Abstract
We have recently developed a bioreactor that can apply both shear and compressive forces to engineered tissues in dynamic culture. In our system, alginate hydrogel beads with encapsulated human mesenchymal stem cells (hMSCs) were cultured under different dynamic conditions while subjected to periodic, compressive force. A customized pressure sensor was developed to track the pressure fluctuations when shear forces and compressive forces were applied. Compared to static culture, dynamic culture can maintain a higher cell population throughout the study. With the application of only shear stress, qRT-PCR and immunohistochemistry revealed that hMSCs experienced less chondrogenic differentiation than the static group. The second study showed that chondrogenic differentiation was enhanced by additional mechanical compression. After 14 days, alcian blue staining showed more extracellular matrix formed in the compression group. The upregulation of the positive chondrogenic markers such as Sox 9, aggrecan, and type II collagen were demonstrated by qPCR. Our bioreactor provides a novel approach to apply mechanical forces to engineered cartilage. Results suggest that a combination of dynamic culture with proper mechanical stimulation may promote efficient progenitor cell expansion in vitro, thereby allowing the culture of clinically relevant articular chondrocytes for the treatment of articular cartilage defects.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis is a prevalent degenerative disease affecting all ages that often results in severe damage to articular cartilage.29 Approximately 70% of the senior population has osteoarthritis at varying levels of severity.13 The most common occurrence of osteoarthritis is in the knee due to excessive and constant mechanical loading. To date, there is no known cure for osteoarthritis and the resulting cartilage defects. Commonly used conventional therapies include biomechanical or behavior interventions, pain killers, anti-inflammatory drugs and physical treatments,6 which mainly aim to control the pain and improve the quality of life, not to overcome the disease. Severe cases must rely on surgeries such as meniscus transplant, microfracture and autologous chondrocyte implantation (ACI); however, the success rates vary greatly.31 Unexpected immune response is a major concern of allograft transplantation. Although microfracture has shown positive therapeutic effects for cartilage regeneration, the repaired tissue often becomes fibrocartilaginous with increased amounts of type I collagen and decreased type II collagen as long-term results. For ACI, it requires dual-surgery for chondrocytes expansion. Major obstacles for ACI to help cartilage regeneration include the lack of an abundant cell source, poor interaction with native tissues, and the dedifferentiation of implanted chondrocytes.26 Therefore, a more abundant and phenotypically stable cell source is necessary for efficient clinical treatment.
To solve these problems with advanced technologies, tissue engineering approaches to restore the biological functions of damaged cartilage have been explored. However, cartilage has an intrinsically limited ability for self-repair due to a low cell population and a lack of blood supply; which are significant challenges for tissue engineering strategies.9 Biologically active, cell-based therapies with appropriate signaling molecules and supporting matrix are promising techniques for treating osteoarthritis.7,18,19 Recently, mesenchymal stem cells (MSCs) have been considered an attractive cell source for cartilage engineering. In addition to their availability and well documented isolation protocols, implanting undifferentiated MSCs may result in a better integration with in situ differentiated chondrocytes when compared to the implantation of differentiated chondrocytes.8,17,22 However, the use of human MSCs (hMSCs) is limited as studies have shown the reduced formation of extracellular matrix which is a major contributor to the mechanical function of cartilage. To improve upon current methods, mechanical stimuli can be introduced to engineered tissues to better mimic the natural environment of articular cartilage and to induce differentiation of MSCs in 3D scaffolds.2,15
It is well known that native articular cartilage is exposed to mechanical stimuli such as dynamic compression, fluid shear, and hydrostatic pressure. Considering the mechanical function of articular cartilage which transmits and distributes loads in joints, proper mechanical stimulation plays an important role in altering cell response for in vitro tissue culture.16 Our laboratory previously developed a tubular perfusion system (TPS) that creates a dynamic environment for cell culture.33 In this system, cells are encapsulated in 3D scaffolds and cultured in a tubular growth chamber. Nutrient and oxygen supply can be enhanced by perfusing media through the scaffolds. Concurrently, cells at the interface of the applied flow are exposed to shear forces. Previously published studies have demonstrated an improvement in osteogenic differentiation using the TPS. However, the influence of the TPS bioreactor on chondrogenic differentiation of MSCs still remains unclear.37 To recapitulate the native mechanical environment that facilitates chondrogenesis, we have recently developed a new approach wherein both shear and compressive forces are applied to an engineered 3D cell-scaffold construct. Our design incorporates a metal roller, which applies mechanical compression in combination with the dynamic culture conditions provided by the TPS. The overall goal of this study is to develop an advanced in vitro chondrogenesis protocol with proper chemical and mechanical stimulation. We hypothesize that our compression perfusion bioreactor would enhance chondrogenic differentiation by providing mechanical stimulation that mimics the in vivo environment. There are three main objectives to comprehensively examine the proposed hypothesis. First, to evaluate the effect of dynamic culture on hMSC proliferation. Second, to evaluate the effect of dynamic culture on hMSC chondrogenic differentiation. Third, to evaluate the influence of an additional, periodic compression on hMSC differentiation.
