Abstract
Neuroscience is of emerging importance along with the contributions of Operational Research to the practices of diagnosing neurodegenerative diseases with computer-aided systems based on brain image analysis. Although multiple biomarkers derived from Magnetic Resonance Imaging (MRI) data have proven to be effective in diagnosing Alzheimer’s disease (AD) and mild cognitive impairment (MCI), no specific system has yet been a part of routine clinical practice. This paper aims to introduce a fully-automated voxel-based procedure, Voxel-MARS, for detection of AD and MCI in early stages of progression. Performance was evaluated on a dataset of 508 MRI volumes gathered from the Alzheimer’s Disease Neuroimaging Initiative database. Data were transformed into a high-dimensional space through a feature extraction process. A novel 3-step feature selection procedure was applied. Multivariate Adaptive Regression Splines method was used as a classifier for the first time in the field of brain MRI analysis. The results were compared to those presented in a previous study on 28 voxel-based methods in terms of their ability to separate control normal (CN) subjects from the ones diagnosed with AD and MCI. It was observed that our method outperformed all of the others in sensitivity (83.58% in AD/CN and 78.38% in MCI/CN classification) with acceptable specificity values (over 85% in both cases). Furthermore, the method worked for discriminating MCI patients which converted to AD in 18 months (MCIc) from non-converters (MCInc) with a sensitivity outcome better than 27 of 28 methods. Overall, it was shown that the proposed method is promising in early detection of AD.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Alzheimer’s disease (AD) is the most common form of dementia that causes problems with thinking, memory and behavior. It gradually affects all cognitive functions and eventually causes death. This neurodegenerative disease is known to be related to structural atrophy, pathological amyloid depositions, and metabolic alterations in the brain (Jack et al. 2010). The underlying mechanism causing the deformations has not yet been completely explained and it remains to be a topic of interest in the fields of neurology and mathematical modeling (Álvarez-Miranda et al. 2016). Since histopathological confirmation of amyloid plaques and neurofibrillary tangles is required for definite diagnosis, it can only be made through a postmortem examination (Tiraboschi et al. 2004). However, early detection of AD has increasingly gained importance along with efforts to delay the onset or prevent progression of the disease and develop an effective treatment.
Unfortunately, no single test that proves a person has AD or mild cognitive impairment Footnote 1 (MCI) currently exists. Diagnosis is made through a complete assessment including medical history, physical examination and diagnostic tests (e.g., clinical, laboratory, and genetic tests—such as APOE-e4 gene), neurological examination (of reflexes, coordination, muscle tone and strength, eye movement, speech, and sensation), and mental status tests (such as mini-mental state exam (MMSE), mini-cog, and mood assessment). A medical workup for AD often includes structural imaging with Magnetic Resonance Imaging (MRI) or Computed Tomography (CT) (and functional imaging, too). However, brain imaging techniques are primarily used to eliminate other probable conditions such as tumors, stroke, and damage from trauma or fluid collection in intracranial compartments, which may cause symptoms similar to AD but require different kinds of treatment.
Multiple biomarkers have been shown to be sensitive to existence of AD and MCI; i.e., structural MRI for brain atrophy measurement, functional imaging (such as FDG-PET: Fluorodeoxyglucose Positron Emission Tomography) for hypometabolism quantification, and cerebrospinal fluid (CSF) for quantification of specific proteins (Zhang et al. 2011). Computer-aided diagnosis (CAD) systems for AD and MCI have been proposed and tested for PET and SPECT (Single-Photon Emission Computed Tomography) (Padilla et al. 2010; Ramìrez et al. 2013; Salas-Gonzalez et al. 2010, 2009), structural MRI (Davatzikos et al. 2008; Gerardin et al. 2009; Klöppel et al. 2008; Savio and Graa 2013), and Diffusion Tensor Imaging (DTI) (Graa et al. 2011).
T1-weighted (W) MRI is a very well-known imaging technique to provide high-resolution and high-contrast anatomical brain images in three dimensions. T1-W MRI technique uses T1 relaxation time (a measure which is related to the recovery time of longitudinal magnetization created by a strong magnetic field, after the excitation pulse is applied) as a physical property to form images. Resulting images are composed of voxels (i.e., volumetric pixels) where each voxel is associated to an intensity value varying between 0 and 1 (where 0 is black, 1 is white, and any rational number between these bounds is mapped into a gray-scale color). “Voxel index” is defined as a three dimensional (3D) vector defining a specific position in a volumetric image, and “voxel intensity” is the numeric value representing the gray-scale color of that specific position. T1-W MR images provide a good contrast between the anatomical structures forming central nervous system, such as white matter (seeming brighter with a high content of fat) and cerebrospinal fluid (seeming darker because of a high content of water). It has already been proven that both the features based on specific anatomical structures such as hippocampus, amygdale and entorhinal cortex (Boutet et al. 2014; Chupin et al. 2009a, b; Colliot et al. 2008; Morra et al. 2010; Westman et al. 2011), and the voxel-based features derived from whole-brain anatomy (Adaszewski et al. 2013; Davatzikos et al. 2008; Li et al. 2014; Ye et al. 2008; Zhang and Davatzikos 2011) may serve as biomarkers to detect AD.
There exist many recent classification methods which allow individual class prediction. In particular, Support Vector Machine (SVM) classifiers are widely used to help differentiating subjects with AD from the healthy ones. An SVM constructs hypersurfaces in a high-dimensional space, which can be used for classification and regression. In Klöppel et al. (2008), an SVM classifier is directly applied, whereas in Chaves et al. (2009), Magnin et al. (2009), Misra et al. (2009), Padilla et al. (2012), Vemuri et al. (2008) and many other Alzheimer’s CAD systems, the dimensionality of the feature space relying on different information sources (e.g. MRI/PET/SPECT images, laboratory results, genetic information) is firstly reduced. This is a crucial step in order not to suffer from a well-known common issue in machine learning; the so-called “curse of dimensionality” or, equivalently, “peaking phenomenon” (Jain et al. 2000). There also exist studies which use machine learning methods other than SVM classification; i.e., Principal Component Analysis (PCA)-based techniques (Alvarez et al. 2009; López et al. 2011; Park et al. 2012), Linear Discriminant Analysis (LDA)-based approaches (Salas-Gonzalez et al. 2010; Savio and Graa 2013), Random Forest Classifiers (Chincarini et al. 2011; Ramìrez et al. 2010), Multiple Kernel Learning (MKL) approaches (Ye et al. 2011; Zhang et al. 2011), Artificial Neural Networks (Segovia et al. 2012), and Deep Learning (Liu et al. 2014; Suk et al. 2014).
In this study, a fully-automated machine learning routine to solve the problem of early detection of AD is introduced and the performance of the proposed method is evaluated. Main contribution of our approach is the utilization of the Multivariate Adaptive Regression Splines (MARS) method for classification of structural brain MRI images for detection of AD. Employing MARS made flexible models available for the domain, since this method does not obligate making preliminary assumptions on the characteristics of the input data. A raw feature set was derived from the voxel-wise tissue probabilities for 3 tissue classes (TCs) which form the brain; namely, white matter (WM), gray matter (GM), and cerebrospinal fluid (CSF). Unified Segmentation (Ashburner and Friston 2005) and DARTEL Image Registration (Ashburner 2007) methods—followed by modulation and Gaussian smoothing—were applied to transform the original MRI data into the form of tissue probability maps. Moreover, a novel 3-step decision-making procedure for determining the final subset of significant features was proposed.
The remaining parts of this paper are organized as follows: In Sect. 2, materials and the methods used in this study are explained. Information associated with the studied data is provided. Data preparation procedure involving tissue segmentation, bias field correction, registration, modulation and smoothing is introduced. Additionally, a brief subsection is reserved for the MARS method and its use in solving classification problems. Details of the experimental setup, performance evaluation results, a qualitative discussion, and conclusions together with an outlook to future studies can be found in Sects. 3 and 4, respectively.
