Abstract
The advances in improved fluorescent probes and better cameras in collaboration with the advent of computers in imaging and image analysis, assist the task of diagnosis in microscopy imaging. Based on such technologies, we introduce a computer-assisted image characterization tool based on fractal analysis and fuzzy clustering for the quantification of degree of the Idiopathic Pulmonary Fibrosis in microscopy images. The implementation of this algorithmic strategy proved very promising concerning the issue of the automated assessment of microscopy images of lung fibrotic regions against conventional classification methods that require training such as neural networks. Fractal dimension is an important image feature that can be associated with pathological fibrotic structures as is shown by our experimental results.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
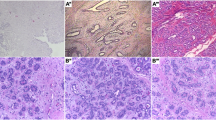
Idiopathic pulmonary fibrosis (IPF), also referred to as cryptogenic fibrosing alveolitis, is a chronic, progressive and usually lethal lung disorder of unknown aetiology. Median survival of newly diagnosed patients with IPF is about 3 years, similar to that of clinical non-small cell lung cancer. At present there are no proven therapies for IPF (Selman and Pardo 2002). IPF usually affects patients aged 50–70 years, with a male preponderance, evidenced by a male-to-female ratio of 2:1. The incidence of IPF has been estimated at 10.7 cases per 100,000 men and 7.4 cases per 100,000 women and the incidence appears to be rising (Coultas et al. 1994). The anatomy of IPF is shown in Fig. 1, which depicts typical salient regions of corresponding lung sections microscopy images.
Advances in improved fluorescent probes (Chalfie et al. 1994) and better cameras (Inoué and Spring 1997; Shotton 1998) have expanded the capabilities of the light microscope and its usefulness in biologic and medical research. More specifically a light microscope, also called an optical microscope, is an instrument to observe small objects using visible light and lenses. The advancements in optical technology brought lenses with better characteristics and capturing devices (i.e. CMOS chips adapted to microscopes) allowing the acquisition of high-resolution digital images. In addition, improved understanding of chemical and physical properties of fluorescence markers has led to the optimization of cell imaging applications and limited undesired experimental side effects. Thus, automatic classification and assessment of microscopy images is possible in an attempt to assist the task of diagnosis or measuring therapeutic procedures.
In the present study, we employ one of the most widely used fuzzy clustering algorithms; namely the fuzzy c-means (Bezdek 1981). The goal is to identify the presence and the degree of lung pathology caused by idiopathic pulmonary fibrosis (IPF), which is a chronic, progressive and usually lethal lung disorder of unknown etiology (Antoniou et al. 2007), whose variability concerning the severity of the lesions it incurs in the lung is great, when assessed by microscopy histological images (Izbicki et al. 2002). Fuzzy c-means clustering has been applied in many real life problems, e.g. in environmental cases (Iliadis et al. 2010) and here we extend the approach presented in (Tasoulis et al. 2012) in an attempt to achieve better results. Our motivation is to create a tool for the quantification of the disease and monitoring of therapeutical procedures and schemes.
The Fuzzy c-means clustering method was applied to digital images of sections, captured using a Nikon ECLIPSE E800 microscope and a Nikon digital camera DXM1200 at a magnification of 4\(\times \). Each image was partitioned into windows of specified size and features were extracted from each window. The main contribution of this study is the introduction of fractal dimension (FD) as a feature for analysing the microscopy images. The box-counting method was used for the estimation of the fractal dimension of each image window. To extend that approach, we use the clustering result of the Fuzzy c-means algorithm to introduce a new scoring system that defines the degree of lung pathology at each corresponding image. More specifically the scoring is based on the membership values of each cluster given by the fuzzy clustering algorithm.
The rest of the paper is structured as follows. In Sect. 2, we briefly review the Fuzzy c-means clustering algorithm and we outline the fractal dimension and the box-counting method for its computation. Next, in Sect. 3, we introduce the proposed methodology and finally in Sect. 4 we present the experimental analysis and results. The paper ends with concluding results and pointers for future work.
2 Background information
The continuously growing medical data necessitates the development of new methods that can extract information in several applications. The most popular approach used for analyzing data is Data Mining. In particular, clustering can be used to help physicians to cope with the information overload by assisting on reducing resources used for pattern recognition.
