Abstract
Identification of Bacillus cereus sensu stricto is a challenge for the food industry since it is being increasingly reported as implicated in many foodborne outbreaks. So far no conclusive microbiological or biochemical traits have been described for their specific differentiation. Here a polyphasic approach aiming at identification of new isolates is presented. It was conducted on a total of 75 strains, 59 Bacillus cereus group (29 reference strains and 30 food and environmental isolates) and 16 other Bacillus species. It includes biochemical traits (API 50CH and API 20E) and genetic profiles: PCR amplification of the internal spacer region (ISR) between 23S and 16S rRNA genes (ISR-PCR), randomly amplified polymorphic DNA (RAPD-PCR) with three universal primers (M13, T3, and T7), and PCR amplification using specific primers directed to genes encoding hemolysin BL (hbl), cytotoxin K (cytK) and cereulide (ces). As expected, PCR-enterotoxin profiles revealed the toxigenic potential of strains within the B. cereus group irrespective of the species. Cluster analysis combining the three RAPD fingerprints (RAPD-M13, RAPD-T3 and RAPD-T7) allowed almost a complete separation of strains within the B. cereus group. As a result, the ISR-PCR profile is proposed for the rapid assignation of isolates to B. cereus group with the advantage over the API profile of being a specific and culture-independent technique. Following, differentiation at species level can be obtained by RAPD profiles analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Bacillus cereus group is a taxonomically ambiguous group that includes the food poisoning Bacillus species, B. cereus and B. weihenstephanensis; B. mycoides and B. pseudomycoides, common and ubiquitous soil organisms; B. thuringiensis, an insect pathogen widely used as a bioinsecticide, and B. anthracis, the causative agent of anthrax (for a review see reference Vilas-Boas et al. 2007). These species share many phenotypic properties and a high level of DNA sequence similarity which difficult a proper identification either by biochemical traits or molecular techniques such as rRNA gene sequencing. Studies on the genomics of the B. cereus group revealed that plasmids are mainly responsible for pathogenicity-related characteristics while chromosomes exhibit limited differences in gene content. These findings have questioned their taxonomic inter-relationship and even the status of these species as separate species (Helgason et al. 2000; Rasko et al. 2005).
Among the B. cereus group, the species B. cereus is being increasingly reported as implicated in many foodborne outbreaks (European Food Safety Authority 2009; Fricker et al. 2007), i.e. in the latest EFSA report it was the causative agent in 97 outbreaks and caused 24.5% of the cases due to bacterial toxins. B. cereus strains have been identified in two different types of food poisoning, both can occasionally be fatal (Dierick et al. 2005). The emetic type is due to the synthesis of cereulide, the emetic toxin encoded by the ces genes (Ehling-Schulz et al. 2004). The diarrheal type is caused by several heat-labile enterotoxins, i.e. haemolysin BL (Hbl), nonhaemolytic enterotoxins (Nhe) and the cytotoxin K (CytK), but other virulence factors including phosphatidylinositol and phosphatidylcholine specific phospholipases C (PI-PLC and PC-PLC), cereolysine O (Clo), sphingomyelinase (SMase) and enterotoxin FM (EntFM) can be involved (Granum 2002). Besides, the HBL enterotoxin production is broadly distributed among species of the B. cereus group (Pruss et al. 1999) and a large proportion of B. thuringiensis, B. mycoides and B. weihenstephanensis strains have also been reported to contain at least one out of the virulence genes indicating their pathogenic potential (Beattie and Williams 1999; Gaviria et al. 2000; Stenfors et al. 2002; Yang et al. 2005). The opportunistic nature of B. cereus group organisms, and the fact that isolates responsible for disease outbreaks and contamination of foodstuffs can originate from various genetic backgrounds has recently been underlined by a global and extended phylogenetic study (Tourasse et al. 2010).
Detection of B. cereus in food-borne outbreaks is usually performed by culture techniques that include the use of selective media, biochemical confirmation and other parameters (e.g. cell and spore morphology and presence of the crystal protein) for bacterial identification. Currently, two international norms are recommended for the isolation, identification and enumeration of B. cereus in food, ISO 7932:2004 (Anonymous 2004) and ISO 21871:2006 (Anonymous 2006), both standard methods rely on polymyxin B resistance, lack of mannitol fermentation and lecithinase activity. Further, identification is usually completed by the API system which is not sufficiently discriminative for the species within the B. cereus group (Logan and Berkeley 1984; Choma et al. 2000; Valero et al. 2002). As an alternative to the traditional procedures, diagnostic PCR has been applied for the detection of emetic B. cereus strains (Fricker et al. 2007) or B. cereus group isolates (Martínez-Blanch et al. 2009). But the lack of species specific signatures, as well as the presence of toxigenic genes in other Bacillus spp., hampers species identification by PCR. On the other hand, PCR-based fingerprinting methods offer the possibility to overcome this drawback, since the whole genome is screened. They are commonly used to analyze genetic relatedness of strains allowing identification of organisms that might not possess the target for specific primers as well as in the typing of isolates. A number of them have been applied to the B. cereus group for species identification and/or typing like 16S–23S rRNA genes interspacer PCR analysis (PCR-ISR) (Daffonchio et al. 2000), Restriction Fragment Length Polymorphism (PCR-RFLP) (Manzano et al. 2003), Arbitrary Primed Polymerase Chain Reaction (AP-PCR) (Torkar and Mozina 2000), Repetitive Sequence-based PCR (REP-PCR) (Cooper and Mckillip 2006), Enterobacterial Repetitive Intergenic Consensus-PCR analysis (ERIC-PCR) (Shangkuan et al. 2000), Randomly Amplified Polymorphic DNA (RAPD) (Svensson et al. 1999), Amplified Fragment Length Polymorphism (AFLP) (Hill et al. 2004) or Multi Locus Sequence Typing (MLST) (Helgason et al. 2004). All these techniques agreed in clearly separating B. anthracis from the other species in the B. cereus group while revealing high intraespecific diversity, mainly in B. cereus and B. thuringiensis. Among them, RAPD-PCR is quite simple and fast to test a large number of strains and it has been successfully applied to identify B. cereus group isolates from milk powders and fermented traditional foods (Ronimus et al. 2003; Abriouel et al. 2007; Kwon et al. 2009) and in the monitoring of contaminants or commercial biopesticide strains (Jensen et al. 2002; Svensson et al. 1999).
