Abstract
The Gulf of California is a coastal marine ecosystem characterized as having abundant biological resources and a high level of endemism. In this work we report the isolation and characterization of Actinobacteria from different sites in the western Gulf of California. We collected 126 sediment samples and isolated on average 3.1–38.3 Actinobacterial strains from each sample. Phylogenetic analysis of 136 strains identified them as members of the genera Actinomadura, Micromonospora, Nocardiopsis, Nonomuraea, Saccharomonospora, Salinispora, Streptomyces and Verrucosispora. These strains were grouped into 26–56 operational taxonomic units (OTUs) based on 16S rRNA gene sequence identities of 98–100 %. At 98 % sequence identity, three OTUs appear to represent new taxa while nine (35 %) have only been reported from marine environments. Sixty-three strains required seawater for growth. These fell into two OTUs at the 98 % identity level and include one that failed to produce aerial hyphae and was only distantly related (≤95.5 % 16S identity) to any previously cultured Streptomyces sp. Phylogenetic analyses of ketosynthase domains associated with polyketide synthase genes revealed sequences that ranged from 55 to 99 % nucleotide identity to experimentally characterized biosynthetic pathways suggesting that some may be associated with the production of new secondary metabolites. These results indicate that marine sediments from the Gulf of California harbor diverse Actinobacterial taxa with the potential to produce new secondary metabolites.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In an effort to discover new natural products that can be developed for the treatment of human disease and other biotechnological applications, research has moved towards the study of microorganisms from unexplored habitats such as the ocean. After almost two decades of research in marine microbiology, we now know that marine Actinobacteria are a prolific source of secondary metabolites with antibacterial, immunosuppressive and antitumor activity (Bull and Stach 2007; Fenical and Jensen 2006; Olano et al. 2009). Marine-derived Actinobacteria include new genera, as demonstrated by the descriptions of Salinibacterium, Salinispora, Sciscionella, Serinicoccus and Marinactinospora (Han et al. 2003; Maldonado et al. 2005; Tian et al. 2009a; Yi et al. 2004; Tian et al. 2009b), and new species that belong to genera that also occur on land (Helmke and Weyland 1984; Hozzein and Goodfellow 2007; Liu et al. 2010). These discoveries highlight the potential of the marine environment to yield new Actinobacterial taxa and secondary metabolites.
The Gulf of California, also known as the Sea of Cortez, is a marginal sea located between the Mexican mainland and the Baja California peninsula. This 1,100 km long coastal marine ecosystem covers 210,000 km2 and includes 37 named islands (Carreño and Helenes 2002). The peninsular coastal zone has little fresh water input due to the sub-desert climate, while the continental shore is characterized by large amounts of fresh water input (Roden and Groves 1959). A past study of marine sediments from the Gulf of California yielded nearly 300 actinomycetes belonging to the genera Actinomadura, Dietzia, Gordonia, Micromonospora, Nonomuraea, Rhodococcus, Saccharomonospora, Saccharopolyspora, Salinispora, Streptomyces, “Solwaraspora” and Verrucosispora (Maldonado et al. 2009). These genera expand upon those traditionally recovered from marine sediments, such as Micromonospora, Rhodococcus and Streptomyces, and demonstrate the potential of the Sea of Cortez as a source of diverse actinomycete taxa.
The marine actinomycete genus Salinispora has been an important source of bioactive secondary metabolites (Fenical and Jensen, 2006). Two species have been formally described, Salinispora tropica and Salinispora arenicola (Maldonado et al. 2005) while “S. pacifica” has been proposed (Jensen and Mafnas 2006). Biogeographical studies have revealed that S. tropica is restricted to the Caribbean, “S. pacifica” occurs worldwide except for the Caribbean, and S. arenicola is broadly distributed and co-occurs with both species (Jensen and Mafnas 2006; Freel et al. 2012). Previous studies have also revealed relationships between Salinispora taxonomy and secondary metabolite production (Jensen et al. 2007) and correlations between where the strains were derived and the biosynthetic genes they maintain (Edlund et al. 2011). These results suggest that culturing new Salinsipora species or known species from new locations is a potentially productive approach to natural product discovery.
In this study, a culture dependent approach was undertaken to better characterize Actinobacterial diversity in the Gulf of California. The results reveal considerable levels of diversity including the new “S. pacifica” 16S rRNA sequence type “K”. In addition, an analysis of polyketide synthase gene sequences suggests that these strains have considerable potential for the production of secondary metabolites not previously reported from the genus.