2 Materials and methods
2.1 Data description
Data used for training, validation, and testing purposes were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database.Footnote 2 ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public/private partnership. The primary goal of ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessments can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of a very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
Cuingnet et al. (2011) evaluated the performance of 10 different approaches of other researchers (5 voxel-based methods, 3 methods based on cortical thickness and 2 methods based on the hippocampus), using 509 subjects from the ADNI archive. 3 classification experiments were performed: (i) CN versus AD, (ii) CN versus MCIc (MCI who had converted to AD within 18 months, MCI converters), and (iii) MCIc versus MCInc (MCI who had not converted to AD within 18 months, MCI non-converters). They only used T1-W MR images for the experiments. MRI acquisition had been done according to the ADNI acquisition protocol in Jack et al. (2008). For each subject, MRI scan—when available from the baseline visit, and otherwise from the screening visit—was chosen. To enhance standardization across centers and platforms of images acquired in the ADNI study, pre-processed images which were exposed to some processes for correction of certain image artifacts were used. These preprocessing steps involve image geometry corrections for gradient nonlinearity and magnetic field intensity non-uniformity corrections due to non-uniform receiver coil sensitivity, both of which can be directly applied on the MRI console. All subjects were scanned twice at each visit. MR scans were graded qualitatively by the ADNI investigators for artifacts and general image quality. Each scan was graded considering several criteria, such as, blurring, ghosting, homogeneity, flow artifact, intensity, etc. For each subject, the MRI scan which was considered as the “best” quality scan by the ADNI investigators is used. Here, “best” is defined as the one which was used for the complete preprocessing steps in ADNI methods web page.Footnote 3
In this study, it was decided to work on the same set of training and testing images as used in Cuingnet et al. (2011), in consideration of the objectivity of the comparative performance evaluation. Information involving the demographic characteristics of the training and the test subjects is presented in Table 1. The whole set was formed by exactly the same images except for a single one belonging to a subject with AD in the test group (the group is indicated with “*” in the table). Since it was observed that the SPM12 Toolbox could not produce meaningful results at the end of the image processing routine, the image is left out and the group statistics are updated accordingly.
2.2 Voxel-wise tissue probability maps
2.2.1 Unified segmentation
Segmentation of the brain images for extraction of different tissue classes usually takes two forms: The first one is classification of voxels in terms of probabilities of belonging to specified tissue, and the second one is directly registering the images to pre-determined templates. Unified Segmentation (Ashburner and Friston 2005) can be defined as a probabilistic framework which combines tissue classification and image registration—with addition of bias field correction—operations in an iterative scheme within the same generative model. In Ashburner and Friston (2005), it is marked that brain tissue segmentation by estimating Unified Segmentation model parameters produces more accurate results than sequential applications of each component separately.
The objective function minimized by the optimum parameters is derived from a Mixture of Gaussians (MoG) model. Equation (1) shows the standard MoG model. Overall probability distribution can be modelled through a mixture of K Gaussians. The kth Gaussian is modelled by its mean (\(\mu _k\)), variance (\(\sigma _k^2\)), and mixing proportion (\(\gamma _k\)), where \(\gamma _k\ge 0\) \((k=1,\ldots ,K)\) and \(\sum _{k=1}^{K} \gamma _k = 1\):
After the insertion of bias field (modelled by additive noise and scaling) and the information coming from the priors (TC templates), and taking the logarithm of both sides, the objective function (to be minimized) takes the final form:
where \(\rho _i(\beta )\) represents the bias field with parameter vector \(\beta \), and \(b_{i,k}(\alpha )\) stands for the prior spatial information with parameter vector \(\alpha \) originating from TC templates \((i=1,\ldots ,I;~k=1,\ldots ,K)\). Maximization of the probabilities defined by the MoG function in Eq. (1) is accomplished when the right-hand side of Eq. (2) is minimized (with respect to \(\mu \), \(\sigma \), \(\gamma \), \(\beta \), and \(\alpha \)), because these two functions are monotonically related. The reader is referred to the original work of Ashburner and Friston (2005) for a detailed explanation of the method and derivation of the objective function in Eq. (2). The optimization problem is solved iteratively by using a scheme based on Expectation-Maximization (EM) algorithm, where the E-step involves computation of the tissue probabilities, and the M-step involves computation of the cluster and the non-uniformity field parameters (Frackowiak et al. 2003).
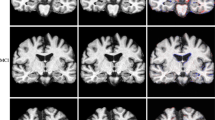
Unified Segmentation implementation in SPM12 Toolbox (Ashburner 2009) was used for producing Tissue Probability Maps (TPMs). Spatial prior information for each one of the TCs was provided by ICBM Tissue Probabilistic Atlas,Footnote 4 2D axial cross-section images of which are shown in Fig. 1. The numbers of Gaussians for each TC were assigned as 3 for GM, 2 for WM and 2 for CSF based on the typical values given in Ashburner and Friston (2005). The procedure for selecting the optimal number of Gaussians per class was declared to be a model-order selection issue and was not addressed in the referenced study. It is observed that typical K values which we used in this paper are equal to the ones that are given as default unified model parameters provided by multiple versions of SPM Toolbox.
Warping a set of images to a template image causes differences in total tissue volumes. This negative effect of spatial normalization was eliminated by applying a modulation operation for updating the voxel intensities to compensate the volume differences introduced by warping.
2.2.2 Nonlinear image registration
Images belonging to different subjects and acquired at different time frames must be spatially aligned and resampled in order to make the dataset ready for further analysis with increased localization and, thus, with higher sensitivity. For this purpose, we took advantage of the DARTEL (Diffeomorphic Anatomical Registration through Exponentiated Lie Algebra) Toolbox of SPM, which had been implemented on the grounds of the study by Ashburner (2007).
First of all, DARTEL templates were created with a setup of 6 outer iterations including 3 inner loops per each. At the end of every outer iteration, each individual image in the training set was warped to match the existing template, and warped images were averaged to regenerate a new template. The starting version of the template was assumed to be the average of the original tissue probability maps. The number of inner iterations indicates the number of Gauss–Newton iterations within the outer loop. The whole process starts with a coarse registration and iteratively evolves into a relatively more accurate state with higher detail and less amount of regularization. Deformations were parametrized by and stored in the form of flow-fields.
Afterwards, the test set was processed for nonlinear registration by matching each of the individual images to the existing templates generated in the former step. It is essential at this step not to allow the images in the test set to share information among each other for the objectivity and the extensibility of the tests. More specifically, DARTEL flow-fields belonging to the images in the test set were computed using templates which were acquired using the training images. If new templates based on test data were generated and used, the registration process would have caused misleading classification results. Moreover, it would have been impossible to obtain the same result for a particular image when it is processed together with other groups of test images or alone. Additionally, insertion of a new image into the test set would have required repeating the top–down procedure for registration, causing updated deformations in other images.
Finally, train and test images were normalized to the MNI-spaceFootnote 5 paying attention to the considerations stated in previous paragraph. The final template and the flow-fields generated at the previous step were used for this purpose. Consequently, a modulation operation was performed to preserve the total amount of information encapsulated by the 3 TCs. Finally, the images were smoothed with a FWHM Gaussian filter and saved to the disk in NIfTIFootnote 6 file format. At the end of this process, each individual tissue probability map was aligned with the MNI-space with dimensions of (121–145–121) and resolution of 1.5 mm in all three orthogonal directions. Figure 2 shows the final (segmented, warped, modulated, and smoothed) version of 3 tissue probability maps belonging to an image randomly chosen from the AD group of the training set.
Since we treated each voxel as a feature set containing the tissue probabilities belonging to 3 separate brain TCs as elements, the overall dimensionality of the raw input space \(\mathrm{I}\mathrm{R}^p\) appeared to be:
For example, in the AD/CN classification case, only 150 training images were used. This means that we have 150 samples, each one of them being represented as a point in a p-dimensional space, where each dimension of this feature space is called a tissue probability index. Evidently, a classifier with that proportion of sample and variable sizes in training data is highly likely to have a poor generalization ability, due to the very well-known phenomenon of Curse of Dimensionality (Jain et al. 2000). Furthermore, high-dimensional vector operations directly cause high costs for memory use and computational time. In consideration of these reasons, a sequence of algebraic operations for determination of an optimal set of significant features was designed and applied. Steps involved in this procedure are explained in the following subsection.
2.3 Determination of the subset of significant features: a 3-step approach
In machine learning problems, prediction accuracy of a classifier does not necessarily increase with increasing number of features, unless class-conditional densities are completely known (i.e., we have made all possible observations) (Jain et al. 2000). This phenomenon is the aforementioned Curse of Dimensionality and is well-recognized in most problem domains associated with pattern classification.