The correct separation of B. cereus group strains from other Bacillus species and their reliable identification at species level is of utmost importance because of several reasons: the implication of this group of strains as pathogens, with reports on highly pathogenic B. cereus strains; their use as food additives and as biological control agents; the ease with which B. cereus and B. mycoides can be isolated from natural habitats, and also the finding that B. thuringiensis strains can produce B. cereus-specific enterotoxin.
To accomplish this objective, we carried out a polyphasic approach using the standard culture based-procedures and PCR-based profiles on a comprehensive collection of reference strains, environmental and food isolates. We report the comparative evaluation of several molecular techniques and biochemical profiles in order to search for rapid and valid methods for the routine identification of B. cereus group isolates in the food industry.
Materials and methods
Bacterial strains and growth conditions
A collection of 75 Bacillus strains, including 29 reference strains of B. cereus group, 30 food and environmental isolates most of them kindly provided by Dr. Tomás (AINIA, Valencia, Spain) and 16 other Bacillus strains were used in this study (Table 1). Reference strains were provided by the Spanish Type Culture Collection (CECT). Strains were grown on Tryptone Soya Agar (TSA, Oxoid, Basingstoke, UK) or Nutrient Agar (NA, Oxoid) for 2–3 days at 30 or 37°C and were stored in growth liquid medium containing 20% (v/v) glycerol at −80°C.
Phenotypic characterization
Phenotypic characterization of isolates was carried out following the standard procedures ISO 7932:2004 and ISO 21871:2006 (Anonymous 2004, 2006) which included colony morphology on B. cereus selective agar (polymyxin-egg yolk-mannitol-bromothymol blue, PEMBA, Oxoid), glucose fermentation, nitrate reduction, Voges-Proskauer and hemolytic activity on TSS agar (Tryptone Soya, including 5% defibrinated sheep blood, BioMérieux). Further biochemical characterization was carried out using the API CH50/B and API 20E identification kits (BioMèrieux, Geneve, Switzerland) according to the manufacturer’s instructions. The strips were incubated at 30°C and checked against the API identification index and database (APIweb version 4.0, BioMèrieux) after 16, 24, 40 and 48 h. The API 50CH/B and API 20E profiles were analyzed using the BioNumerics software version 2.5 (Applied Maths, Kortrijk, Belgium). Comparisons were carried out using the Jaccard coefficient and the data were clustered by using the un-weighted pair group method with averages (UPGMA) (Sneath and Sokal 1973).
The presence of the crystal protein (δ-endotoxin or parasporal body) of B. thuringiensis was investigated on smears obtained from colonies grown on Nutrient Agar (NA, Merck) for 3 days at 30°C. They were stained with basic fuchsine solution, and parasporal body production was verified by microscopic examination. Low temperature-tolerance, characteristic for B. weihenstephanensis, was determined on Nutrient Agar plates incubated at 7°C for 15 days.
DNA isolation
Genomic DNA was extracted following the guanidium thiocyanate method described by (Pitcher et al. 1989). DNA was spectrophotometrically quantified using an Ultrospec 2100 pro spectrophotometer (Amersham Biosciences UK Limited, Buckinghamshire, UK) and adjusted to a final concentration of 200 ng μl−1 in ultra pure water (Sigma-Aldrich, Madrid, Spain).
Screening of genes encoding B. cereus enterotoxins and virulence factors
The reference strains and food isolates within the B. cereus group and related Bacillus species (Table 1) were previously tested for the presence of virulence factor genes, i.e. phosphatidylcholine-specific phospholipase C (pc-plc), sphingomyelinase (sph), cereolysine O (clo), nonhaemolytic enterotoxin (nhe) and enterotoxin FM (entFM) (Martínez-Blanch et al. 2009). In the present study the presence of the genes encoding hemolysin BL (hbl), cytotoxin K (cytK) and cereulide (ces) was investigated by conventional PCR using the primer pairs HD2 F (5′-GTAAATTAIGATGAICAATTTC-3′)/HA4 R (5′-AGAATAGGCATTCATAGATT-3′), CK F2 (5′-ACAGATATCGGICAAAATGC-3′)/CK R5 (5′-CAAGTIACTTGACCIGTTGC-3′) and CesF1 (5′-GGTGACACATTATCATATAAGGTG-3′)/CesR2 (5′-GTAAGCGAACCTGTCTGTAACAACA-3′), respectively, as previously described (Ehling-Schulz et al. 2006). PCR mixture consisted of 1 μl of genomic DNA, 100 μmol l−1 of each deoxynucleotide triphosphate, 1.5 mmol l−1 MgCl2, 1 μmol l−1 concentration of each primer, 0.5 U of thermostable DNA polymerase (Genotaq polymerase, Genotek, Sabadell, Spain), and 1× PCR buffer (67 mmol l−1 Tris–HCl, pH 8.8; 16 mmol l−1 (NH4)2SO4; 0.1% Tween 20) in a final volume of 50 μl. Amplification conditions were: 5 min at 94°C, 35 cycles of 30 s at 94°C, 45 s at 56°C and 60 s at 72°C, followed by a final extension of 2 min at 72°C. Reactions were carried out in a GeneAmp PCR System 9700 thermal cycler (PE Biosystems, Foster City, USA). Five microliter of PCR products were visualized by electrophoresis in a 1.8% agarose (Pronadisa, Hispanlab, Madrid, Spain) gel in TAE buffer at 80 V for 90 min and ethidium bromide staining.