Methods
Sediment collection
A total of 126 marine sediment samples were collected during two expeditions. The first took place in November 2007 off San Felipe, B.C. The second took place in April 2008 and targeted three sites along the west coast of the Gulf of California (Fig. 1) under collection permit DGOPA.06472.110909.3745 (marine protected areas from Los Ángeles Bay to Loreto Bay). Sediments were obtained from depths of 20–300 m using a modified Kahlisco grab sampler (model # 214WA110). Samples from 0 to 15 m were collected by hand. The specific sampling sites were S1: San Felipe (31°62′N, 114°48.95′W to 31°59.74′N, 114°49.04′W); S2: Los Ángeles Bay (28°58.93′N, 113°29.54′W to 28°57.19′N, 113°25.85 W); S3: Concepción Bay (26°34.17′N, 111°47.11′ W to 26°45.73′N, 111°53.50′W). The Loreto Bay sampling sites were S4: mainland Beach (25°50.2′N, 111°19.4′W and 25°43.3′N, 111°14.6′W); S5: island beach (25°48′N, 111°15.35′W); S6: around Danzante Island (25°57.0′N, 111°18.37′W to 25°48′N 111°15.42′W) and S7: around Danzante Island (25°48.91′N, 111°14.36′W to 25°47.95′N, 111°14.32′W). Each sample was transferred to a previously labeled sterile bag (Nasco whirl-pack) and stored at 4 °C for transport to the Universidad Autónoma de Baja California (UABC) Ensenada, Mexico for processing.
Actinobacterial isolation
All sediment samples were dried in a laminar flow hood for 24 h. Once dried, they were ground and inoculated using the plate stamping technique (Mincer et al. 2002) on 6 different culture media: M1 (18 g agar, 1 l natural sea water), M2 (18 g agar, 0.5 g mannitol, 0.1 g peptone, 1 l natural sea water), M3 (18 g agar, 1.0 g starch, 0.2 g peptone, 0.4 g yeast extract, 1 l natural sea water), M4 (18 g agar, 2.5 g mannitol, 1 g peptone, 1 l natural sea water), M5 (18 g agar, 0.5 g mannitol, 0.1 g casamino acids, 1 l natural sea water), and M6 (10 g agar, 0.6 g tryptone, 1.0 g casitone, 0.8 g glucose, 1 l natural sea water). Cyclohexamide (100 μg ml−1 final concentration) was added to all media to reduce fungal contamination and the antibiotic rifamycin or gentamicin (5 μg ml−1 final concentration) was added to select for Actinobacteria. In total, 12 different media and antibiotic combinations were used for the isolation of Actinobacteria.
After 2–12 weeks of incubation at 25–28 °C, all well separated Actinobacterial colonies observed by eye or using a stereomicroscope were removed from the original isolation plate and repeatedly sub-cultured on medium A1 (18 g agar, 10 g starch, 2 g peptone, 4 g yeast extract, 1 l natural sea water) until pure cultures were obtained as judged by uniform colony morphology. Colony characteristics were observed using a stereomicroscope and all pure isolates were tested for Gram reaction following the non-staining KOH method (Powers 1995).
Effects of seawater on growth
All strains were examined to determine the effects of replacing seawater with deionized water in the growth medium. Using a sterile loop, cells from a well-defined colony were removed from an A1 plate prepared with 100 % natural seawater and streaked onto plates of A1 prepared with seawater and deionized water. Plates were incubated at 25–28 °C and growth was monitored at up to 60× magnification for 8 weeks.
16S rRNA gene amplification and sequencing
The strains were primarily grouped based on the presence or absence of aerial mycelium and the effects of seawater on growth. Within these categories, strains were additionally grouped according to colony color, morphology, and diffusible pigment production. Representatives from each group were selected for 16S rRNA gene sequencing and phylogenetic analysis. Strains were cultured in 25 ml of medium A1 and shaken at 215 rpm and 28 °C for 7 days. Cells were pelleted by centrifugation and genomic DNA extracted using previously described methods (Gontang et al. 2007). 16S rRNA genes were PCR amplified using the primers FC27 (5′–3′AGAGTTTGATCCTGGCTCAG) and RC1492 (5′–3′TACGGCTACCTTGTTACGACTT) and the following conditions: initial denaturalization at 95 °C for 15 min followed by 32 cycles at 95 °C for 1 min, 61 °C for 1 min and 72 °C for 1 min, followed by a final extension at 72 °C for 7 min. PCR products were purified with a Qiagen QIAquick PCR cleanup kit, using protocols provided by the manufacturer (Qiagen Inc., Chatsworth, Calif.). Sequences were obtained using the primers listed above at SeqXcel, INC (http://www.seqxcel.com/) using the BigDye Terminator Cycle Sequencing Chemistry 3.1 technique and a Genetic Analyzer ABI Prism 3100 (Applied Biosystem).