According to the recent review study of Mwangi et al. (2014), in a majority of the neuroimaging studies, the sample size is smaller than 1000 and the number of non-zero voxels (voxels which point to the positions included in the brain tissue, thus, carrying a non-zero intensity value) contained in a 3D brain scan is larger than 100,000. This statement is consistent with our case, in which the number of non-zero features is in the order of 1,000,000 and training set sample sizes lie between 100 and 150. Consequently, a model has a high potential of overfitting (which is also referred as overlearning or memorization in the literature) to the training data, if it is built without elimination of the irrelevant features. Additionally, it is obvious that for computational issues such as solvability and reasonable time and memory costs, the number of features should be reduced to a more narrow range. For these reasons, application of a feature selection procedure before the model building process is crucial in a neuroimaging study.
If there is no obligation imposing that the remaining feature space must be a subspace of the original one, the number of variables could be reduced by applying linear or nonlinear transformations on the raw data. In this case, the remaining features are not the ones selected among the originals, but new ones derived from the original data. This concept involves many linear and nonlinear methods such as Principal Component Analysis (PCA), Multidimensional Scaling (MDS), Kernel PCA, Diffusion Maps, Laplacian Eigenmaps, and Generalized Discriminant Analysis (GDA). A remarkably good comparative review on many of the methods falling into this group can be seen in van der Maaten et al. (2009).
In our study, feature transformation based approaches were not employed because of the following reasons: (i) Feature selection (without transformation) provides a clearer understanding of the main question of interest; for example, with revealing significant regions in the brain scans. (ii) Those transformations usually require operations in spectral domain such as solution of eigenvalue problems, which are usually expensive at such high dimensions. Also, inherent constraints of those approaches are generally not in consistence with the domain. (For example, PCA is not able to provide a set of transformed vectors, of which the number of elements are greater than the number of training samples, which is not adequate in our case.) Shih et al. (2014), stated this fact as follows: “Conventional variable selection techniques are based on assumed linear model forms and cannot be applied in this ‘large p and small N’ problem.” (iii) It was observed that the insertion of the domain knowledge via a more heuristic approach affects the overall performance in the positive direction (see Sect. 3.1).
Instead of working with space-transformative dimensionality-reduction techniques, we preferred to develop a methodology to choose optimal subsets of features from the raw dataset constrained to pre-determined parameters. Our methodology consists of three steps, namely, Statistical Analysis, Tissue Probability Criteria, and Within-class Norm Thresholding. It is essential to note that each step is applied on the raw data (after removal of the zero-voxels) and the union set of eliminated features are specified by superposing individual sets, at the end of the procedure. Therefore, the proposed procedure does not impose carrying out the three steps in this specific order. It should also be clarified that while we are claiming our 3-step feature selection procedure to be novel, we are not implying novelty for each individual step. In particular, Step I (Statistical Analysis) -on its own- is not novel with respect to other GLM-based feature selection methods for MRI.
Effects of reducing the dimensionality by utilization of the aforementioned well-known techniques were briefly examined and the performances of these methods became compared with the performance of our procedure. Implementation details and results were introduced in Sect. 3.1.
Individual steps of the procedure were introduced in the following subsections.
2.3.1 Step I: statistical analysis
For statistical analysis of the gray matter tissue probabilities at each voxel, a multivariate General Linear Model (GLM) (cf. Eq. (4)) is considered. A GLM could mathematically be expressed as:
where \(\mathbf {y}\) is an N-dimensional response vector (where N is the total number of MR images), \(\mathbf {X}\) is an \(N \times (p/3)\) matrix containing gray matter tissue probability valuesFootnote 7 of each image as feature vectors, and \(\mathbf {b}\) is a vector of (p / 3) unknown parameters to be estimated. The errors comprised in the vector \(\mathbf {u}\) are independent and identically distributed (i.i.d.) random variables with mean value 0. A GLM explains the response variable (in our case, the class label indicating patient and healthy subject) in terms of a linear combination of the explanatory variables (in our case, normalized and modulated gray matter tissue probabilities).
The matrix \(\mathbf {X}\) in Eq. (4) is also referred as design matrix, which was constructed using two-sample t-test for comparing the means of the two populations (namely, sets of MR images belonging to patient and healthy subjects) at each voxel index and discriminating the statistically significant voxel positions from others, in our case. Using voxel-level height threshold and cluster-level extent threshold parameters as measures of significance, different masks could be acquired with different threshold levels. In other words, different subsets of columns of the matrix \(\mathbf {X}\) could be specified as the set of significant feature vectors, depending on the two threshold values.Footnote 8
2.3.2 Step II: tissue probability criteria
Vemuri et al. (2008) introduced a novel methodology to assign values according to discriminative power to the voxels of a structural MRI segmented into the three tissue classes. Their metric is called STAND-score (Structural Abnormality Index Score).
In order to calculate the STAND-score, first the volumetric data were down-sampled to an isotropic voxel size of 8 mm (\(22 \times 27 \times 22\) voxels). After this, the voxels carrying less than 10% tissue density, and the images with CSF in half or more of the voxels were eliminated. Finally, a linear SVM was applied to the remaining data, and weights corresponding to each tissue density were estimated. These weights were used for further elimination of irrelevant voxels (the definition of “irrelevant” differs among distinct tissue classes). The main motivation of the procedure arises from the fact that existence and absence of GM and WM in certain positions of the brain have anatomical relevance to diagnose the Alzheimer’s disease.
This step of our procedure was suggested being inspired by the second step of the STAND-score approach (which puts forward a direct interpretation of the domain knowledge), except for two slight modifications: (i) The lower bound constraint on the ratio of tissue content to eliminate a voxel was not directly assumed as 10%. Instead, various—higher—threshold values were tested. (ii) An additional constraint on the first moment (sample mean) of the ratio of tissue content was introduced. The idea here was to avoid losing a substantial amount of information at a single pass with a very small lower boundary, and to compensate the effect of increasing the threshold by an additional operation, using the sample means this time.
First, the voxels, at which the sum of the probability of being GM and of the probability of being WM is smaller than a certain first threshold, \(\tau _1\), in all of samples were marked to be eliminated. Secondly, the voxels at which the sum of the sample means of probability of being GM and of being WM is smaller than a certain second threshold, \(\tau _2\), were marked to be eliminated. Both of the operations were applied on the raw data. Thus, the order of the two operations could be reversed. Finally, the union set of variables which were marked to be eliminated were removed from the data. In closer terms:
or,
The expressions given in Eqns. (5) and (6) state the elimination criteria for the jth feature mathematically, where i and j are indices of samples and features, \(P_{{ GM}}\) and \(P_{{ WM}}\) are gray matter and white matter probabilities at specified indices, \(\overline{P_{{ GM}}}\) and \(\overline{P_{{ WM}}}\) are sample means of tissue probabilities calculated across observations, respectively.
In this paper, the value of the two thresholds were determined manually by observation among successive experiments. MARS parameters were kept constant (see Sect. 3.2 for the definition of the MARS parameters), while different combinations of \(\tau _1\) and \(\tau _2\) values were used for feature selection. The resulting classifier was applied on the blind test data and the thresholds giving the highest AUC (Area Under the ROC Curve) were chosen. For example, in the AD/CN classification case, appropriate values for the two thresholds appeared to match the highest AUC value at: \(\tau _1=0.5\) and \(\tau _2=0.7\). In the future, a further study involving adaptive selection of the threshold values will be conducted.
2.3.3 Step III: within-class norm thresholding
The final step could be basically explained as a comparison of the Euclidean norm of a variable among samples with the mean value of the norms belonging to the tissue class of that variable. Here, the underlying rule is based on the idea that in case of a significant variable, intensity differences between samples having different labels should create a higher vector norm (when compared with the case of an insignificant variable). First of all, the Euclidean norm of samples at each tissue probability index is computed by:
Afterwards, within-class averages of the computed norms are calculated for the three tissue classes. Namely, \({\hat{\mu }}_{{ GM}}\), \({\hat{\mu }}_{{ WM}}\), and \({\hat{\mu }}_{{ CSF}}\) are computed by:
where c stands for the tissue class \((c \in \{{ GM}, { WM}, { CSF}\})\), and \(j_c\) is the number of variables belonging to the specified tissue class. The rule for a variable to be eliminated is defined as its norm \(\left\| P(j) \right\| _2\) being smaller than a fraction of the within-class norm mean (\({\hat{\mu }}_{c}\)) of the corresponding class. That fraction could be called as \(\varepsilon \), where the value of \(\varepsilon \) must be between 0 and 1. In order to determine the \(\varepsilon \) value, the same method as explained in Sect. 2.3.2 was used. In the AD/CN classification case, setting \(\varepsilon =0.9\) provided the best classification performance among several trials.