2.1 Applications
The field of microscopy image analysis has occupied several research teams and significant research work may be found in the literature of this field. In Maglogiannis et al. (2008) a tool that classifies biological microscopy images of lung tissue sections with idiopathic pulmonary fibrosis was presented. Similar tools have also been proposed for the assessment of liver fibrosis (Bedossa et al. 2003; Caballero et al. 2001; Masseroli et al. 2000; Yagura et al. 2000), the study of micro vascular circulating leukocytes (Hussain et al. 2004), the assessment of testicular interstitial fibrosis (Shiraishi et al. 2002, 2003), or that of lung fibrosis (Izbicki et al. 2002). The use of pattern recognition or classification methods like Support Vector Machines or Neural Networks has enabled the design of decision-making algorithms, appropriate to microscopy data. Within this context, a method for evaluation of electron microscopy images of serial sections based on the Gabor wavelets and the construction of a mapping between the model and the target image has been proposed in König et al. (2001).
2.2 Clustering of medical data
Data clustering is the process of partitioning a set of data vectors into disjoint groups (clusters), so that objects of the same clusters are more similar to each other than objects in different clusters. Clustering is related to classification in the sense that it creates a labeling of the data points, however it derives these labels only for the data. On the other hand, classification uses information from data with known class labels to assign a class label to unlabeled data. For this reason, in some cases data clustering is referred as “supervised or automatic classification”. In addition more rarely data clustering is also referred as “numerical taxonomy” and “typological analysis”.
A complete clustering process is constituted by the following steps:
-
1.
Representation of data (may include the feature extraction or the feature selection process).
-
2.
Definition of the similarity measure between the elements.
-
3.
Clustering.
-
4.
Evaluation of the results.
The term “representation of data” refer to the number and the type of features that describe each one of the data elements. Feature selection is the process that tries to select the most important features. On the other hand, feature extraction refers to the transformation of features to others that are more important with respect to the further clustering procedure. Different measures of similarity may be used in the clustering process, where the similarity measure controls how the clusters are formed. Some examples of measures that can be used in clustering include distance, connectivity, and intensity.
The final clustering results can be exclusive, where each data element is assigned to a single cluster, or fuzzy where every element belongs to every cluster with a membership weight. Moreover, a clustering result can be describe as a hierarchy, which is a set of nested clusters that are organized as a tree, or just as a division of the data set into non overlapping clusters.
In traditional (hard) clustering, data are divided into distinct clusters, where each data vector belongs to exactly one cluster. On the other hand, in fuzzy (soft) clustering, data points can belong to more than one cluster, and associated with each element is a set of membership levels. The membership level that defines the association of a data point with a cluster is between \(0\) (where the data point absolutely does not belong to the cluster) and \(1\) (where the data point absolutely belongs to the cluster), with the constraint that the sum of the weights for each data point must equal \(1\). These indicate the strength of the association between that data point and a particular cluster. Fuzzy clustering is a process of assigning these membership levels and then using them to assign data points to one or more clusters. In the same way, probabilistic clustering techniques can be used to compute the probability with which each data point belongs to each cluster with the similar constraint that the probabilities must sum to \(1\). Since both the probabilities and the membership weights must sum to \(1\), these types of clustering differs from the non-exclusive clustering, where a data point simultaneously belong to multiple clusters. Instead, these approaches are mostly used to avoid the arbitrariness of assigning an object to only one cluster, when it may be close to several. In many cases, fuzzy clustering is converted to hard clustering by assigning each data point to the cluster for which its membership weight is highest.
2.3 Fuzzy c-means algorithm
One of the most widely used fuzzy clustering algorithms is the fuzzy c-means (FCM) algorithm. This technique was originally introduced by Bezdek (1981) as an improvement on earlier clustering methods and attempts to partition a finite collection of data vectors into a collection of fuzzy clusters with respect to some given criterion. A theoretical discussion of FCM can be found in Cox (2005).
Given a finite set of data, the algorithm returns a list of cluster centers and a partition matrix indicating the degree to which each element belongs to a given cluster. Like the \(k\)-means algorithm, the FCM aims to minimize an objective function, that is:
where \(m\) is any real number greater than 1, \(u_{ij}\) is the degree of membership of \(x_i\) in the cluster \(j, x_i\) is the \(i\)th of \(d\)-dimensional measured data, \(c_j\) is the \(d\)-dimension center of the cluster, and \(\Vert \cdot \Vert \) is any norm expressing the similarity between any measured data and the center. Fuzzy partitioning is carried out through an iterative optimization of the objective function shown above, with the update of membership \(u_{ij}\) and the cluster centers \(c_j\) by:
where
This iteration stops when
where \(k\) is the iteration number and \(\epsilon \) is a constant between \(0\) and \(1\) that controls the termination of algorithm. This procedure converges to a local minimum or a saddle point of \(J_m\). The steps of the algorithm are shown in Algorithm 1.