Phylogenetic analysis
The forward and reverse 16S rRNA sequences obtained from each strain were assembled and analyzed using BLAST (Basic Local Alignment Search Tool) (Altschul et al. 1990) available on the NCBI website (http://www.ncbi.nlm.nih.gov/). Sequences were aligned using ClustalX and imported into the Bioedit program (Hall and Brown 2001) for manual alignment. A neighbor-joining phylogenetic tree was constructed from 1300 base pairs using the program MEGA4 (Tamura et al. 2007) with 1000 bootstrap replicates. Sequences were deposited in GenBank (http://www.ncb.nlm.nih.gov/genbank/index.html) under the accession numbers HQ873926-HQ873952 and HQ877423-HQ877448.
Operational taxonomic units (OTUs)
Aligned 16S rRNA gene sequences (1300 bp) were grouped into OTUs based on 98, 99 and 100 % sequence identity using the program Clusterer (www.bugaco.com/mioritir/cluster_jlp.php). An OTU was considered ‘new’ if all of the members shared ≤98 % sequence identity with any previously cultured strain for which sequences had been deposited in GenBank. The OTU was designated ‘marine’ if all GenBank sequences that shared >98 % sequence identity had been cultured from marine sources. The strain in each 98 % OTU with the lowest similarity to the closest type strain was selected for the construction of a phylogenetic tree.
KS domain amplification and sequencing
Ketosynthase (KS) sequences were PCR amplified from 20 to 50 ng of genomic DNA using the degenerate primers PKSRb (5′-GTSCCSGTSCCGTGSGCCTCSA-3′) and PKSFa (5′-CCSCAGSAGCGCSTSTTSCTGGA-3′) (Edlund et al. 2011). The conditions consisted of an initial denaturalization at 95 °C for 15 min, followed by one cycle of 95 °C for 1 min, 65 °C for 1 min and 72 °C for 1 min, and then 31 cycles of 1 min at 95 °C, 62 °C for 1 min and 72 °C for 1 min, followed by a 7 min final extension at 72 °C. The PCR products were purified using the Qiagen QIAquick MinElute Gel Extraction kit following the manufacturer protocols.
The purified PCR products were cloned using the Topo TA cloning kit (Invitrogen, Catalog nos. C4040-10) following the manufacturer’s protocol. Transformed clones were inoculated into 10 ml Falcon tubes containing 3 ml of Luria-Bertoni culture media with kanamycin (50 μg ml−1). Plasmid DNA was extracted using the QiaPrep® MiniPrep extraction kit, following the manufacturer’s instructions (Qiagen, Cat. No: 27106). Plasmid DNA was subjected to a restriction digest using BstX I (New England BioLabs) and the product run on a 1 % agarose gel to confirm the presence of the appropriate size insert. Select plasmids were sequenced at SeqXcel (La Jolla) using the M13 primer. KS sequences were deposited in GenBank (http://www.ncb.nlm.nih.gov/genbank/index.html) under the accession numbers HQ877411-HQ877422.
KS sequences were analyzed using BLAST and the program NaPDoS (http://napdos.ucsd.edu). Sequences were aligned using ClustalX and imported into the Bioedit program (Hall and Brown 2001) for manual alignment. A neighbor-joining phylogenetic tree was constructed from 86 amino acids using the program MEGA4 (Tamura et al. 2007) with 1000 bootstrap replicates.
Fermentation and chemical analysis
Strains AMS515, AMS22 and AMS300 were inoculated from frozen stocks into 25 ml of medium A1 and shaken at 215 rpm and 28 °C for 3 days. The cultures were then transferred to 2.8-l Fernbach flasks containing 1 l of the same medium. On days 3, 5 and 7, 25 ml of the culture was removed and extracted with 50 ml of ethyl acetate. The organic layers were separated, dried over anhydrous sodium sulfate, decanted and concentrated under vacuum. The crude extracts were dissolved in methanol and analyzed in a Hewlett-Packard MSD 1100 liquid chromatography-mass spectrophotometer (LC–MS) system using a C-18 reversed-phase column and a 10–100 % acetonitrile in water gradient. Fractions that showed masses characteristic of compounds in the rifampicin class were analyzed by high-resolution (HR) electrospray ionization (ESI)-mass spectrometry (MS) in positive mode using a Thermo Scientific LTQ Orbitrap XL Mass Spectrometer.