It should be noted that, the tissue probability values could also have been used directly instead of Euclidean norms for comparison with the within-class means. Although this decision would have affected the value of \(\varepsilon \), the overall results would not have changed since the tissue probabilities are guaranteed to be positive real numbers and the Euclidean norm is monotonic for positive numbers. In this paper, we preferred using norms for providing a more general notation to represent the vectorial distances.
Table 2 shows the amount of reduction in the dimensionality of the problem space after removal of the zero-voxels and after applying the feature selection procedure. The positive effects of reducing the dimensionality on the overall classification performance were observed; however, a comprehensive analysis was not included in this paper. A brief demonstration can be found in Sect. 3.1.
In MCI and MCIc detection cases, the General Linear Models constructed with MCI data were visually examined and it was seen that the masks include meaningless (or less meaningful) regions (such as air and CSF). This originated from the smallness of the intensity difference between MCIs and normal controls, when compared to that of ADs and normal controls. This concept can be thought of as a low signal-to-noise ratio problem. In order to overcome this problem, we chose to use AD/CN GLM as the initial mask in the other two cases, too. This movement provided a good boost in the classifier performances.
In Fig. 3, the resulting mask consisting of the voxels which were marked as significant at the end of the feature selection procedure is highlighted. It is noteworthy that the resulting region appeared to be asymmetric between brain lobes.
2.4 Multivariate adaptive regression splines
Multivariate Adaptive Regression Splines (MARS) (Friedman 1991) is a nonparametric regression technique which aims to build adaptive models. The method is nonparametric since it involves a procedure with no specific assumptions on the relations between the variables. It is adaptive because its basis-function selection process depends completely on the data themselves. Main ability of MARS is to construct flexible models by allowing piecewise linear regression functions through the utilization of hinge functions defined by:
and
The two functions given by Eqns. (9) and (10) are called a reflected pair. The idea is to form reflected pairs for each input variable \(X_j\) with knots at each observed value \(x_{ij}\) of that variable. Therefore, the collection of basis functions is:
MARS builds models of the form
where M is the number of basis functions in the model and each \(h_m(X)\) is a function (or a product of two or more such functions) in C, multiplied by a factor of \(\beta _m\).
MARS builds a model in two phases: the forward pass and the backward pass. In fact, MARS starts with a model which consists of only the intercept term (which is estimated by the mean of the response values). MARS then repeatedly adds basis function in pairs to the model. At each step it finds the pair of basis functions that gives the maximum reduction in an objective function, such as sum-of-squares residual error. The forward pass usually builds an overfit model. To build a model with a preferable generalization ability, the backward pass prunes the model function. It removes terms one by one, deleting the least effective term at each step until it finds the best submodel. Basis functions are removed in the order of least contribution, using the Generalized Cross-Validation (GCV) criterion. A measure of importance of a variable is determined by observing the decrease in the recomputed GCV values when a variable is removed from the model (Yao 2009).
The GCV criterion is defined by:
In Eq. (13), N is the number of samples; \({\hat{f}}_\alpha \) is the optimally estimated submodel; \(M_\alpha = u_\alpha + dk\), where \(u_\alpha \) is the number of linearly independent basis functions in the submodel, \(\alpha \); k is the number of knots in the forward process; and d is the cost coefficient for optimization—also operating as a smoothing parameter for the procedure.
2.4.1 Classification with MARS
The MARS algorithm can be extended to handle classification problems (Hastie et al. 2009). In two-class cases, the output can be coded as 0 and 1 (for example, 0 for healthy controls and 1 for Alzheimer’s disease patients) and the problem can be treated as a regression problem. The only major deficiency of a MARS classification model is that it is built using a least-squares loss function directly interpreting the class label as a continuous random variable with two realizations (0 and 1) everywhere. Therefore, it is not guaranteed for the predictions to be constrained to the interval [0, 1]. Thus, they cannot be thought of as probabilities. They should instead be treated as generalized probability scores. In order to make a class decision from the continuous output response, a threshold value is required. However, the value of this threshold is not definite as the maximum and minimum values of the predictions are not known. Automatic optimal threshold selection for image segmentation method of Otsu (1975) was used to determine a valid class-separating threshold.
It was also observed that statistically normalizing the data with standard score provided a better classification performance. Therefore, in each experiment, mean and standard deviation values of the training group were computed at each dimension and used for normalization of both the training and the test group input vectors. ARESLab,Footnote 9 which is an open-source MATLAB toolbox for building piecewise-linear and piecewise-cubic regression models using MARS technique, was selected and used for the model building and testing processes.
2.5 Performance evaluation
Resulting class labels of each classification experiment were compared with the known class labels of the test data in order to assess the number of true positives (TP; i.e., individuals with disease which are correctly classified), true negatives (TN; i.e., healthy individuals which are correctly classified), false positives (FP; i.e., healthy individuals which are classified as diseased) and false negatives (FN; i.e., individuals with disease which are classified as healthy). Sensitivity defined as \({ SEN}={ TP}/({ TP}+{ FN})\), specificity defined as \({ SPE}={ TN}/({ TN}+{ FP})\), positive predictive value defined as \({ PPV}={ TP}/({ TP}+{ FP})\) and negative predictive value defined as \({ NPV}={ TN}/({ TN}+{ FN})\) were computed as the performance evaluation metrics. Since the groups do not contain equal numbers of subjects, the overall accuracy (ratio of number of correctly classified sample to the number of all samples) does not provide meaningful information to compare classifier performances. Therefore, it was not included as a measure of performance.
3 Results and discussion
3.1 Dimensionality reduction
Our 3-step procedure for feature selection was compared with six widely-used methods which are mentioned in Sect. 2.3, namely, PCA, MDS, Laplacian Eigenmaps, Kernel PCA, Diffusion Maps, and GDA, in terms of SEN, SPE, PPV, and NPV outcomes (see Table 3). All of the methods were applied on the AD/CN dataset. MARS models were constructed employing all of the training samples for each technique. Performance outcomes were acquired through testing the resulting models on the blind test data.
Before the application of each method, the first step (Statistical Analysis via GLM) of our procedure was applied on the raw training and testing data. In this way, the number of dimensions were reduced to a value (26,448) which is appropriate for the constraints induced by the methods. After that, the intrinsic dimension of the training data was computed through Maximum Likelihood Estimation (MSE), which appeared to be 11 for the AD/CN group. The methods listed in Table 3 (from PCA to GDA) were appliedFootnote 10 both on the training and on the test data to reduce dimensionality of the two values to the value computed as the intrinsic dimension. Keeping the input parameters constant (\(\mathbf{max }\_\mathbf{fnc } = 11\), \(\mathbf{max }\_\mathbf{interact } = 1\), the reader may refer to Sect. 3.2 for definitions of these input parameters), MARS models were built. Finally, the models were tested using the blind test data. (In the case labeled as “None”, model building was performed just after the “Statistical Analysis” step, without further reduction in the dimension. In the case labeled as “Voxel-MARS”, our 3-step procedure was run and the number of dimensions was reduced to 3320 before the model building process.)
According to the results shown in Table 3, only three of the six methods (PCA, MDS, and Laplacian Eigenmaps) were able to provide feature vectors with discriminative strength necessary for classification. It is seen that they provided greater sensitivity compared to the “None” case, in which the number of dimensions were not reduced. However, our procedure for feature selection outperformed all of the other methods listed on the table in terms of sensitivity, specificity, positive predictive value, and negative predictive value.
A deeper research on the effects of dimensionality reduction methods to the classification performance is planned to be conducted in the future.
