchen (Fagus sylvatica L.). Eur J Forest Pathol 17:411–425
Sieber T (2007) Endophytic fungi in forest trees: are they mutualists? Fungal Biol Rev 21:75–89
Sim HJ, Khoo CH, Lee LH, Cheah YK (2010) Molecular diversity of fungal endophytes isolated from Garcinia mangostana and Garcinia parvifolia. J Microbiol Biotechn 20:651–658
Simpson GG (1964) Species density of North American recent mammals. Syst Zool 13:57–73
Stierle A, Strobel G, Stierle D (1993) Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260:214–216
Sun G, Luo P, Wu N, Qiu PF, Gao YH, Chen H, Shi FS (2009) Stellera chamaejasme L. increases soil N availability, turnover rates and microbial biomass in an alpine meadow ecosystem on the eastern Tibetan Plateau of China. Soil Biol Biochem 41:86–91
Sun X, Guo LD, Hyde KD (2011) Community composition of endophytic fungi in Acer truncatum and their role in decomposition. Fungal Divers 47:85–95
Sun X, Ding Q, Hyde KD, Guo LD (2012) Community structure and preference of endophytic fungi of three woody plants in a mixed forest. Fungal Ecol 5:624–632
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol biol evol 28:2731–2739
Tao G, Liu ZY, Hyde KD, Lui XZ, Yu ZN (2008) Whole rDNA analysis reveals novel and endophytic fungi in Bletilla ochracea (Orchidaceae). Fungal Divers 33:101–122
Vega FE, Simpkins A, Aime MC, Posada F, Pehner SA, Infante F, Castillo A, Arnold AE (2010) Fungal endophyte diversity in coffee plants from Colombia, Hawai’i, Mexico and Puerto Rico. Fungal Ecol 3:122–138
Wang Y, Guo LD, Hyde KD (2005) Taxonomic placement of sterile morphotypes of endophytic fungi from Pinus tabulaeformis (Pinaceae) in northeast China based on rDNA sequences. Fungal Divers 20:235–260
Wang Y, Wnag G, Wang L, Xu X, Xia J, Huang X, Wu Y, Zhang C (2010) Isolation and identification of an endophytic fungus of Polygonatum cyrtonema and antifungal metabolites. Acta Microbiol Sinica 50:1036–1043 (in Chinese with English abstract)
Wearn JA, Sutton BC, Morley NJ, Gange AC (2012) Species and organ specificity of fungal endophytes in herbaceous grassland plants. J Ecol 100:1085–1092
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322
Widler B, Müller E (1984) Untersuchungen über endophytische Pilze von Arctostaphylos uva-ursi (L.) Sprengel (Ericaceae). Bot Helv 94:307–337
Yang S, Zhang W, Shi JS, Shi GY, Qin SG, Chang ZL, Chen Y, Wang ZH, Huang JP (2010) Preliminary analyses of the characteristics of black carbon in the semi-arid area. Clim Environ Res 15:756–764 (in Chinese with English abstract)
Yang JY, Yan ZQ, Xu R, Liu Q, Jin H, Cui HY, Qin B (2011) Isolation of plant growth inhibitory components from the root of Stellera chamaejasme and function mechanism. Acta Botanica Boreali-Occidentalia Sinica 31:0291–0297 (in Chinese with English abstract)
Yuan Z, Zhang C, Lin F, Kubicek CP (2010) Identity, diversity, and molecular phylogeny of the endophytic mycobiota in the roots of rare wild rice (Oryza granulate) from a nature reserve in Yunnan, China. Appl Environ Microbiol 76:1642–1652
Zamora P, Martínez-Ruiz C, Diez JJ (2008) Fungi in needles and twigs of pine plantations from northern Spain. Fungal Divers 30:171–184
Zhang ZQ, Zhang YH, Sun H (2011) The reproductive biology of Stellera chamaejasme (Thymelaeaceae): a self-incompatible weed with specialized flowers. Flora 206:567–574
Zhou X, Wang Z, Jiang K, Wei Y, Lin J, Sun X, Tang K (2007) Screening of taxol producing endophytic fungi from Taxus chinensis var. mairei. Appl Biochem Microbiol 43:439–443
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (No. 310703863 and No. 21102154), Basic Research Program of Lanzhou Institute of Chemical Physics, Chinese Academy of Sciences (080423SYR1), and the associate scholar program for talents cultivation plan of “Western Light” of Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jin, H., Yan, Z., Liu, Q. et al. Diversity and dynamics of fungal endophytes in leaves, stems and roots of Stellera chamaejasme L. in northwestern China. Antonie van Leeuwenhoek 104, 949–963 (2013). https://doi.org/10.1007/s10482-013-0014-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-013-0014-2