Results
Cultivable Actinobacteria
One hundred 26 sediment samples were collected at depths ranging from 0 to 300 m from four different locations in the Gulf of California (Fig. 1). From these samples, 1497 bacterial strains with colony morphologies indicative of the Order Actinomycetales were obtained in pure culture by repeated streaking on agar media. Many of the actinomycete colonies were visible by the fourth week of incubation however some took as long as 12 weeks to appear. The largest number of strains per sample originated from sediments collected at depths from 200 to 300 m at sites S6 and S7 (Table 1) around Danzantes Island, Loreto Bay (Table 1). Los Ángeles Bay site S2 and Loreto Bay mainland beach site S4 also yielded high numbers of strains per sample. On average, 3.1–7.2 strains were isolated from sediments collected at sites S1, S3, and S5 while 12.0–38.3 were obtained from sites S2, S4, S6 and S7 (Table 1). It should be mentioned that sites S1, S3 and S4 are all <1 km from the mainland and yielded many Gram-negative bacteria that may have out-competed slower growing Actinobacteria for space and nutrients. This was not the case for sites S2, S5–S7, which were 5–10 km from shore and yielded relatively few Gram-negative bacteria.
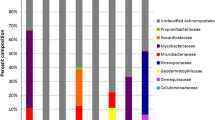
Interestingly, 44–91 % of the Actinobacteria cultured from Los Angeles Bay and Loreto Bay sites S2, S5, S6 and S7 possessed colony morphologies characteristic of the genera Micromonospora or Salinispora. These include orange-pigmented colonies that lack areal hyphae and spores that blacken the colony surface. The majority of these strains failed to grow when seawater was replaced with deionized water in the growth medium providing preliminary evidence to support their inclusion in the genus Salinispora (Maldonado et al. 2005). The relative abundance of seawater dependent strains was greatest at sites S2 and S5–S7 (Fig. 2). Overall, only 16 of 625 Actinobacteria cultured from shallow (0–15 m) sediment samples collected from near-shore Baja California sites (S1, S3 and S4) failed to grow when seawater was replaced with deonized water in the growth medium. While near-shore sites S1 and S2 yielded many isolates (48 and 40 % of total, respectively) with colony morphologies characteristic of the genera Micromonospora or Salinispora, all of these grew in the absence of seawater as is typical of the genus Micromonospora. Sites S1, S3, and S4 also yielded large numbers of actinomycetes with well-developed aerial hyphae that did not require seawater for growth (52, 90 and 91 % of total respectively) as might be expected from strains that were introduced from land (Fig. 2).
Actinobacterial colony morphology and seawater requirements. GI = substrate mycelium only. GI* = substrate mycelium only and requires seawater for growth. GII = aerial hyphae. S1 = San Felipe Bay, S2 = Los Angeles Bay, S3 = Concepción Bay, S4 = Mainland beach, S5 = Danzantes Island beach, S6 = Around Danzante Island site a and S7 = Around Danzante Island site b
Actinobacteria diversity
Actinobacterial diversity was evaluated based on the partial 16S rRNA gene sequences of 136 strains that were selected to represent a diversity of seawater requirements and colony morphologies from each location. These strains included a large number that failed to grow on media prepared with deionized water. In total, 85 of the strains sequenced originated from 200 to 300 m (sites 6 and 7) while the remaining strains were obtained from 0 to 20 m (sites 1–5). BLAST analyses revealed they were most closely related to 8 genera (Table 2). Those strains with closest BLAST matches to Micromonospora, Nonomuraea, Salinispora and Verrucosispora all lacked aerial hyphae and produced orange colonies and black spores, yet among these, only the strains with top matches to Salinispora spp. failed to grow when seawater was replaced with deionized water in the growth medium. Bacteria with well-developed aerial hyphae (53 strains) were related to four genera: Actinomadura (7.1 %), Saccharomonospora (4.2 %), Streptomyces (76.8 %) and Nocardiopsis (1.7 %). Of these, only strains related to Streptomyces spp. failed to grow when seawater was replaced with deionized water in the growth medium.