3.2 Parameter optimization with grid search and ROC analysis
There are two main MARS parameters, based on which we can determine values to control the model building process. The first parameter is the “maximum allowed number of basis functions in the forward model” (max_fnc), and the second one is the “maximum allowed degree of interactions between variables” (max_interact). Increasing max_fnc introduces a rise in the amount of flexibility—therefore, complexity—of the resulting model, where increasing max_interact provides ability to model nonlinearities and statistical dependencies between variables.
AD/CN, MCIc/CN, and MCIc/MCInc classification experiments were performed with two iterations of grid search to find the optimal values for these two variables. In the first iteration, max_fnc was varied taking the values {11, 21,..., 101} one by one, and max_interact was varied taking {1, 2, 3} for each value that max_fnc takes. In the second iteration, max_fnc was varied with smaller steps (\(\hbox {step size} = 2\)) around the parameter value giving the maximum Area Under the ROC Curve (AUC) in the first iteration. In this way, a coarse-to-fine selection of the model parameters was aimed at.
\(\tilde{{{\varvec{N}}}}\) -times replicated \(\tilde{{{\varvec{k}}}}\) -fold cross-validation (Wendy and Martinez 2002) technique was used to validate the model parameters.Footnote 11 In this approach, the original training set is randomly divided into \({\tilde{k}}\) subsets (folds). While each subset is utilized for testing the model, the remaining \({\tilde{k}}-1\) are used for building the models. This process is repeated \({\tilde{N}}-1\) more times with randomly updated partitions and the \({\tilde{k}}\) multiplied by \({\tilde{N}}\) results—in our case, AUC (area under the ROC curve), sensitivity, and specificity values–are averaged. The parameters providing the maximum AUC value were determined as the final model parameters. MARS models built using these final parameters and all of the training samples were applied on the blind test data to obtain final results.
However, the difference in the sample sizes of the training sets for the three different experiments (see Table 1) posed a problem in determination of the \({\tilde{N}}\) and \({\tilde{k}}\) values for cross-validation (the AD/CN group contains 150 samples, the MCI/CN group contains 120 samples, and the MCIc/MCInc group contains 104 samples for training). Also, it has been observed that when the sample size used in the model building process is reduced under 100, the classification performance drastically decreases. Therefore, the value of \({\tilde{k}}\) was assigned to be 3 in the AD/CN case, 6 in the MCI/CN case, and 18 in the MCIc/MCIcn case, by taking the difference in sample sizes into consideration. The reason for this approach was to keep the number of samples in each partition used for training balanced (\( N \times ({\tilde{k}}-1)/{\tilde{k}} \approx 100 \), where N is the total number of samples) during the cross-validation procedure, independent from the test case. The \({\tilde{N}}\) values are selected to compensate the differences in overall repetition counts (introduced by using different \({\tilde{k}}\) values), accordingly (\({\tilde{N}} = 18 \) for AD/CN group, \( {\tilde{N}} = 9 \) for MCI/CN group, and \( {\tilde{N}} = 3 \) for MCIc/MCInc group).
Classification experiments were performed by building MARS models with the parameters acquired through the parameter optimization procedure involving grid search and cross-validation as explained in this subsection. Obtained models became applied on the blind test data and the results achieved at the end of our decision process are introduced with the confusion matrix given in Table 5 (see Sect. 3.3). The individual results for AD/CN, MCI/CN, and MCIc/MCInc cases are presented in Figs. 4, 5, and 6, respectively. Table 4 shows the maximum average AUC value acquired through the search procedure, corresponding parameter values, and the averages of the performance outcomes for each case.
Optimization of the MARS model parameters with grid search and cross validation for the AD/CN group. Images in the first row show the coarse search and the ones in the second row show the fine tuning of the optimization parameters. Columns represent different values (1, 2, and 3) for max_interact (see Sect. 3.2 for the definitions of max_fnc and max_interact). The dashed black line points the maximum AUC acquired in each process. The overall maximum AUC is indicated by the yellow rectangle (\(\textit{AUC} = 0.8656\) with \(\mathbf{max }\_\mathbf{fnc }=11\) and \(\mathbf{max }\_\mathbf{interact }=1\))
Optimization of the MARS model parameters with grid search and cross validation for the MCI/CN group. Images in the first row show the coarse search and the ones in the second row show the fine tuning of the optimization parameters. Columns represent different values (1, 2, and 3) for max_interact (see Sect. 3.2 for the definitions of max_fnc and max_interact). The dashed black line points the maximum AUC acquired in each process. The overall maximum AUC is indicated by the yellow rectangle (\(\textit{AUC} = 0.7025\) with \(\mathbf{max }\_\mathbf{fnc } = 11\) and \(\mathbf{max }\_\mathbf{interact } = 1\))
Optimization of the MARS model parameters with grid search and cross validation for the MCIc/MCInc group. Images in the first row show the coarse search and the ones in the second row show the fine tuning of the optimization parameters. Columns represent different values (1, 2, and 3) for max_interact (see Sect. 3.2 for the definitions of max_fnc and max_interact). The dashed black line points the maximum AUC acquired in each process. The overall maximum AUC is indicated by the yellow rectangle (\(\textit{AUC} = 0.5477\) with \(\mathbf{max }\_\mathbf{fnc } = 71\) and \(\mathbf{max }\_\mathbf{interact } = 1\))
3.3 Classification results
Classification experiments were performed by building MARS models with the parameters acquired through our parameter optimization procedure involving grid search and cross-validation as explained in the preceding subsection. The obtained models were applied on the blind test data, and the results achieved at the end of our decision process are introduced with the confusion matrix given in Table 5.
SEN and SPE values were computed assuming the diseased label as positive and the healthy label as negative. Thus, TPs were defined as diseased samples which our algorithm labeled as diseased, TNs were defined as healthy ones which our algorithm labeled as healthy, FPs were defined as healthy ones which our algorithm labeled as diseased, and FNs were defined as diseased ones which our algorithm labeled as healthy. In AD/CN classification case, ADs were assumed diseased and CNs were assumed healthy. Similarly, in MCI/CN case, the diseased set was composed of subjects with MCI. In the final case, diseased ones were assumed to be the converter MCIs (MCIc), and the MCInc subjects were assumed healthy (Table 5).
3.3.1 AD/CN classification
Classification results for AD/CN groups are presented in Table 7. Our method is entitled as Voxel-MARS (ID: 0) referring the naming convention used in Cuingnet et al. (2011). All 28 methods are sorted in descending order by their sensitivity outcomes and the uppermost five are chosen. The columns show the unique hierarchical IDs given in the original paper, names of the methods, sensitivity, specificity, positive predictive value, and negative predictive value outcomes. In the table, it is seen that our method (Voxel-MARS) produced the highest-ranking sensitivity (SEN) and negative predictive value (NPV) among all 28 methods, with an acceptable specificity (86.42%). On the other hand, Voxel-MARS ranks 22th at specificity (SPE) and 21th at positive predictive value (PPV) among 28 other methods. Looking at the rankings, the proposed method may seem to underperform in terms of specificity. However, deviation of sensitivity in the positive direction (\(+\)12.12) is greater than that of specificity in the negative direction (−2.97) when compared with the average outcomes of all of the other methods (see Table 7).
3.3.2 MCI/CN classification
Performance metrics for MCI/CN classification case are given in Table 8. All 28 methods are sorted in descending order by their sensitivity outcomes and the uppermost five is chosen. Similarly to the first case, our method outperformed all 28 voxel intensity-based methods of sensitivity (SEN) and negative predictive value (NPV). Voxel-MARS ranks 17th at specificity (SPE) and 12th at positive predictive value (PPV) among all of the 28 other methods; these two ranks are both higher than the averages of others (see Table 6). In AD/CN and MCI/CN cases, optimal values for max_fnc and max_interact were 11 and 1, respectively, and the resulting number of basis functions was 7 for both.
3.3.3 MCIc/MCInc classification
In Cuingnet et al. (2011), only 15 out of 28 methods had been observed to produce meaningful results (where both of the SEN and SPE outcomes are different from 100% or 0) in MCIc/MCInc case. Similarly to the previous two cases, methods are sorted by their sensitivity outcomes and the uppermost five of them are listed in Table 9. Our technique is ranked 2nd in terms of sensitivity. Voxel-MARS ranks 16th at specificity (SPE), 12th at positive predictive value (PPV), and 7th at negative predictive value (NPV) among 28 other methods. Very similar to the AD/CN classification case, the amount of gain in SEN is greater than the amount of loss in SPE (see Table 7).