Fingerprinting analysis
ISR-PCR amplification
Amplification of the internal spacer region (ISR) between the 16S and 23S rRNA genes was carried out using bacterial specific primers: 1387V: 5′-GCCTTGTACACWCCGCCC-3′ and 118R: 5′-GTTNCCCCATTCRGA-3′ as previously described (Chenoll et al. 2003). PCR amplifications were conducted in a solution containing 1× PCR buffer (10 mmol l−1 Tris–HCl, pH 8.8; 1.5 mmol l−1 MgCl2, 50 mmol l−1 KCl, 0.1% Triton X-100), 100 μmol l−1 of each dNTP, 1 μmol l−1 of each primer, 0.5 U of thermostable DNA polymerase (Dynazyme II DNA polymerase, Finnzymes Oy, Finland), and 5 μl of DNA template, in a final volume of 50 μl. Amplification conditions were: 5 min at 94°C, 35 cycles of 30 s at 94°C, 45 s at 56°C and 45 s at 72°C and a final extension of 10 min at 72°C. Reactions were carried out in a GeneAmp PCR System 9700 thermal cycler (PE Biosystems).
RAPD analysis
Universal primers M13 (5′-GAAACAGCTATGACCATG-3′), T7 (5′-AATACGACTCACTATAGG-3′) and T3 (5′-ATTAACCCTCACTAAAGG-3′) were used as previously described (Pinto et al. 2005). PCR reaction was conducted in a total volume of 50 μl containing: 5 μmol l−1 universal primer, 0.5 U of thermostable DNA polymerase (Dynazyme II DNA polymerase), 5 μl of 1× PCR buffer, 3 mmol l−1 MgCl2 (Promega, Madison, USA), 0.2 mmol l−1 of each dNTP, and 5 μl of DNA template. Amplification was performed for one cycle of 94°C for 5 min, one cycle of 40°C for 5 min, one cycle of 72°C for 5 min and 33 cycles of 94°C for 20 s, 45°C (for M13 and T7 primers) or 41°C (for T3 primer) for 30 s and 72°C for 45 s in a GeneAmp PCR System 9700 thermal cycler (PE Biosystems).
Electrophoresis and banding pattern analysis
Fifteen microliter of amplification products (ISR and RAPD) and the molecular weight markers were electrophorezed on 1.8% agarose (low EEO; Pronadisa) gel in TAE buffer at 80 V for 90 min. Gels were stained with ethidium bromide and photographed under UV light. Gel images were recorded using a video camera (GelPrinter Plus, TDI, Madrid, Spain) and stored as TIFF files. Digitized images were converted, normalized, analyzed and combined with the Software package BioNumerics 2.5 (Applied Maths). In order to normalize the banding patterns, molecular weight markers were included every seven tracks. The levels of similarity between pairs of traces were computed by using the Jaccard coefficient for ISR and the Pearson correlation coefficient for M13/T7/T3-PCR, and the data were clustered by using the UPGMA algorithm (Sneath and Sokal 1973).
Cluster analysis
Clustering of multiple data sets, combining banding patterns (ISR, RAPD-M13, T3, and T7) and adding phenotypic traits (API profiles) and enterotoxin genotype, were calculated taking the similarity values from experiments with the aid of the “Composite data set” tool in the BioNumerics software. The consistence of the clusters obtained was estimated by calculating the cophenetic correlation values corresponding to the dendrograms. The concordance of groups formed with the techniques used was calculated by comparing the similarity matrices obtained from the different experiment types, using the Pearson product-moment correlation coefficient (Sneath and Sokal 1973).
Sequencing of the 16S rRNA gene
Genomic DNA of bacterial strains was used for amplification of the almost full-length 16S rRNA gene fragment using primers 616V (5′-AGAGTTTGATYMTGGCTCAG-3′) and 630R (5′-CAKAAAGGAGGTGATCC-3′) as previously described (Chenoll et al. 2003). Amplification products were purified using the UltraClean PCR clean-up kit (MoBio, Solana Beach, USA) and sequenced using a ABI PRISM 377 automated sequencer and the ABI PRISM BigDye Terminator Cycle Sequencing Ready Re-action Kit (PE Applied Biosystems). The 16S rRNA sequences of about 1400 bp were compared using the Blast-N tool (National Center for Biotechnology Information; www.ncbi.nlm.nih.gov).