2.4 Fractals and fractal dimension
It is known that a fractal designates a rough or fragmented geometric shape that can be subdivided into parts, each of which is a reduced-size copy of the whole (Foroutan-pour et al. 1999). A series of complex objects found in nature such as coastlines or clouds can be analyzed based on a mathematical model provided by fractal geometry (Mandelbrot 1983; Peitgen et al. 1993; Pentland 1984). In general, nature conforms to fractals much more than it does to classical shapes and hence fractals can serve as models for many natural phenomena. In most cases, this kinds of objects are hard to describe based on the Euclidean geometry.
Fractal dimension is used to quantify self-similarity, which is the basic property of the fractals since fractals are generally self-similar and independent of scale. Contrary to classical geometry, fractals are not regular and may have a non-integer dimension. Fractal concepts have provided a new approach for quantifying the geometry of complex or noisy shapes and objects. Fractal geometry has been proven capable of quantifying irregular patterns, such as tortuous lines, crumpled surfaces and intricate shapes, and estimating the ruggedness of systems (Mandelbrot 1983). There are several scientific fields on which FD has been applied such as image analysis (Asvestas et al. 1998; Li et al. 2009) and texture analysis and segmentation (Chaudhuri and Sarkar 1995; Ida and Sambonsugi 1998; Liu and Chang 1997).
There exists many methods for estimating FD but they can be classified into three major categories (Balghonaim and Keller 1998). These are the box counting methods, the variance methods and the spectral methods. The most widely used method is the box counting dimension. That is because it is very simple and automatically computable (Peitgen et al. 1993). Among a number of techniques (Mandelbrot 1983), it was found to be the most appropriate method of fractal dimension estimation.
In this study, we incorporate the fractal dimension in an attempt to guide the FCM algorithm to construct meaningful clusters and for the fractal dimension estimation we use the box-counting approach. The box-counting method is easy, automatically computable, and applicable for patterns with or without self-similarity (Foroutan-pour et al. 1999; Peitgen et al. 1993). In the box counting procedure, each input image is covered by a sequence of grids of descending sizes and for each of the grids, two values are recorded. These are the number of square boxes intersected by the image, \(N(s)\), and the side length of the squares, \(s\).
Then the fractal dimension is calculated by the regression slope \(D\) (\(1 \le D \le 2\)) of the straight line formed by plotting \(\log (N(s))\) against \(\log (1/s)\) (Mandelbrot 1983). An image having a fractal dimension of \(1\), or \(2\), is considered as completely differentiable, or very rough and irregular, respectively. The linear regression equation used to estimate the fractal dimension is
where \(K\) is a constant and \(N(s)\) is proportional to \((1/s)D\).
3 Proposed methodology
As long as the microscopy images are captured the procedure continues as follows. Initially, the input microscopy images are being binirized using the Otsu’s method (Otsu 1979). This method is one of many binarization algorithms, i.e. it automatically performs reduction of a graylevel image to a binary one. The algorithm assumes that the image contains two classes of pixels (e.g. foreground and background) and calculates the optimum threshold separating those two classes so that their combined spread (intra-class variance) is minimal. At the next step we segment the binary image to \(n\times n\) windows and then we calculate FD of each window. Fractal dimension is a useful feature proposed to characterize roughness and self-similarity in a picture, which has been used for texture segmentation, shape classification, and graphic analysis in many fields. It can be defined as a ratio providing a statistical index of complexity comparing how detail in a pattern changes with the scale at which it is measured. Finally, based on the Fractal Dimension, the mean value of the intensity and the standard deviation of the intensity of the grayscale window image, we perform clustering using FCM clustering algorithm. The fuzzification parameter \(m\) is set to the default value \(1.25\). The complete scheme of the proposed method is shown in Fig. 2. Next, in Fig. 3 an example of an image is illustrated in various steps of the procedure. From left to right, we exhibit the input microscopy image, the greyscale image, the binary image, and the image that shows the clustering result.