An overall quantitative comparison of the performance of our method with other methods is introduced in Table 6. Performance statistics of other methods are presented in the format: “average (%) ± standard deviation (%) [range (%)]”. In both of the AD/CN and AD/MCI classification cases, all 28 methods had produced reasonable results, whereas in MCIc/MCInc classification case, only 15 of them had. Therefore, for the 3rd case, 13 methods producing “zero sensitivity” were not included in computations. The column “Difference” compares the results gathered using our method and the averages of other methods as percentage.
3.4 Visualization of the model function
The pseudo-code shown in Fig. 7 is partially expressing the model function constructed for AD/CN separation. The model is composed of 7 terms—each representing a MARS basis function and, here, only the 1st term is expressed explicitly. The lowermost line of the code shows the model function in terms of those basis functions.
Visualization of problem spaces with such a high dimensionality is always a difficult issue. In order to offer a visual sense of the hypersurface which separates the problem space into two subspaces, projections were used. Figure 8 shows two exemplary models projected onto two knot dimensions each. Figure 8a, b show training data points and test data points (output response), and the MARS model of Exp. 1, projected onto knot dimensions (voxels) with indices 2553 and 2195 (which correspond to BF1 and BF4 and were chosen randomly among all BFs included in the model), respectively. Similarly, Fig. 8c, d show projections of an example model produced for Exp. 2 onto knot dimensions with indices 2369 and 2617, respectively. The value for the parameter max_interact was selected to be 2 for visualizing nonlinear BFs. Aforementioned voxel indices were chosen since their corresponding hinge functions were appearing in a nonlinear BF in multiplication form. In both cases, a clear separation of structural brain MRI images belonging to the healthy and diseased subjects was observed.
In both of the cases shown in Fig. 8, the voxel indices -to project the model onto- were chosen according to the contribution of the basis functions (coinciding with these voxel indices) to the overall discriminative power of the model function. Residual Sum of Squares (RSS) was used as a measure to choose the binary combination of basis functions among all of them. The two voxel indices, the corresponding basis functions of which provided the minimum RSS on the blind test data, were chosen for model visualization.
3.5 Discussion
A fully-automated machine learning method was proposed to help for diagnosis of Alzheimer and MCI at early stages of diseases by analyzing 3D T1-W structural MRI. The proposed technique includes novel contributions in feature selection and classification procedures; namely, a 3-step approach for determination of the subset of significant features, and utilization of MARS method as a classifier for the first time in the field of OR in Neuroscience.
Training and test data were gathered from the ADNI database with permission, and data groups were composed referring to the comparative study of Cuingnet et al. (2011). A set of experiments was composed in order to compare the overall performances under different conditions, and our best (in terms of maximum AUC) results were presented side by side with the ones gathered in the referred study. It was seen that our method (Voxel-MARS) outperformed all 28 methods compared within the study in terms of sensitivity and NPV, with acceptable specificity values in early detection of AD and MCI. Additionally, our algorithm managed to produce meaningful results in the MCIc/MCInc classification case and worked better than 27 of 28 other methods in terms of sensitivity. Although the specificity outcomes produced by the proposed method were below the averages provided by the other methods in AD/CN and MCIc/MCInc classification experiments, the amount of gain in sensitivity turned out to be greater than the amount of loss in specificity in both cases.
MARS is an adaptive regression method, which has the ability to model nonlinearities and dependencies between variables automatically. GCV criterion of MARS constitutes an equilibrium between flexibility and generalization ability of the MARS model function. It is known that the aforementioned characteristics of the method may better be observed with larger datasets. However, the method was completely proved to be working for our problem with relatively simpler models provided (for example, models consisting of 7 basis functions without interactions among them).
4 Conclusions
In this study, a complete, fully-automated computer-aided diagnosis system for early detection of AD and MCI based on structural brain MRI was introduced. A thorough image processing scheme was applied to 3D T1-W MRI data downloaded from the ADNI archive, in order to obtain voxel-based feature vectors. The choice of treating all voxel intensities forming the three main brain tissues as potential features (i.e., voxel-as-feature approach) effectively yields a “small N—large P” problem, which calls for a method capable to mitigate the challenges arising from the coexistence of data sparsity and high dimensionality.
A novel 3-step feature selection approach was elaborated for the determination of the features with significantly higher discriminative power. The feature selection procedure starts with generation of an initial mask using a General Linear Model, proceeds with injection of domain rules into the model for elimination of some features according to the tissue probability distributions, and ends with an application of the within-class norm thresholding—a method offering elimination of features whose Euclidean sample norms are not large enough with respect to the mean of norms of feature vectors within their tissue class.
Multivariate Adaptive Regression Splines (MARS) is a non-parametric, adaptive extension of decision trees (specifically, of Classification and Regresion Trees—CART) which is able to produce nonlinear models for regression and classification. In Hastie et al. (2009), a comparative analysis involving 5 of the “off-the-shelf” procedures for data mining is presented. According to this analysis, MARS provides better support for handling of data of mixed-type and missing values, computational scalability, dealing with irrelevant inputs, and interpretability than Support Vector Machines (SVM), the most widely-used technique for AD classification, does. In Strickland (2014), it is stated that MARS is able to make predictions quickly compared to SVM, where “every variable has to be multiplied by the corresponding element of every support vector”. Furthermore, a special advantage of MARS lies in its ability to estimate contributions of some basis functions, which are allowed to determine the output response through additive and interactive effects of the input variables (Weber et al. 2012). This final ability of MARS -which SVM lacks- is of particular importance in our study, whereby we employ all of the voxel intensities (forming the brain tissue) as potential predictor variables at the beginning, and do not have the conclusive anatomical knowledge of significant brain regions to be affected in the very early phases of AD.
When compared with another commonly used method in the field, Artificial Neural Networks (ANN), MARS is reported to be more computationally efficient (Zhang and Goh 2016). An additional drawback of ANN is that it provides models in form of a “black box”. This is explained by Francis (2003) as follows: “The functions fit by neural networks are difficult for the analyst to understand and difficult to explain to management. One of the very useful features of MARS is that it produces a regression like function that can be used to understand and explain the model.” MARS, like ANN, is also effective in modeling the interactions among variables. Additionally, in Moisen and Frescino (2002), using MARS is reported to be tremendously advantageous over using Linear Models (LM), Generalized Additive Models (GAM), and CART for prediction.
Keeping these qualities in mind, we have chosen MARS as the method to be utilized for classification. MARS was employed to construct nonlinear models to function as class-separating hypersurfaces. Employment of MARS as a classifier with such high degrees of space dimensionality is one of the major contributions of this study, as it is the first time use of the method in the field of structural brain MRI analysis. It has resulted in a remarkable success, compared to the alternative approaches which correspondingly addressed voxel intensities of structural brain MRI as the source of information. It was proved that MARS can excellently perform as a nonlinear classifier having the ability to learn from high-dimensional data and to construct complex—and yet stable—models without pre-assigning any constant parametric form. Utilization of the method enabled for the resulting system to detect the effects of microscopic changes occurring at the early phases of AD and converter-MCI in the intensity distributions on T1-W brain MRI.
Recent technological improvements of all kinds of measurement devices (which is an MRI scanner in our case) create a gigantic and continuously growing supply of information (i.e., big data) to analyze. This situation forces us, scientists, to position the machine learning issues within the areas of data mining and model optimization, and to elaborate our work in the area of Operational Research within near future. Automatized detection of AD (and MCI) at early phases with image analysis involves employment of techniques from mathematical sciences such as statistical analysis, mathematical modeling, and decision theory. Therefore, improvement of computer-aided AD and MCI diagnosis tools can be considered as an open OR problem in the field of Neuroscience. This study showed that our decision-making approach is one of the promising advances in the domain.