Results
Phenotypic analysis
Identification by API profiles
Results on the phenotypic characterization are summarized in Table 1. Regarding the screening tests, B. cereus group reference and environmental strains showed the typical reaction on B. cereus selective agar (PEMBA), with the only exception of B44 which was negative for lecithin hydrolysis and mannitol assimilation. The remaining strains corresponding to other species were clearly differentiated by colony morphology on PEMBA agar. Growth on PEMBA and β-haemolysis test were the most discriminative assays for B. cereus group ascription with a 100% of positives, while Voges-Proskauer reaction, nitrate reduction and glucose fermentation scored, 98, 94, 88% of positives, respectively. According to API identification (ID > 80%), only three (CECT 148T, CECT 4094 and CECT 4387) out of 13 B. cereus reference strains were successfully identified as B. cereus 1 (11 strains) and B. cereus 2 (2 strains). The eight B. thuringiensis reference strains were classified as B. cereus 1 (seven strains) and B. cereus 2 (one strain), and the presence of the crystal protein was observed in three of them (Table 1). B. mycoides and B. pseudomycoides reference strains showed the B. mycoides API profile except CECT 4123 and CECT 4128T, but they were confirmed by exhibiting rhizoidal growth. B. weihenstephanensis showed a 90% identity with the B. mycoides API-profile but did not show rhizoidal growth. Of the remaining 13 non-“B. cereus group” reference strains, nine were successfully identified at species level, strain INRA P1 198, provided as B. circulans, was identified as B. pumilus, B. lentus CECT 18T showed 70.2% identity with the API profile for the species, and B. sphaericus resulted non-reactive since no growth was observed in any of the API 50CH substrates.
Regarding environmental isolates, only seven out of 30, B. cereus (B46, B47, B77), B. thuringiensis (B40, B48) and B. mycoides (B72, B74), were identified at species level because they showed an ID > 80% and/or the corresponding characteristic trait. Twenty-three strains showed ID < 80% but the significant taxons corresponded to species within the B. cereus group and were identified at “group level” (19 strains) or the significant taxon corresponded to other species in the genus and they were identified as Bacillus sp. (CECT 40, B54, B55, B64). These strains were confirmed as members of the B. cereus group by 16S rRNA gene sequencing showing a high degree of homology (>98%) with sequences of the reference strains in this group (data not shown).
Cluster analysis of API profiles
A UPGMA clustering was performed with the results of the similarity matrix obtained from the API50CH/20E profiles corresponding to 59 characters and 75 Bacillus strains (Cluster not shown). The cophenetic correlation value was 93%. The resulting dendrogram showed three clusters at 58% that grouped all but the type strains of the species B. lentus, B. firmus and B. sphaericus. Twenty-nine reference strains and 30 isolates of the B. cereus group (B. cereus, B. thuringiensis, B. weihenstephanensis, B. mycoides and B. pseudomycoides) clustered together. A second cluster included reference strains and environmental isolates belonging to the species B. megaterium, B. subtilis, B. licheniformis, B. circulans and P. polimyxa. B. pumilus type strain and environmental isolates belonging to this species formed a third cluster.
PCR screening of enterotoxin and virulence genes
A high incidence of virulence genes was previously reported for this panel of strains (Martínez-Blanch et al. 2009). Additionally the presence of other virulence factor genes, i.e. hbl, cytK and ces was investigated by conventional PCR in the present study. These genes were detected in 81, 56, and 3% of strains belonging to B. cereus group, respectively. Taking into account our previous results (Martínez-Blanch et al. 2009) most of the strains (90%) contained more than two genes, displaying 20 toxigenic profiles. Of them, the most frequent were hbl, nhe, cytK, pc-plc, sph, clo, entFM (22%), hbl, nhe, pc-plc, sph, clo, entFM (15%) and hbl, cytK, pc-plc, sph, clo, entFM (14%). Only two strains, B. mycoides CECT 4125 and B. pseudomycoides CECT 7065T, showed no amplification for the genes encoding food-related toxins (hbl, nhe, cytK, ces).
Genotypic analysis
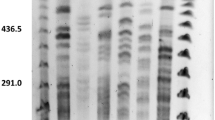
The 16S–23S ISR amplification products with the universal primers 1387V and 118R, yielded between two and five bands of molecular sizes ranging from 435 to 1078 bp. Two bands of 660 and 435 bp were present in all the B. cereus group strains, allowing differentiation from the rest of Bacillus species included in the study. Species specific profiles can be observed, i.e. a band of 594 bp was present in B. mycoides, the band of 660 bp was very faint in B. pseudomycoides, a band of 941 bp was present in B. cereus, and B. thuringiensis showed two or more extra bands of 941, 769 or 594 bp. The ISR profiles of food and environmental isolates included bands of different sizes in addition to the ones of 660 and 435 bp. However, differentiation between species within the B. cereus group was not supported by the cluster analysis with food, environmental and reference strains. Figure 1 shows a simplified dendrogram obtained with the ISR patterns corresponding to reference strains, food and environmental isolates, after the UPGMA clustering with a cophenetic correlation value of 94%. At 86% similarity three clusters were formed. Cluster 1 grouped B. cereus group strains; strains belonging to other Bacillus species grouped in Cluster 2 and 3 or remained unclustered.