4 Experiment setup and results
For this study, Age- and sex- matched, 6–8 week-old mice were used for the induction of pulmonary fibrosis by a single intravenous injection with a dose of 100 mg/kg of body weight (100 mg/kg body weight; 1/3 LD50; Nippon Kayaku Co. Ltd., Tokyo). Bleomycin administration initially induces lung inflammation that is followed by a progressive destruction of the normal lung architecture. To monitor disease initiation and progression, mice were sacrificed at 7, 15 and 23 days after bleomycin injection. Mice injected with saline alone and sacrificed 23 days post injection, served as the control group. For pathology assessment, at each time point, bronchoalveolar lavage (BAL) has been performed (3\(\times \) 1 ml Saline) for the estimation of total and differential cell populations. Finally after perfusion of lungs via the heart ventricle with 10 ml Phosphate Buffer Saline, lungs were then removed, weighed dissected and collected, for histology. Sagittal sections from the right lung were used for Hematoxylin and Eosin staining and histopathologic analysis.
The results that are presented in this section are based on the images that correspond to 7, 15 and 23 days after bleomycin administration in mice (see Fig. 4). The lung images were initially separated to windows of size \(100\times 100\) pixels and the box-counting method was applied at each window to calculate the corresponding fractal dimension. Next, the images were separated into windows of smaller sizes (\(5, 10\), and \(20\), respectively). Each of these windows corresponds to a data vector, with attributes the fractal dimension of the initial \(100\times 100\) window that contains it and the mean value of the intensity of the grayscale window image. Finally, to cluster each of the datasets constituted by such data points, we employed the Fuzzy c-Means clustering algorithm, for \(3\) and \(4\) clusters, respectively.
Figures 5 and 6 illustrate the clustering results with respect to the window size for all images, where windows of the same cluster are colored equally for the 4 and 3 clusters case, respectively. In the 4 clusters case, the red cluster represents the severe pathology, the orange cluster represents the mild pathology and green cluster the normal lung. Finally, the blue cluster denotes the image background. In the 3 cluster case, the mild pathology class is omitted and the colors red, green and blue represent the severe pathology, the normal lung and the background, respectively. In the case of black and white printing, the background corresponds to the black color and the other classes varies from dark gray to light gray in the presented order.
The results are quite encouraging with respect to their medical content. The clustering algorithm has satisfactory performance in distinguishing pathologic from normal. Thus, the majority of the blocks that are clustered into pathological clusters (i.e. severe and mild) belong into the regions indicated by the expert pathologists as fibrotic.
Clustering result for the 4 clusters case with respect to the window sizes (rows) for the images retrieved after 7, 15 and 23 days (columns), respectively. The colors red, orange, green and blue corresponds to the severe pathology, mild pathology, normal lung and background (non informative), respectively. (Color figure online)
Tables 1 and 2 report the percentage of the image that belongs to each cluster found by the algorithm. The results are presented with respect to the different window sizes given as input for each of the images used (images retrieved after 7, 15 and 23 days). The computed percentages coincide with manual annotation and scoring performed by our collaborating experts pathologists. The clustering algorithm seems to achieve high performance in pathological from normal in most cases.
At this point it should be mentioned that the importance of FD is not only to enhance the clustering results of the method since there are no significant improvement when using FD as a feature , but also to use FD as a pathology index. More specifically, using the FD we can locally quantify the extent of fibrosis in the pathological areas and assist the physicians to understand better the pathologic mechanisms behind fibrosis. An indicated example is illustrated in Fig. 7. This figure depicts a sample fibrotic image retrieved after 15 days with the pathological areas denoted by experts with green color (top left) along with the visualization of the corresponding FD values of each window (top right). The black color represents the highest FD value, while the white color the lowest. In addition, we can see the clustering result based on the window size we used to compute FD with respect to FD (bottom left) and the with respect to mean intensity (bottom right). As shown the FD can identify better the local extent of sever fibrosis.
Next, in Table 3 the scores of the assessment of a group of expert pathologists (denoted by \(P1,P2,P3\)) are presented. These scores are set according to the score guidelines provided in Maglogiannis et al. (2008). We have the following states: healthy [0–4], mild pathology (4–11] and severe pathology (11–15]. In our study, we introduce two new scores based on the membership values of the produced clusters given by the Fuzzy c-means algorithm and we examine if they follow the trend of the score given by the experts. An example of the membership values of the severe class along with the original image and the corresponding clustering result is given in Fig. 8. Membership values are illustrated with the help of a greyscale contour plot. A value close to \(1\) corresponds to the white color while values close to \(0\) correspond to black [see Fig. 8 (right)].