Inclusion of data provided by other modalities (both the imaging modalities such as PET and other biomarkers such as CSF proteins and MSE scores) into the model, a quantitative analysis of the effects of feature selection and dimensionality reduction on overall classification performance, a deeper comparison between performances of SVM-based algorithms, Artificial Neural Networks and MARS method in the field, and expansion of the system with a wider learning set to be used in clinics are potential future studies that are motivating our group. Moreover, a deeper research will be conducted by the help of recent methods which are mathematically more integrated variants of the algorithmic part of MARS, with less heuristic elements, and supported by modern optimization theory, such as CMARS (Weber et al. 2012) -being reported to provide more complex models enabling higher classification accuracy than MARS does- and its robust counterpart RCMARS (Özmen et al. 2011)—shown to be even more successful than CMARS in handling various forms of uncertainty (e.g., by existence of noise, by outliers, and by missing values) in the data.
Notes
A brain function syndrome which causes a slight decline in cognitive abilities and an increased risk of converting into AD.
All 452 ICBM subject T1-weighted scans were aligned with the atlas space, corrected for scan inhomogeneities, and classified into gray matter, white matter, and cerebrospinal fluid. The 452 tissue maps were separated into their separate components and each component was averaged in atlas space across the subjects to create the probability fields for each tissue type. These fields represent the likelihood of finding gray matter, white matter, or cerebrospinal fluid at a specified position for a subject that has been linearly aligned to the atlas space (http://www.loni.usc.edu/atlases/Atlas_Methods.php?atlas_id=7).
Standard brain space defined by Montreal Neurological Institute (MNI).
The total number of the raw data features containing the probabilities for 3 tissue classes at each voxel is p. Therefore, considering only the gray matter tissue probabilities, the dimensionality is equal to the total number of voxels, which is (p / 3).
An explanatory example for the use of height threshold and extent threshold in fMRI analysis is provided in Friston et al. (1996).
Jekabsons G., ARESLab: Adaptive Regression Splines toolbox for MATLAB/Octave, 2011, available at http://www.cs.rtu.lv/jekabsons/.
The Matlab Toolbox for Dimensionality Reduction van der Maaten et al. (2009) was used for computations.
Here, the letters N and k are used with a tilde (\(\sim \)) sign over them in order not to be confused with the N used for expressing the sample size and the k appearing in Eq. (13) as the number of knots, respectively.
References
Adaszewski, S., Dukart, J., Kherif, F., Frackowiak, R., & Draganski, B. (2013). How early can we predict Alzheimer’s disease using computational anatomy? Neurobiology of Aging, 34(12), 2815–2826.
Alvarez, I., Gorriz, J., Ramirez, J., Salas-Gonzalez, D., Lopez, M., Puntonet, C., et al. (2009). Alzheimer’s diagnosis using eigenbrains and support vector machines. Electronics Letters, 45(7), 342–343.
Álvarez-Miranda, E., Farhan, H., Luipersbeck, M., & Sinnl, M. (2016). A bi-objective network design approach for discovering functional modules linking Golgi apparatus fragmentation and neuronal death. Annals of Operations Research, 1–26. http://springerlink.bibliotecabuap.elogim.com/article/10.1007/s10479-016-2188-2.
Ashburner, J. (2007). A fast diffeomorphic image registration algorithm. Neuroimage, 38(1), 95–113.
Ashburner, J. (2009). Computational anatomy with the SPM software. Magnetic Resonance Imaging, 27(8), 1163–1174. (Proceedings of the international school on magnetic resonance and brain function).
Ashburner, J., & Friston, K. J. (2005). Unified segmentation. Neuroimage, 26(3), 839–851.
Boutet, C., Chupin, M., Lehricy, S., Marrakchi-Kacem, L., Epelbaum, S., Poupon, C., et al. (2014). Detection of volume loss in hippocampal layers in Alzheimer’s disease using 7T MRI: A feasibility study. Neuroimage: Clinical, 5, 341–348.
Chaves, R., Ramìrez, J., Górriz, J., López, M., Salas-Gonzalez, D., lvarez, I., et al. (2009). SVM-based computer-aided diagnosis of the Alzheimer’s disease using t-test NMSE feature selection with feature correlation weighting. Neuroscience Letters, 461(3), 293–297.
Chincarini, A., Bosco, P., Calvini, P., Gemme, G., Esposito, M., Olivieri, C., et al. (2011). Local MRI analysis approach in the diagnosis of early and prodromal Alzheimer’s disease. Neuroimage, 58(2), 469–480.
Chupin, M., Grardin, E., Cuingnet, R., Boutet, C., Lemieux, L., & Lehricy, S., et al. (2009a). Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild cognitive impairment applied on data from ADNI. Hippocampus, 19(6), 579–587.
Chupin, M., Hammers, A., Liu, R., Colliot, O., Burdett, J., & Bardinet, E., et al. (2009b). Automatic segmentation of the hippocampus and the amygdala driven by hybrid constraints: Method and validation. Neuroimage, 46(3), 749–761.
Colliot, O., Chtelat, G., Chupin, M., Desgranges, B., Magnin, B., Benali, H., et al. (2008). Discrimination between Alzheimer disease, mild cognitive impairment, and normal aging by using automated segmentation of the hippocampus. Radiology, 248(1), 194–201.
Cuingnet, R., Gerardin, E., Tessieras, J., Auzias, G., Lehricy, S., Habert, M. O., et al. (2011). Automatic classification of patients with Alzheimer’s disease from structural MRI: A comparison of ten methods using the ADNI database. Neuroimage, 56(2), 766–781.
Davatzikos, C., Fan, Y., Wu, X., Shen, D., & Resnick, S. M. (2008). Detection of prodromal Alzheimer’s disease via pattern classification of magnetic resonance imaging. Neurobiology of Aging, 29(4), 514–523.
Frackowiak, R., Friston, K., Frith, C., Dolan, R., Price, C., Zeki, S., et al. (2003). Human Brain Function (2nd ed.). Cambridge: Academic Press.
Francis, L. (2003). Martian chronicles: Is MARS better than neural networks? In: Casualty Actuarial Society Forum (pp. 75–102).
Friedman, J. H. (1991). Multivariate adaptive regression splines. The Annals of Statistics, 19(1), 1–67.
Friston, K., Holmes, A., Poline, J. B., Price, C., & Frith, C. (1996). Detecting activations in PET and fMRI: Levels of inference and power. Neuroimage, 4(3), 223–235.
Gerardin, E., Chtelat, G., Chupin, M., Cuingnet, R., Desgranges, B., Kim, H. S., et al. (2009). Multidimensional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. Neuroimage, 47(4), 1476–1486.
Graa, M., Termenon, M., Savio, A., Gonzalez-Pinto, A., Echeveste, J., Prez, J., et al. (2011). Computer aided diagnosis system for Alzheimer disease using brain diffusion tensor imaging features selected by Pearson’s correlation. Neuroscience Letters, 502(3), 225–229.
Hastie, T., Tibshirani, R., & Friedman, J. (2009). The elements of statistical learning: Data mining, inference and prediction (2nd ed.). Berlin: Springer.
Jack, C. R., Bernstein, M. A., Fox, N. C., Thompson, P., Alexander, G., Harvey, D., et al. (2008). The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging, 27(4), 685–691.
Jack, C. R., Knopman, D. S., Jagust, W. J., Shaw, L. M., Aisen, P. S., Weiner, M. W., et al. (2010). Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. The Lancet Neurology, 9(1), 119–128.
Jain, A., Duin, R. P. W., & Mao, J. (2000). Statistical pattern recognition: A review. IEEE Transactions on Pattern Analysis and Machine Intelligence, 22(1), 4–37.
Klöppel, S., Stonnington, C. M., Chu, C., Draganski, B., Scahill, R. I., Rohrer, J. D., et al. (2008). Automatic classification of MR scans in Alzheimer’s disease. Brain, 131(3), 681–689.
Li, M., Qin, Y., Gao, F., Zhu, W., & He, X. (2014). Discriminative analysis of multivariate features from structural MRI and diffusion tensor images. Magnetic Resonance Imaging, 32(8), 1043–1051.
Liu, S., Liu, S., Cai, W., Pujol ,S., Kikinis, R., & Feng, D. (2014). Early diagnosis of Alzheimer’s disease with deep learning. In: 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI) (pp. 1015–1018).
López, M., Ramìrez, J., Górriz, J., Álvarez, I., Salas-Gonzalez, D., Segovia, F., et al. (2011). Principal component analysis-based techniques and supervised classification schemes for the early detection of Alzheimer’s disease. Neurocomputing, 74(8), 1260–1271. (Selected papers from the 3rd international work-conference on the interplay between natural and artificial computation (IWINAC 2009)).