Dendrograms derived from UPGMA cluster analysis based on: a the similarity matrix obtained from ISR-PCR profiles of the 75 Bacillus strains, including 29 reference strains of B. cereus group, 30 food isolates and 16 belonging to other Bacillus spp; b the combined similarity matrix obtained from RAPD-PCR profiles (M13, T3, and T7) of the 59 B. cereus group strains, including 29 reference strains and 30 food isolates. Tracks corresponding to the type strains of the species in the B. cereus group are marked: a B. weihenstephanensis CECT 5894T, b B. mycoides CECT 4128T, c B. pseudomycoides CECT 7065T, d B. cereus CECT 148T and e B. thuringiensis CECT 197T. The scale measures the similarity percentage. Clusters are numbered
Further genotypic differentiation was approached by RAPD-PCR using M13, T3 and T7 coliphage-based universal primers (RAPD-M13, RAPD-T7 and RAPD-T3). A new database was created in this study that includes the RAPD-PCR fingerprinting of 75 strains, together with their biochemical traits derived from API 50CH/API 20E profiles and their toxigenic genotype. The cluster analysis was calculated individually for the M13, T3 and T7 RAPD profiles and the corresponding cophenetic correlation values obtained for the dendrograms were 76, 86 and 75%, respectively. The clustering was calculated also based on the combined similarity matrix obtained from the three RAPD profiles and the cophenetic correlation value obtained for the dendrogram was 72% (Fig. 1). At 36% similarity four clusters were formed. Cluster I grouped reference strains belonging to B. mycoides, B. pseudomycoides and B. weihenstephanensis together with an environmental isolate (B69) that clustered near to these strains in the phenotypic analysis, as well; Cluster II grouped two food and two environmental isolates that had been identified as B. mycoides by API profile (B44, B66) or rhizoidal growth (B72, B74). Interestingly, B44 showed an atypical reaction on PEMBA. Cluster III grouped 26 strains, ten out of 12 B. cereus reference strains and B. thuringiensis CECT 4454 (crystal toxin positive), strain CECT 40, identified as B. cereus by 16S rRNA genes sequencing, and 14 food-related isolates. B. cereus CECT 40 and three isolates (B68, B64, B65) formed a subcluster, and they had in common the inability to hydrolyze starch and glycogen. Cluster IV grouped 20 strains, seven out of eight B. thuringiensis reference strains, two B. cereus reference strains (CECT 5148, CECT 193), ten food isolates and one environmental isolate, among them, strains B48 and B40 were crystal toxin positive. Ascription of food isolates into species within the B. cereus group was based on the clusters derived from RAPD M13, T3, T7, assessed by the API results, and 16S rRNA gene sequencing of phenotypically discordant isolates.
Comparison of clusters derived from the techniques used
The comparison of the groups formed with the different techniques used, in separate or combined, showed a high concordance between the grouping based on the Global data (RAPD M13, T3, and T7 fingerprints, ISR pattern, API 20E and API 50CH profiles and virulence/enterotoxin genotype) and the API 20E/50CH profiles (81.3%), followed by RAPD M13, T3, and T7 fingerprints (69.7%). Figure 2 shows the matrix of congruence values between techniques and a dendrogram derived from that matrix. The highest concordance obtained by a single technique corresponds to API 50CH profiles (75.3%), followed by ISR patterns (70.3%) and virulence/enterotoxin genotype (69.8%). Of them, ISR-PCR consisting in a single PCR reaction is easier to perform than virulence/enterotoxin genotyping, and it is not affected by growth of the strain as it is the case for API 50CH. Concordance between groupings obtained with the combined RAPD M13, T3 and T7 profiles and each of them on separate were 64.5, 63.6 and 62.3%, respectively.
Matrix of congruence values between the techniques applied calculated by using the Pearson product-moment correlation coefficient and dendrogram derived from that matrix. Shading corresponds to percentage of similarity and shows at a glance the concordance of groupings obtained between two techniques, i.e. groupings obtained with Global data (RAPD M13, T3, and T7 fingerprints, ISR pattern, API 50CH and API 20E profiles and virulence/enterotoxin genotype) have a 81.3% concordance with the ones obtained including API 50CH/20E profiles and 69.7% with the one corresponding to RAPD M13, T3, and T7. The highest concordance obtained by a single technique corresponds to API 50CH profiles (75.3%)
Discussion
So far no conclusive microbiological or biochemical media/reactions have been described for the identification of B. cereus “sensu stricto”. The key diagnostic features considered in the ISO7234:2004 for the enumeration of B. cereus strains are their ability to provoke hemolysis and to hydrolyze lecithin, but an inability to ferment mannitol. Although these tests have highly improved the identification of “presumptive B. cereus”, atypical strains for these two traits have been described that would lead to misidentifications. For instance, around 1% of the B. cereus strains are positive for mannitol fermentation according to the API profiles (API manual, BioMèrieux). Moreover, transcription of several extracellular proteins, including phospholipases, proteases, and hemolysins in B. cereus and B. thuringiensis is under the control of a pleiotropic regulator encoded by the plcR gene. Mutations in this gene have been associated to the hemolysis negative phenotype in B. anthracis or hemolysis and lecithinase negative phenotype in B. cereus group (Agaisse et al. 1999). For example, Slamti et al. (2004) reported this latter phenotype in 1% of the tested strains (four out of 400). Furthermore, haemolysis on blood agar has also been described in food isolates belonging to species that are not included within the B. cereus group like B. subtilis, B. licheniformis and B. pumilus (Ouoba et al. 2008).
We have included the API profiles because identification of presumptive colonies at the food industry is usually carried out using the API 50CH and API 20E biochemical tests which in most cases identify only at “B. cereus group” level, or even only at genus level. In fact, in the present work 3% of food isolates and strain CECT 40 were identified by API as belonging to genus Bacillus. This fact has been also stated by other authors in the identification of Bacillus strains from marrow purée (Guinebretière et al. 2001), rice (Sarrías et al. 2002) and cardboard packages (Priha et al. 2004). Guinebretière et al. (2001) observed discrepancies between API identification and molecular methods (16S rRNA gene sequencing) in 23% of strains. The cluster analysis of API profiles obtained in the present work failed to separate the strains of different species or even between B. cereus 1 and B. cereus 2 biotypes. However, it revealed a group of isolates that shared the inability to hydrolyze starch, glycogen and gluconate, or to ferment salicin. This pattern had been previously described in some B. cereus strains and was related to the production of emetic toxin (Ehling-Schulz et al. 2004). Accordingly, the only two ces positive isolates (B68, B77) were situated in this group. Since this group includes the isolates that could only be identified at genus level, it stresses the difficulty to detect these potentially toxigenic organisms using only biochemical tests. Nevertheless, in our study, the combined use of API20E and 50 CH has showed a good concordance with the groupings obtained with the global data.