The image retrieved after 15 days with the pathological areas denoted by experts with green color (top left), along with the visualization of the corresponding FD values of each window (top right), and the clustering result with respect to FD (bottom left) along with the result with respect to mean intensity (bottom right) when cluster number is set to 3. (Color figure online)
The first score (score 1), corresponds to the weighted mean of the membership values of the mild and severe classes for the 4 cluster case and the severe and normal classes for the 3 cluster case respectively. We have
where \(n\) is the number of the windows that constitute the image, \(S_m(n)\) is the membership value of the severe class for the window \(n\) and \(M_m(n)\) is the membership value of the mild class for the window \(n\) respectively. Next, score 2 is given by the following equation:
where
To further examine the behavior of the two scores when noise is reduced, we apply the median filter and a \(50\,\%\) cutoff to the membership values of each class, respectively. The resulting scores are summarized in Tables 4 and 5 for the 3 and 4 cluster cases, respectively. As shown, in most of the cases, the computed scores coincide with manual scoring performed by our collaborating experts pathologists. As expected, only in the cases where the window size is set to \(20\) the scores have a different behavior due to the lack of information.
An example plot of the membership values of the severe class for the image retrieved after 7 days and when the window size is set to \(10\) is illustrated in Fig. 9, along with the plots of the membership values after the median filter and the \(50\,\%\) cutoff respectively. The reduced noise in Fig. 9(center) and (right) that correspond to the median filter and the \(50\,\%\) cutoff respectively is obvious. In addition, the clearly visible circular shapes in these images motivate us to further develop our technique in the future for the recognition of such shapes. Finally a three dimensional plot of the membership values of the severe class with median filter is illustrated in Fig. 10.
5 Conclusions
In the present paper, a computer-assisted image characterization tool based on a Fuzzy clustering method was introduced for the quantification of degree of IPF in medical images. The implementation of this algorithmic strategy is very promising, concerning the issue of the automated assessment of microscopy images of lung fibrotic regions. The results obtained so far show that the proposed strategy addresses the vital biological issues concerning their imaging part as this is contained in the specific type of microscopy images.
The majority of the blocks that are clustered according to their fractal dimension into pathological clusters (i.e. severe and mild) belong into the regions indicated by the expert pathologists as fibrotic. In addition, the computed percentages coincide with manual annotation by our collaborating experts pathologists. Thus, the FD was proved an important image feature that can be associated with pathological fibrotic structures.
In a future study, we intent to use the degree of membership to postprocess the clustering results in an attempt to eliminate small isolated regions and increase further the accuracy of the proposed approach.
References
Antoniou KM, Pataka A, Bouros D, Siafakas NM (2007) Pathogenetic pathways and novel pharmacotherapeutic targets in idiopathic pulmonary fibrosis. Pulm Pharmacol Ther 20(5):453–461
Asvestas P, Matsopoulos G, Nikita K (1998) A power differentiation method of fractal dimension estimation for 2-d signals. J Vis Commun Image Represent 9(4):392–400
Balghonaim AS, Keller JM (1998) A maximum likelihood estimate for two-variable fractal surface. Trans Image Process 7(12):1746–1753
Bedossa P, Dargére D, Paradis V (2003) Sampling variability of liver fibrosis in chronic hepatitis C. Hepatology 38(6):1449–1457
Bezdek JC (1981) Pattern recognition with fuzzy objective function algorithms. Kluwer, Norwell, MA
Caballero T, Pérez-Milena A, Masseroli M, O’Valle F, Salmerón F, Del Moral R, Sánchez-Salgado G (2001) Liver fibrosis assessment with semiquantitative indexes and image analysis quantification in sustained-responder and non-responder interferon-treated patients with chronic hepatitis C. J Hepatol 34(5):740–747
Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC (1994) Green fluorescent protein as a marker for gene expression. Science 263(5148):802–805
Chaudhuri BB, Sarkar N (1995) Texture segmentation using fractal dimension. IEEE Trans Pattern Anal Mach Intell 17(1):72–77