Operational taxonomic units (OTUs) calculated based on sequence identities of 98 and 99 % revealed 26 and 37 OTUs, respectively. Within the 98 % group, there are three new OTUs (Table 2) that potentially represent new taxa within the genera Streptomyces and Actinomadura. Forty-one of the 56 OTU’s identified at 100 % sequence identity had not previously been reported (Table 2). The most distinct OTU (no. 20, Supplemental Table 1) is represented by strain AMS37 and shared only 95.4 % 16S rRNA sequence identity to the nearest Streptomyces type strain. All three of the strains in this OTU originated from site S6 (200 m). Additionally, all of the strains in OTUs 13–21 share <98.5 % sequence identity with the nearest type strains and thus may also represent new taxa (Rossello-Mora and Amann 2001). Eight of these new OTUs originated from samples collected at depths of 200–300 m.
Many of the OTUs were designated as marine if they had previously been isolated from marine sources and all publically available sequences in the OTU were from marine sources (Table 2). As the number of new OTUs decreases with decreasing levels of OTU 16S sequence identity, the relative proportion of known marine OTUs increases indicating the existence of diverse lineages that have only been reported from marine sources. To date, eight of these lineages have been formally described as new genera (Salinispora) or species (Micromonospora krabiensis, Saccharomonospora marina, Streptomyces fenhuangensis, Streptomyces marinus, Streptomyces nanshensis, Verrucosispora maris and Verrucosispora sediminis).
A phylogenetic tree reveals that the five strains belonging to OTU number one (Supplemental Table 1), which represent the 60 seawater-requiring strains that possessed Micromonospora or Salinispora-like morphologies, all claded with Salinispora spp. (Fig. 3). Fifty-three of the 60 Salinispora strains were identified as S. arenicola and the remaining seven as “S. pacifica”. All of the S. arenicola strains were identical to those previously reported as sequence types “A” or “B” (Jensen and Mafnas 2006). Interestingly, the “standard” S. arenicola sequence type (i.e., the first 16S sequence type reported for the species and represented by strain CNS-205 in Fig. 3) was not observed. S. arenicola sequence type “A” was the most abundant Salinispora sequence type, representing 86.6 % (52 strains) of the 60 Salinispora strains examined in this study while only one strain belonging to sequence type “B” was recovered. The seven strains identified as “S. pacifica” fell into three OTUs at 100 % sequence identity (Fig. 3) and included the “standard” “S. pacifica” sequence type (AMS365), sequence type “C” (AMS178) and one sequence type that has not previously been reported (AMS301). We have designated this new sequence type, which shares 99.84 % sequence identity with the standard “S. pacifica” sequence type, as sequence type “K”.
A Phylogenetic tree based on 16S rRNA gene sequences from 26 Actinomycetales OTUs (calculated using a sequence identity value of ≥98 %) observed in this study (AMS numbers) and the nearest type strains. The isolates include 9 OTUs that appear to represent new taxa and 8 OTUs with nearest type strains isolated from marine sources. Rubrobacter radiotolerans was used to root the tree. B Phylogenetic tree based on 16S rRNA gene sequences from 5 Salinispora OTUs (calculated using a sequence identity value of 100) observed in this study (AMS numbers) and previously reported “S. pacifica”, S. arenicola and S. tropica strains. Letters after the species name designate 16S “sequence types”. Those without letters represent the “standard” sequence type (i.e., the first sequence type observed for the species). The trees were constructed using the neighbor-joining method and the program MEGA4 (1000 bootstrap replicates). Accession numbers (in parentheses). M = marine source, * Actinobacteria that require seawater for growth
Effects of seawater on growth
Six OTUs identified based on 100 % sequence identity failed to grow when seawater was replaced with deionized water in the culture medium. Five of these OTUs were affiliated with the genus Salinispora (strains AMS300, AM360, AMS301, AMS365 and AMS178) and the sixth with the genus Streptomyces (strain AMS37, Supplemental Table 1). The Salinispora isolates were recovered from Los Angeles Bay to Loreto Bay and were collected from depths of 20–300 m demonstrating a broad distribution in the Gulf of California.