Our previous results on virulence and enterotoxin genotypes showed a high prevalence within the members of B. cereus group in contrast to strains not belonging to this group (Martínez-Blanch et al. 2009). The presence of genes encoding toxins such as HBL, NHE, EntFM, PI-PLC and SPH had been reported in other species of the genus Bacillus (From et al. 2005; Ngamwongsatit et al. 2008; Taylor et al. 2005), but the presence of cytK in B. pumilus and B. subtilis is reported here for the first time.
Prevalence of hbl, nhe and cytK genes within the B. cereus group isolates was observed in agreement to previously reported results (Guinebretière et al. 2002; Ouoba et al. 2008; Schoeni and Lee Wong 2005). Besides, the association hbl/nhe/cytK, that had been emphasized by Guinebretière et al. (2002) in outbreak isolates, was reported in the present study in 44% of the B. cereus group strains, indicating their toxigenic potential. The ces gene related to the vomiting syndrome, showed a low presence (4%) which is in line with other studies carried out in strains from ground, vegetables or industries of milky products and farms (Altayar and Sutherland 2006; Ehling-Schulz et al. 2006; Kim et al. 2009; Svensson et al. 2007).
Regarding identification by molecular techniques, ISR-PCR fingerprinting of whole genomes was selected according to the presence of a high number of ribosomal operons (from eight to 12) in the species of B. cereus group (Lechner et al. 1998). They can differ in the ISR sequence which might lead to detect interspecies and intraspecies differences based on agarose patterns following amplification with universal primers. ISR-PCR fingerprinting of whole genomes had previously been applied for the identification of species of the genus Bacillus revealing different patterns in B. cereus and B. licheniformis (Daffonchio et al. 1998), but an identical profile for the six species of the B. cereus group (Daffonchio et al. 2000). In the present study, we used universal primers (Chenoll et al. 2003), different from those used by Daffonchio et al. (2000), for the ISR fingerprinting analysis of our collection of 75 Bacillus strains. It revealed as very useful for the rapid ascription of isolates to the “B. cereus group”, although similarly to the results obtained with phenotypic studies, some B. thuringiensis and B. cereus strains appeared dispersed in different branches within the “B. cereus group” main cluster.
We further analyzed the genetic diversity of isolates and reference strains within the B. cereus group by RAPD-PCR to approach the potential contribution of extrachromosomal or modular mobile elements to the genome plasticity and biodiversity of the members of B. cereus group. In this study, RAPD-M13, RAPD-T3 and RAPD-T7 profiles on separate did not succeed in species separation, which confirmed the great diversity of B. cereus group strains. However, when their profiles were combined, strains belonging to the species B. mycoides/B. pseudomycoides/B. weihenstephanensis clustered apart from B. cereus and B. thuringiensis, although some strains were trans-located between the last two branches. The intermixed position of B. cereus and B. thuringiensis has been reported in different phylogenetic studies based on MLEE, MLST and AFLP profiles (reviewed by Tourasse et al. 2006) and more recently, in an extended and global study combining MLST, AFLP, and MLEE genotyping data (Tourasse et al. 2010). These results corroborate that B. cereus and B. thuringiensis cannot be distinguished. Derived from the present study, a new database with the ISR-PCR, RAPD-PCR and enterotoxin profiles was built which provided the identification of the isolates under study at species level.
In conclusion, after a comparative evaluation of several phenotypic and molecular techniques, a PCR-based schema has been established for the rapid identification of potentially toxigenic B. cereus group isolates that sequentially includes: PCR-ISR profile, a simple and efficient test without requiring expensive equipment (at group level), and RAPD profile (at species level). The unequivocal identification of B. cereus sensu stricto at species level would require more sophisticated and/or cumbersome techniques, such as DNA/DNA hybridization, only possible in specialized laboratories and not suitable for routine analysis. Therefore, the approach proposed here is a valid tool for addressing quality control at the food industry.