Coultas DB, Zumwalt RE, Black WC, Sobonya RE (1994) The epidemiology of interstitial lung diseases. Am J Respir Crit Care Med 150(4):967–972
Cox E (2005) Fuzzy modeling and genetic algorithms for data mining and exploration. Elsevier, Amsterdam
Foroutan-pour K, Dutilleul P, Smith D (1999) Advances in the implementation of the box-counting method of fractal dimension estimation. Appl Math Comput 105(23):195–210
Hussain M, Merchant S, Mombasawala L, Puniyani R (2004) A decrease in effective diameter of rat mesenteric venules due to leukocyte margination after a bolus injection of pentoxifylline digital image analysis of an intravital microscopic observation. Microvasc Res 67(3):237–244
Ida T, Sambonsugi Y (1998) Image segmentation and contour detection using fractal coding. IEEE Trans Circuits Sys Video Technol 8(8):968–975
Iliadis L, Vangeloudh M, Spartalis S (2010) An intelligent system employing an enhanced fuzzy c-means clustering model: application in the case of forest fires. Comput Electron Agric 70(2):276–284
Inoué S, Spring K (1997) Video microscopy. Plenum, New York
Izbicki G, Segel M, Christensen T, Conner M, Breuer R (2002) Time course of bleomycin-induced lung fibrosis. Int J Exp Path 83(3):111–119
König P, Kayser C, Bonin V, Würtz R (2001) Efficient evaluation of serial sections by iterative gabor matching. J Neurosci Methods 111(2):141–150
Li J, Du Q, Sun C (2009) An improved box-counting method for image fractal dimension estimation. Pattern Recognit 42(11):2460–2469
Liu SC, Chang S (1997) Dimension estimation of discrete-time fractional Brownian motion with applications to image texture classification. Trans Image Process 6(8):1176–1184
Maglogiannis I, Sarimveis H, Kiranoudis CT, Chatziioannou AA, Oikonomou N, Aidinis V (2008) Radial basis function neural networks classification for the recognition of idiopathic pulmonary fibrosis in microscopic images. IEEE Trans Inf TechnolBiomed 12(1):42–54
Mandelbrot B (1983) The fractal geometry of nature. W.H. Freeman and Co., New York
Masseroli M, Caballero T, O’Valle F, Del-Moral R, Pérez-Milena A, Del-Moral R (2000) Automatic quantification of liver fibrosis: design and validation of a new image analysis method: comparison with semi-quantitative indexes of fibrosis. J Hepatol 32(3):453–464
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybern 9(1):62–66
Peitgen HO, Jürgens H, Saupe D (1993) Chaos and fractals: new frontiers of science. Springer, Berlin
Pentland AP (1984) Fractal-based description of natural scenes. IEEE Trans Pattern Anal Mach Intell 6(6): 661–674
Selman M, Pardo A (2002) Idiopathic pulmonary fibrosis: an epithelial/fibroblastic cross-talk disorder. Respir Res 3(1):3
Shiraishi K, Takihara H, Naito K (2002) Influence of interstitial fibrosis on spermatogenesis after vasectomy and vasovasostomy. Contraception 65(3):245–249
Shiraishi K, Takihara H, Naito K (2003) Quantitative analysis of testicular interstitial fibrosis after vasectomy in humans. Aktuelle Urologie 34(4):262–264
Shotton DM (1998) Image resolution and digital image processing in electronic light microscopy. In: Celis JE (ed) Cell biology, a laboratory handbook, vol 3, 2nd edn. Academic Press, San Diego, pp 85–98
Tasoulis S, Maglogiannis I, Plagianakos V (2012) Unsupervised detection of fibrosis in microscopy images using fractals and fuzzy c-means clustering. In: Iliadis L, Maglogiannis I, Papadopoulos H (eds) Artificial intelligence applications and innovations, IFIP advances in information and communication technology, vol 381. Springer, Berlin, pp 385–394
Yagura M, Murai S, Kojima H, Tokita H, Kamitsukasa H, Harada H (2000) Changes of liver fibrosis in chronic hepatitis c patients with no response to interferon-alpha therapy: including quantitative assessment by a morphometric method. J Gastroenterol 35(2):105–111
Acknowledgments
The authors would like to thank the European Union (European Social Fund ESF) and Greek national funds through the Operational Program “Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF)—Research Funding Program: “Heracleitus II. Investing in knowledge society through the European Social Fund.” for financially supporting this work. Part of this work has been also funded by Program: Thalis—Interdisciplinary Research in Affective Computing for Biological Activity Recognition in Assistive Environments/Operational Program “Education and Lifelong Learning”.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tasoulis, S.K., Maglogiannis, I. & Plagianakos, V.P. Fractal analysis and fuzzy c-means clustering for quantification of fibrotic microscopy images. Artif Intell Rev 42, 313–329 (2014). https://doi.org/10.1007/s10462-013-9408-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10462-013-9408-9