Analysis of biosynthetic genes sequences
Ketosynthase (KS) domains were cloned and sequenced from three S. arenicola (AMS300, AMS22 and AMS515) and two “S. pacifica” (AMS301 and AMS178) strains. These domains are associated with polyketide synthase (PKS) genes and their phylogeny has proven informative in terms of making prediction about secondary metabolite production (Ginolhac et al. 2005; Gontang et al. 2010). The three S. arenicola sequence type “A” strains yielded similar results with the detection of KS sequences that share a high level of identity to those associated with rifamycin biosynthesis (rifA and rifE) and both ten- (Cal05) and nine-membered (NcsE) enediyne biosynthesis (Fig. 4). All of these sequences were observed previously in the S. arenicola strain CNS-205 genome sequence (Penn et al. 2009) or in other related work (Gontang et al. 2010). “S. pacifica” sequence type “K” had not previously been examined for KS sequence diversity. This strain yielded a KS sequence that shared a high level of identity to that associated with lymphostin biosynthesis, as has previously been observed in both S. arenicola and S. tropica (Penn et al. 2009) in addition to a Streptomyces sp. (Nagata et al. 1997). Thus, lymphostin production may be a common feature of the genus. The other three KS sequences shared 55 % similarity with CurI, which is part of the curacin biosynthetic pathway observed from Lyngbya majuscula (Chang et al. 2004). Only one KS domain was detected from “S. pacifica” sequence type “C” (strain AMS178). It possessed 56.57 % identity to EpoC, which is involved in epothilone biosynthesis in Sorangium cellulosum (Walsh et al. 2003) (Supplementary Table 2). The low levels of sequence identity for KS sequences observed in the “S. pacifica” sequence type “C” and “K” suggest that the associated biosynthetic pathways have yet to be characterized.
Salinispora KS phylogeny. Neighbor-joining distance tree constructed using 86 aligned amino acids from strains AMS22, AMS300, AMS515, AMS301 and AMS178. The tree was created using the program MEGA4 (Tamura et al. 2007) with 1000 bootstrap replicates. KS sequences were classified using NaPDoS (http://napdos.ucsd.edu/)
All of the KS domains had an active site cysteine, a conserved histidine residue 135 nucleotides downstream from the active site and the largely conserved active site domain sequence VDTACSSSLVA (Aparicio et al. 1996; Bevitt et al. 1992; Donadio and Katz 1992). Exceptions include clones 2–4 from “S. pacifica” sequence type “K” (strain AMS301) in which the valine at position 94 is replaced with an alanine, and one clone from sequence type “C” (strain AMS178) in which glutamine and threonine are observed at positions 85 and 91, respectively (Supplementary Fig. 1).
Analysis of the LC/MS data
Secondary metabolite production has been successfully predicted in cases where KS amino acid sequence identity is ≥85 % to sequences associated with experimentally characterized biosynthetic pathways (Gontang et al. 2010). Based on this, it was predicted that S. arenicola strain AMS515 produces compounds in the rifamycin class. LC/MS analysis of organic extracts of a culture grown for nine days led to the detection of a compound in the rifamycin class with UV absorbance maxima (λmax) at 220, 300 and 420 nm (Supplementary Fig. 2).
Discussion
Marine Actinobacteria have become an important source of medically relevant secondary metabolites, such as antibiotics, antitumor, anti-inflammatory, and antifungal compounds (Bull and Stach 2007; Fenical and Jensen 2006; Olano et al. 2009). This creates a rationale to explore poorly studied marine ecosystems as a source of new actinobacterial strains and taxa. In the present study, 1497 actinomycete strains were cultured from marine sediments collected at three sites in the Gulf of California. Using a 16S rRNA sequence identity of 98 % to cluster strains into OTUs, three potentially new taxa were found (Table 2). These OTUs belong to the genera Streptomyces and Actinomadura and support further taxonomic studies of marine actinomycetes from the Gulf of California. Of these, OTU 20 is the most unique, sharing only 95.4 % sequence identity with the nearest type strain (Supplemental Table 1). This OTU is comprised of three strains, all of which require seawater for growth, and could potentially represent a new genus-level taxon in the family Streptomycetaceae. In addition to the three potentially new taxa, several previously described marine species were isolated including Micromonospora krabiensis (AMS264, 99.06 % similarity), Saccharomonospora marina (AML899, 99.54 % similarity), Streptomyces fenhuangensis (AML250, 98.71 % similarity), Verrucosispora maris (AMS604, 99.42 % similarity) and Verrucosispora sediminis (AMS180, 98.76 % similarity) suggesting that these species may have broad geographic distributions.
A majority of the strains sequenced (60) were identified as members of the marine actinomycete genus Salinispora. Of these, 53 were identified as S. arenicola sequence types “A” or “B” while the rest were identified as “S. pacifica”. S. arenicola sequence types “A” and “B” were previously reported from the Gulf of California (Jensen and Mafnas 2006; Maldonado et al. 2009) and off the Pacific coast of Costa Rica (Solano et al. 2009). The absence of the “standard” sequence type, which has been reported from the Bahamas, Palau, the Red Sea, Guam and Fiji (Freel et al. 2011; Jensen and Mafnas 2006), but has yet to be reported from the Gulf of California, suggests there are distinct geographical boundaries associated with the three S. arenicola sequence types. While only seven “S. pacifica” strains were identified, they include the “standard” sequence type (Strain AMS363), sequence type “C” (Strain AMS178), and the first report of sequence type “K” (strain AMS301, HQ873948), which represents the twelfth sequence type observed to date for this species (Freel et al. 2012).