References
Abriouel H, Ben Omar N, Lucas R, Martínez-Cañamero M, Ortega E, Gálvez A (2007) Differentiation and characterization by molecular techniques of Bacillus cereus group isolates from Poto Poto and Dégué, two traditional cereal-based fermented foods of Burkina Faso and Republic of Congo. J Food Prot 70:1165–1173
Agaisse H, Gominet M, Okstad OA, Kolsto AB, Lereclus D (1999) PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol Microbiol 32:1043–1053
Altayar M, Sutherland AD (2006) Bacillus cereus is common in the environment but emetic toxin producing isolates are rare. J Appl Microbiol 100:7–14
Anonymous (2004) ISO 7932:2004 Microbiology of food and animal feeding stuffs—horizontal method for the enumeration of presumptive Bacillus cereus—colony-count technique at 30 degrees C. International Organization for Standardization, Geneve
Anonymous (2006) ISO 21871:2006 Microbiology of food and animal feeding stuffs—horizontal method for the determination of low numbers of presumptive Bacillus cereus—most probable number technique and detection method. International Organization for Standardization, Geneve
Beattie SH, Williams AG (1999) Detection of toxigenic strains of Bacillus cereus and other Bacillus ssp. with an improved cytotoxicity assay. Lett Appl Microbiol 28:221–225
Chenoll E, Macián MC, Aznar R (2003) Identification of Carnobacterium, Lactobacillus, Leuconostoc and Pediococcus by rDNA-based techniques. Syst Appl Microbiol 26:546–556
Choma C, Guinebretière MH, Carlin F, Schmitt P, Velge P, Granum PE, Nguyen-The C (2000) Prevalence, characterization and growth of Bacillus cereus in commercial cooked chilled foods containing vegetables. J Appl Microbiol 88:617–625
Cooper RM, McKillip JL (2006) Enterotoxigenic Bacillus spp. DNA fingerprint revealed in naturally contaminated non-fat dry milk powder using rep-PCR. J Basic Microbiol 5:358–364
Daffonchio D, Borin S, Consolandi A, Mora D, Manachini PL, Sorlini C (1998) 16S–23S rRNA internal transcribed spacers as molecular markers for the species of the 16S rRNA group I of the genus Bacillus. FEMS Microbiol Lett 163:229–236
Daffonchio D, Cherif A, Borin S (2000) Homoduplex and heteroduplex polymorphisms of the amplified ribosomal 16S–23S internal transcribed spacers describe genetic relationships in the “Bacillus cereus group”. Appl Environ Microbiol 66:5460–5468
Dierick K, Van Coillie E, Swiecicka I, Meyfroid G, Devlieger H, Meulemans A, Hoedemaekers G, Fourie L, Heyndrickx M, Mahillon J (2005) Fatal family outbreak of Bacillus cereus-associated food poisoning. J Clin Microbiol 43:4277–4279
Ehling-Schulz M, Fricker M, Scherer S (2004) Bacillus cereus, the causative agent of an emetic type of food-borne illness. Mol Nutr Food Res 48:479–487
Ehling-Schulz M, Guinebretière MH, Monthán A, Berge O, Fricker M, Svensson B (2006) Toxin gene profiling of enterotoxic and emetic Bacillus cereus. FEMS Microbiol Lett 260:232–240
European Food Safety Authority (2009) The community summary report on food-borne outbreaks in the European Union in 2007. EFSA J 271:1–128
Fricker M, Messelhäuber U, Busch U, Scherer S, Ehling-Schulz M (2007) Diagnostic Real-Time PCR assays for the detection of emetic Bacillus cereus strains in foods and recent food-borne outbreaks. Appl Environ Microbiol 73:1892–1898
From C, Pukall R, Schumann P, Hormazábal V, Granum PE (2005) Toxin-producing ability among Bacillus spp. outside the Bacillus cereus group. Appl Environ Microbiol 71:1178–1183
Gaviria AM, Granum PE, Priest FG (2000) Common occurrence of enterotoxin genes and enterotoxicity in Bacillus thuringiensis. FEMS Microbiol Lett 190:151–155
Granum PE (2002) Bacillus cereus and food poisoning. In: Berkeley R, Heyndrickx M, Logan N, De Vos P (eds) Applications and systematics of Bacillus and relatives). Blackwell Science Ltd, Oxford, pp 141–159
Guinebretière MH, Berge O, Normand P, Morris C, Carlin F, Nguyen-The C (2001) Identification of bacteria in pasteurized zucchini purées stored at different temperatures and comparison with those found in other pasteurized vegetable purées. Appl Environ Microbiol 67:4520–4530
Guinebretière MH, Broussolle V, Nguyen-The C (2002) Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J Clin Microbiol 40:3053–3056
Helgason E, Okstad OA, Caugant DA, Johansen HA, Fouet A, Mock M, Hegna I, Kolsto AB (2000) Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis one species on the basis of genetic evidence. Appl Environ Microbiol 66:2627–2630
Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolstø AB (2004) Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Appl Environ Microbiol 70:191–201
Hill KK, Ticknor LO, Okinaka RT, Asay M, Blair H, Bliss KA, Laker M, Pardington PE, Richardson AP, Tonks M, Beecher DJ, Kemp JD, Kolstø AB, Lee Wong AC, Kleim P, Jackson PJ (2004) Fluorescent amplified fragment length polymorphism analysis of Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis isolates. Appl Environ Microbiol 70:1068–1080
Jensen GB, Larsen P, Jacobsen BL, Madsen B, Wilcks A, Smidt L, Andrup L (2002) Isolation and characterization of Bacillus cereus-like bacteria from faecal samples from greenhouse workers who are using Bacillus thuringiensis-based insecticides. Int Arch Occup Environ Health 75:191–196
Kim SK, Kim KP, Jang SS, Shin EM, Kim MJ, Oh S, Ryu S (2009) Prevalence and toxigenic profiles of Bacillus cereus isolated from dried red peppers, rice, and Sunsik in Korea. J Food Prot 72:578–582
Kwon GH, Lee HA, Park JY, Kim JS, Lim J, Park CS, Kwon DY, Kim YS, Kim JH (2009) Development of a RAPD-PCR method for identification of Bacillus species isolated from Cheonggukjang. Int J Food Microbiol 129:282–287