Magnin, B., Mesrob, L., Kinkingnhun, S., Plgrini-Issac, M., Colliot, O., Sarazin, M., et al. (2009). Support vector machine-based classification of Alzheimers disease from whole-brain anatomical MRI. Neuroradiology, 51(2), 73–83.
Misra, C., Fan, Y., & Davatzikos, C. (2009). Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: Results from ADNI. Neuroimage, 44(4), 1415–1422.
Moisen, G. G., & Frescino, T. S. (2002). Comparing five modelling techniques for predicting forest characteristics. Ecological Modelling, 157(23), 209–225.
Morra, J., Tu, Z., Apostolova, L., Green, A., Toga, A., & Thompson, P. (2010). Comparison of adaboost and support vector machines for detecting Alzheimer’s disease through automated hippocampal segmentation. IEEE Transactions on Medical Imaging, 29(1), 30–43.
Mwangi, B., Tian, T., & Soares, J. (2014). A review of feature reduction techniques in neuroimaging. Neuroinformatics, 12(2), 229–244.
Otsu, N. (1975). A threshold selection method from gray-level histograms. Automatica, 11(285–296), 23–27.
Özmen, A., Weber, G. W., Batmaz, ì, & Kropat, E. (2011). RCMARS: Robustification of cmars with different scenarios under polyhedral uncertainty set. Communications in Nonlinear Science and Numerical Simulation, 16(12), 4780–4787. (sI:Complex Systems and Chaos with Fractionality, Discontinuity, and Nonlinearity).
Padilla, P., Gorriz, J., Ramirez, J., Chaves, R., Segovia, F., Alvarez, I., et al. (2010). Alzheimer’s disease detection in functional images using 2D Gabor wavelet analysis. Electronics Letters, 46(8), 556–558.
Padilla, P., Lopez, M., Gorriz, J., Ramirez, J., Salas-Gonzalez, D., & Alvarez, I. (2012). NMF-SVM based cad tool applied to functional brain images for the diagnosis of Alzheimer’s disease. IEEE Transactions on Medical Imaging, 31(2), 207–216.
Park, H., Yang, J., Seo, J., & Lee, J. (2012). Dimensionality reduced cortical features and their use in the classification of Alzheimer’s disease and mild cognitive impairment. Neuroscience Letters, 529(2), 123–127.
Ramìrez, J., Górriz, J., Salas-Gonzalez, D., Romero, A., López, M., lvarez, I., et al. (2013). Computer-aided diagnosis of Alzheimers type dementia combining support vector machines and discriminant set of features. Information Sciences, 237, 59–72.
Ramìrez, J., Górriz, J., Segovia, F., Chaves, R., Salas-Gonzalez, D., López, M., et al. (2010). Computer aided diagnosis system for the Alzheimer’s disease based on partial least squares and random forest SPECT image classification. Neuroscience Letters, 472(2), 99–103.
Salas-Gonzalez, D., Górriz, J. M., Ramìrez, J., Illn, I. A., López, M., Segovia, F., Chaves, R., Padilla, P., Puntonet, C. G., & Alzheimers Disease Neuroimage Initiative, T. (2010). Feature selection using factor analysis for Alzheimers diagnosis using F18-FDG PET images. Medical Physics, 37(11), 6084–6095.
Salas-Gonzalez, D., Górriz, J. M., Ramìrez, J., López, M., Illan, I. A., Segovia, F., et al. (2009). Analysis of SPECT brain images for the diagnosis of Alzheimer’s disease using moments and support vector machines. Neuroscience Letters, 461(1), 60–64.
Savio, A., & Graa, M. (2013). Deformation based feature selection for computer aided diagnosis of Alzheimers disease. Expert Systems with Applications, 40(5), 1619–1628.
Segovia, F., Górriz, J., Ramìrez, J., Salas-Gonzalez, D., lvarez, I., López, M., et al. (2012). A comparative study of feature extraction methods for the diagnosis of Alzheimer’s disease using the ADNI database. Neurocomputing, 75(1), 64–71.
Shih, D. T., Kim, S. B., Chen, V. C. P., Rosenberger, J. M., & Pilla, V. L. (2014). Efficient computer experiment-based optimization through variable selection. Annals of Operations Research, 216(1), 287–305.
Strickland, J. (2014). Predictive modeling and analytics. LULU Press. https://books.google.com.tr/books?id=1jfXoQEACAAJ.
Suk, H. I., Lee, S. W., Shen, D., Initiative, A. D. N., et al. (2014). Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage, 101, 569–582.
Tiraboschi, P., Hansen, L. A., Thal, L. J., & Corey-Bloom, J. (2004). The importance of neuritic plaques and tangles to the development and evolution of AD. Neurology, 62(11), 1984–1989.
van der Maaten, L. J., Postma, E. O., & van den Herik, H. J. (2009). Dimensionality reduction: A comparative review. Journal of Machine Learning Research, 10(1–41), 66–71.
Vemuri, P., Gunter, J. L., Senjem, M. L., Whitwell, J. L., Kantarci, K., Knopman, D. S., et al. (2008). Alzheimer’s disease diagnosis in individual subjects using structural MR images: Validation studies. Neuroimage, 39(3), 1186–1197.
Weber, G. W., Batmaz, I., Köksal, G., Taylan, P., & Yerlikaya-Özkurt, F. (2012). CMARS: A new contribution to nonparametric regression with multivariate adaptive regression splines supported by continuous optimization. Inverse Problems in Science and Engineering, 20(3), 371–400.
Wendy, L., & Martinez, A. R. M. (2002). Computational statistics handbook with MATLAB. London: Chapman and Hall, CRC.
Westman, E., Simmons, A., Zhang, Y., Muehlboeck, J. S., Tunnard, C., Liu, Y., et al. (2011). Multivariate analysis of MRI data for Alzheimer’s disease, mild cognitive impairment and healthy controls. Neuroimage, 54(2), 1178–1187.
Yao, P. (2009). Hybrid fuzzy SVM model using CART and MARS for credit scoring. In Intelligent Human-Machine Systems and Cybernetics, 2009. IHMSC ’09. International Conference on (Vol. 2, pp. 392–395).
Ye, J., Chen, K., Wu, T., Li, J., Zhao, Z., Patel, R., Bae, M., Janardan, R., Liu, H., Alexander, G., & Reiman, E. (2008). Heterogeneous data fusion for Alzheimer’s disease study. In Proceedings of the 14th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (pp. 1025–1033). ACM: New York, NY, USA, KDD’08.
Ye, J., Wu, T., Li, J., & Chen, K. (2011). Machine learning approaches for the neuroimaging study of Alzheimer’s disease. Computer, 44(4), 99–101.
Zhang, T., & Davatzikos, C. (2011). ODVBA: Optimally-discriminative voxel-based analysis. IEEE Transactions on Medical Imaging, 30(8), 1441–1454.
Zhang, W., & Goh, A. T. (2016). Multivariate adaptive regression splines and neural network models for prediction of pile drivability. Geoscience Frontiers, 7(1), 45–52.
Zhang, D., Wang, Y., Zhou, L., Yuan, H., & Shen, D. (2011). Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage, 55(3), 856–867.
Acknowledgements
This study is based on Alper Çevik’s Ph.D. thesis. B. Murat Eyüboğlu and Gerhard-Wilhelm Weber are the thesis co-supervisors. Kader Karlı Oğuz is a member of the thesis committee. This project has been supported by the Graduate School of Natural and Applied Sciences, METU Scientific Research Fund ‘BAP 07-02-2012-101’. The authors would like to thank Dr. Güçlü Ongun, Dr. Ayşe Özmen, and Dr. Semih Kuter for their valuable comments and suggestions to improve the study. We are also greatly indebted to Ajdan Küçükçiftçi for her proofreading which improved composition of this paper. Data collection and sharing for this study was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimers Association; Alzheimers Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.
Rights and permissions
About this article
Cite this article
Çevik, A., Weber, GW., Eyüboğlu, B.M. et al. Voxel-MARS: a method for early detection of Alzheimer’s disease by classification of structural brain MRI. Ann Oper Res 258, 31–57 (2017). https://doi.org/10.1007/s10479-017-2405-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10479-017-2405-7