The actinomycetes cultured from the Gulf of California provide opportunities to explore these strains for the genetic potential to produce secondary metabolites. Targeting KS domains provides a rapid method to assess PKS diversity and novelty within individual strains. The results revealed evidence of common pathways shared with other Salinispora strains but also sequences that share low levels of identity with any characterized pathways and thus may be associated with the production of new secondary metabolites. It is noteworthy that the new sequence type “K” (strain AMS301) also possesses a KS sequence that has not previously been observed in “S. pacifica”.
This study presents an analysis of the culturable actinomycete diversity obtained from marine sediments collected off the western margin of the Gulf of California. The results add to the diversity observed within the genus Salinispora and to the growing diversity of Streptomyces spp. that have been recovered from marine samples. The unique characteristics of the Gulf of California make it an ideal location to search for new actinomycete diversity that can be incorporated into natural product screening programs.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aparicio JF, Molnár I, Schwecke T, König A, Haydock SF, Khaw LE, Staunton J, Leadlay PF (1996) Organization of the biosynthetic gene cluster for rapamycin in Streptomyces hygroscopicus: analysis of the enzymatic domains in the modular polyketide synthase. Gene 169:9–16
Bevitt DJ, Cortes J, Haydock SF, Leadlay PF (1992) 6-Deoxyerythronolide-B synthase-2 from Saccharopolyspora erythraea. Cloning of the structural gene, sequence analysis and inferred domain structure of the multifuncional enzyme. Eur J Biochem 204:39–49
Bull AT, Stach JEM (2007) Marine Actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 15:491–499
Carreño AL, Helenes J (2002) Geology and ages of the islands. In: Case TJ, Cody ML, Equarra E (eds) A new Island biogeography of the sea of cortés. Oxford University Press, Oxford, pp 14–40
Chang Z, Sitachitta N, Rossi JV, Roberts MA, Flatt PM, Jia J, Sherman DH, Gerwick WH (2004) Biosynthetic pathway and gene cluster analysis of curacin A, an antitubulin natural product from the tropical marine cyanobacterium Lyngbya majuscula. J Nat Prod 67:1356–1367
Donadio S, Katz L (1992) Organization of the enzymatic domains in the multifunctional polyketide synthase involved in erythromycin formation in Saccharopolyspora erythraea. Gene 111:51–60
Edlund A, Loesgen S, Fenical W, Jensen PR (2011) Geographic distribution of secondary metabolite genes in the marine actinomycete Salinispora arenicola. Appl Environ Microbiol 77:5916–5925
Fenical W, Jensen PR (2006) Developing a new resource for drug discovery: marine actinomycete bacteria. Nat Chem Biol 2:666–673
Freel KC, Nam SJ, Fenical W, Jensen PR (2011) Evolution of secondary metabolite genes in three closely related marine actinomycete species. Appl Environ Microbiol 77:7261–7270
Freel KC, Edlund A, Jensen PR (2012) Microdiversity and evidence for high dispersal rates in the marine actinomycete ‘Salinispora pacifica’. Environ Microbiol 14:480–493
Ginolhac A, Jarrin C, Robe P, PerriËre G, Vogel T, Simonet P, Nalin R (2005) Type I polyketide synthases may have evolved through horizontal gene transfer. J Mol Evol 60:716–725
Gontang EA, Fenical W, Jensen PR (2007) Phylogenetic diversity of Gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol 73:3272–3282
Gontang EA, Gaudêncio SP, Fenical W, Jensen PR (2010) Sequence-based analysis of secondary-metabolite biosynthesis in marine Actinobacteria. Appl Environ Microbiol 76:2487–2499
Hall TA, Brown JW (2001) The ribonuclease P family. Methods Enzymol 341:56–77
Han SK, Nedashkovskaya OI, Mikhailov VV, Kim SB, Bae KS (2003) Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microbiol 53:2061–2066
Helmke E, Weyland H (1984) Rhodococcusmarinonascens sp. nov., an actinomycete from the sea. Int J Syst Bacteriol 34:127–138
Hozzein WN, Goodfellow M (2007) Streptomyces synnematoformans sp. nov., a novel actinomycete isolated from a sand dune soil in Egypt. Int J Syst Evol Microbiol 57:2009–2013