Lechner S, Mayr R, Francis KP, Prüß BM, Kaplan T, Wießner-Gunkel E, Stewart GSAB, Scherer S (1998) Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol 48:1373–1382
Logan NA, Berkeley RCW (1984) Identification of Bacillus strains using the API system. J Gen Microbiol 130:1871–1882
Manzano M, Cocolin L, Cantoni C, Comi G (2003) Bacillus cereus, Bacillus thuringiensis and Bacillus mycoides differentiation using a PCR-RE technique. Int J Food Microbiol 81:249–254
Martínez-Blanch JF, Sánchez G, Garay E, Aznar R (2009) Development of a real-time PCR assay for detection and quantification of enterotoxigenic members of Bacillus cereus group in food samples. Int J Food Microbiol 135:15–21
Ngamwongsatit P, Buasri W, Pianariyanon P, Pulsrikarn C, Ohba M, Assavanig A, Panbangred W (2008) Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int J Food Microbiol 121:352–356
Ouoba LI, Thorsen L, Varnam AH (2008) Enterotoxins and emetic toxins production by Bacillus cereus and other species of Bacillus isolated from Soumbala and Bikalga, African alkaline fermented food condiments. Int J Food Microbiol 124:224–230
Pinto B, Chenoll E, Aznar R (2005) Identification and typing of food-borne Staphylococcus aureus by PCR-based techniques. Syst Appl Microbiol 28:340–352
Pitcher DG, Saunders NA, Owen RJ (1989) Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol 8:151–156
Priha O, Hallamaa K, Saarela M, Raaska L (2004) Detection of Bacillus cereus group bacteria from cardboard and paper with real-time PCR. J Ind Microbiol Biotechnol 31:161–169
Pruss BM, Dietrich R, Nibler B, Martlbauer E, Scherer S (1999) The hemolytic enterotoxin HBL is broadly distributed among species of the Bacillus cereus group. Appl Environ Microbiol 65:5436–5442
Rasko DA, Altherr MR, Han CS, Ravel J (2005) Genomics of the Bacillus cereus group of organisms. FEMS Microbiol Rev 29:303–329
Ronimus RS, Parker LE, Turner N, Poudel S, Rückert A, Morgan HW (2003) A RAPD-based comparison of thermophilic bacilli from milk powders. Int J Food Microbiol 85:45–61
Sarrías JA, Valero M, Salmerón MC (2002) Enumeration, isolation and characterization of Bacillus cereus strains from Spanish raw rice. Food Microbiol 19:589–595
Schoeni JL, Lee Wong AC (2005) Bacillus cereus food poisoning and its toxins. J Food Prot 68:636–648
Shangkuan YH, Yang J-F, Lin H-C, Shaio M-F (2000) Comparison of PCR-RFLP, ribotyping and ERIC-PCR for typing Bacillus anthracis and Bacillus cereus strains. J Appl Microbiol 89:452–462
Slamti L, Perchat S, Gominet M, Vilas-Boas G, Fouet A, Mock M, Sanchis V, Chaufaux J, Gohar M, Lereclus D (2004) Distinct mutations in PlcR explain why some strains of the Bacillus cereus group are nonhemolytic. J Bacteriol 186:3531–3538
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman, San Francisco
Stenfors LP, Mayr R, Scherer S, Granum PE (2002) Pathogenic potential of fifty Bacillus weihenstephanensis strains. FEMS Microbiol Lett 215:47–51
Svensson B, Eneroth A, Brendehaug J, Christiansson A (1999) Investigation of Bacillus cereus contamination sites in a dairy plant with RAPD-PCR. Int Dairy J 9:903–912
Svensson B, Monthán A, Guinebretière MH, Nguyen-Thé C, Christiansson A (2007) Toxin production potential and the detection of toxin genes among strains of the Bacillus cereus group isolated along the dairy production chain. Int Dairy J 17:1201–1208
Taylor JMW, Sutherland AD, Aidoo KE, Logan NA (2005) Heat-stable toxin production by strains of Bacillus cereus, Bacillus firmus, Bacillus megaterium, Bacillus simplex and Bacillus licheniformis. FEMS Microbiol Lett 242:313–317
Torkar KG, Mozina SS (2000) Differentiation of Bacillus cereus isolates from milk and milk products with biochemical, immunological, AP-PCR and PCR-RFLP methods. Food Technol Biotechnol 38:135–142
Tourasse NJ, Helgason E, Økstad OA, Hegna IK, Kolstø AB (2006) The Bacillus cereus group: novel aspects of population structure and genome dynamics. J Appl Microbiol 101:579–593
Tourasse NJ, Helgason E, Klevan A, Sylvestre P, Moya M, Haustant M, Økstad OA, Fouet A, Mock M, Kolstø AB (2010) Extended and global phylogenetic view of the Bacillus cereus group population by combination of MLST, AFLP, and MLEE genotyping data. Food Microbiol. doi:10.1016/j.fm.2010.06.014
Valero M, Hernández-Herrero LA, Fernández PS, Salmerón MC (2002) Characterization of Bacillus cereus isolates from fresh vegetables and refrigerated minimally processed foods by biochemical and physiological tests. Food Microbiol 19:491–499
Vilas-Boas GT, Peruca AP, Arantes OM (2007) Biology and taxonomy of Bacillus cereus, Bacillus anthracis, and Bacillus thuringiensis. Can J Microbiol 53:673–687
Yang IC, Shih DYCh, Huang TP, Huang YP, Wang JY, Pan TM (2005) Establishment of a novel multiplex PCR assay and detection of toxigenic strains of the species in the Bacillus cereus group. J Food Prot 68:2123–2130
Acknowledgments
This work was supported by “Comisión Interministerial de Ciencia y Tecnología” (CICYT) grant AGL2000-1462 and the CECT. J.F. Martínez is the recipient of a Ph.D. fellowship UA-BPD2002 and G. Sánchez is the recipient of a JAE doctor grant both from the “Consejo Superior de Investigaciones Científicas” (CSIC). We thank Beatriz Pinto for technical assistance and Dr. Tomás (AINIA, Valencia, Spain) for providing environmental and food B. cereus isolates.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martínez-Blanch, J.F., Sánchez, G., Garay, E. et al. Evaluation of phenotypic and PCR-based approaches for routine analysis of Bacillus cereus group foodborne isolates. Antonie van Leeuwenhoek 99, 697–709 (2011). https://doi.org/10.1007/s10482-010-9545-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-010-9545-y