Jensen PR, Mafnas C (2006) Biogeography of the marine actinomycete Salinispora. Environ Microbiol 8:1881–1888
Jensen PR, Williams PG, Oh D-C, Zeiger L, Fenical W (2007) Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol 73:1146–1152
Liu Z, Li Y, Zheng LQ, Huang YJ, Li WJ (2010) Saccharomonospora marina sp. nov., isolated from an ocean sediment of the East China Sea. Int J Syst Evol Microbiol 60:1854–1857
Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, Ward AC, Bull AT, Goodfellow M (2005) Salinispora arenicola gen. nov., sp nov and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol 55:1759–1766
Maldonado LA, Fragoso-Yañez D, Pérez-García A, Rosellón-Druker J, Quintana ET (2009) Actinobacterial diversity from marine sediments collected in Mexico. Antonie Van Leeuwenhoek 95:111–120
Mincer T, Jensen PR, Christopher A, Kauffman Fenical W (2002) Widespread and persistent populations of a major new marine Actinomycete taxon in ocean sediments. Appl Environ Microbiol 68:5005–5011
Nagata H, Ochiai K, Aotani Y, Ando K, Yoshida M, Takahashi I, Tamaoki T (1997) Lymphostin (LK6-A), a novel immunosuppressant from Streptomyces sp. KY11783: taxonomy of the producing organism, fermentation, isolation and biological activities. J Antibiot 50:537–542
Olano C, Méndez C, Salas JA (2009) Antitumor compounds from marine actinomycetes. Mar Drugs 7:210–248
Penn K, Jenkis C, Nett M, Udwary DW, Gontang EA, McGlinchey RP, Foster B, Lapidus A, Podell S, Allen EE, Morre BS, Jensen PR (2009) Genomic islands link secondary metabolism to functional adaptation in marine Actinobacterias. ISMEJ 3:1193–1203
Powers EM (1995) Efficacy of the Ryu non-staining KOH technique for rapidly determining gram reactions of food-borne and waterborne bacteria and yeasts. Appl Environ Microbiol 61:3756–3758
Roden GI, Groves GW (1959) Recent oceanographic investigations in the Gulf of California. J Mar Res 18:10–35
Rossello-Mora R, Amann R (2001) The species concept for prokaryotes. FEMS Microbiol Rev 25:39–67
Solano G, Rojas-Jimenez K, Jaspars M, Tamayo-Castillo G (2009) Study of the diversity of culturable actinomycetes in the North Pacific and Caribbean coasts of Costa Rica. Antonie Van Leeuwenhoek 96:71–78
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Tian XP, Tang SK, Dong JD, Zhang YQ, Xu LH, Zhang S, Li WJ (2009a) Marinactinospora thermotolerans gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int J Syst Evol Microbiol 59:948–952
Tian XP, Zhi XY, Qiu YQ, Zhang YQ, Tang SK, Xu LH, Zhang S, Li WJ (2009b) Sciscionella marina gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China sea. Int J Syst Evol Microbiol 59:222–228
Walsh CT, O’Connor SE, Schneider TL (2003) Polyketide-nonribosomal peptide epothilone antitumor agents: the EpoA, B, C subunits. Ind Microbiol Biotechnol 30:448–455
Yi H, Schumann P, Sohn K, Chun J (2004) Serinicoccus marinus gen. nov., sp. nov., a novel actinomycete with l-ornithine and l-serine in the peptidoglycan. Int J Syst Evol Microbiol 54:1585–1589
Acknowledgments
We acknowledge financial support from the Universidad Autónoma de Baja California Internal Assembly (grants No. 357 and 369), the Consejo Nacional de Ciencia y Tecnología (México) for the pre-doctorate fellowship given to A.B.E (number 5892). P.J. acknowledges financial support from the National Institutes of Health (grant GM0886261) and the NOAA California Sea Grant College Program Project R/NMP-100 (Grant NA100AR4170060) through NOAA’S National Sea Grant College Program, U.S. Dept. of Commerce. We thank W. Fenical for facilitating the field collections, M. Woolery for LC/MS assistance and D. Guillen, H. Ocampo-Alvarez, C. Barrila, S. Gomez, N. Millán and M. Torres for help with strain preservation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Becerril-Espinosa, A., Freel, K.C., Jensen, P.R. et al. Marine Actinobacteria from the Gulf of California: diversity, abundance and secondary metabolite biosynthetic potential. Antonie van Leeuwenhoek 103, 809–819 (2013). https://doi.org/10.1007/s10482-012-9863-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9863-3