Materials and Methods
hMSC Encapsulation and Culture
Primary hMSCs were purchased (Lonza) and expanded in a monolayer in high glucose Dulbecco’s Modified Eagle Medium (DMEM) (Life Technologies) containing 0.1% penicillin/streptomycin (Life Technologies), 0.1 mM non-essential amino acids (Life Technologies) and 10% fetal bovine serum (Life Technologies). After two passages, hMSCs were trypsinized (Life Technologies) and counted using trypan blue staining and a hemocytometer. Two percent (w/v) alginate was prepared using protocols previously established by our laboratory.36 Briefly, 2.0% (w/v) of alginic acid sodium salt from brown algae (Sigma-Aldrich) was dissolved in a 0.15 M sodium chloride and 0.025 M HEPES (Sigma-Aldrich) buffer, and then autoclaved and filtered for sterilization. The alginate solution was mixed with hMSCs (3 million cells/mL) and injected through a 21-gauge syringe into 0.1 M calcium chloride (Sigma-Aldrich) solution to induce crosslinking for 15 min. The diameter of the formed alginate beads was approximately 2.5 mm. Alginate beads encapsulating hMSCs were loaded into the TPS bioreactor and cultured in chondrogenic media. Chondrogenic media contained: high glucose DMEM, 40 mg/mL proline (Sigma-Aldrich, St. Louis, MO), 0.1% penicillin/streptomycin (Life Technologies), 0.1% sodium pyruvate (Life Technologies), 50 mg/mL ascorbate 2-phosphate (Sigma-Aldrich, St. Louis, MO), 0.1 mM dexamethasone, 1% ITS + premix (BD Biosciences, Bedford, MA), and 10 ng/mL TGF-β3 treatment (R&D systems, Minneapolis, MN).
The TPS bioreactor was set up as described previously.33 Media flow was driven by an L/S Multichannel Pump System (Cole Parmer, Vernon Hills, IL) at a flow rate of 1 mL/min. The media was changed every 3 days. For the study comparing static and dynamic culture, alginate beads were fabricated and divided into two groups, one cultured in a petri dish (Fisher Scientific) and the other in the TPS bioreactor. For the compression study, alginate beads (n = 50 to 55) were loaded into five 12 cm growth chambers for collections at different time points. All beads with encapsulated hMSCs were exposed to dynamic flow until the end of each time point. Beads cultured for 7 days in one chamber were collected as a baseline for future assays. Two of the chambers were introduced to additional compression (group 1) while the other two were under dynamic culture only (group 2). The cyclic pressure was applied at 0.5 Hz frequency.24 At each time point, hMSCs were isolated from alginate beads by dissolution in ethylenediaminetetraacetic acid (EDTA) for 20 min at 37 °C. A cell pellet was formed by centrifugation and used for subsequent analyses.
Bioreactor Set-Up and Pressure Detection
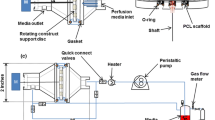
The TPS bioreactor was set-up as described previously.33 Particularly, for the compression study, a customized metal roller was manufactured to apply cyclic compressive force onto tubular chambers that were confined in the supporting box (Fig. 3a). We utilized a 3D printed reducing connector to connect the growth chamber to the supply line. Into this connector, we inserted the pressure sensor with a 21-gauge needle as a guide to insert the probe 1.5 inches from the connector into an alginate bead.
Pressure Sensor Fabrication
Custom Fabry–Perot pressure sensors were constructed by adapting fabrication methods previously established.4 A length of fiber optic cable (OD = 150 µm) was spliced at one end to a capillary tube (ID = 30 µm, OD = 150 µm) ensuring the fiber core was aligned with the hollow center of the capillary. The spliced capillary was cleaved approximately 15 µm from the fiber interface and a 500 nm thick, UV curable polymer layer was added to the freshly cleaved capillary thereby enclosing the capillary cavity. A titanium metal layer was sputtered onto the surface of the cured polymer to complete a flexible, water-resistant diaphragm atop the capillary cavity (Fig. 3a). Pressure changes at the capillary terminus cause the diaphragm to mechanically deflect allowing for changes in cavity length to be recorded to quantify pressure at the sensor tip. All sensors were calibrated with a reference pressure gauge (SSI Technologies Inc, Janesville, WI) in a custom calibration chamber. A fiber optic system (USB-4000 Spectrometer and HL-2000 Light Source, Ocean Optics, Dunedin, FL; Beam Splitter, Gould Fiber Optics) was used to record cavity length data at a frequency of 14.3 Hz in LabView 2012 (National Instruments, Austin, TX). Signal optimization and processing were completed using MATLAB R2012b (MathWorks, Inc., Natick, MA).
DNA Isolation and Quantification
Total hMSC DNA was isolated from alginate beads using a DNeasy Blood and Tissue Kit (Qiagen, Frederick, MD). A Quant-iT PicoGreen dsDNA Assay kit (Life Technologies) was used to quantify DNA following the manufacturer’s protocol. Total DNA was calculated as ng/bead.
RNA Isolation and qRT-PCR
The total RNA was isolated from the hMSCs using an RNeasy Plus Mini Kit (Qiagen, Frederick, MD) and then reverse transcribed to complementary DNA (cDNA) using a High Capacity cDNA Archive Kit (Life Technologies). Quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) was performed by combining the cDNA solution with a Universal Master Mix (Life Technologies), as well as oligonucleotide primers and Taqman probes for Sox 9, type I collagen (COL1A1), type II collagen (COL2A1), aggrecan (AGC), and the endogenous gene control glyceraldehyde 3 phosphate dehydrogenase (GAPDH; Life Technologies). The reaction was performed using a 7900HT real-time PCR System (Applied Biosystems) at thermal conditions of 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. The relative gene expression level of each target gene was normalized to the mean of GAPDH in each group then the fold change was determined relative to the day 7 pre-culture gene expression in the bioreactor without compression. Fold change was calculated using the ΔΔCT relative comparative method as described previously.35 Samples were completed in technical triplicates and mean ± standard deviations were reported.
Immunohistochemistry
Samples were fixed in 4% paraformaldehyde on slides for 10 min. After washing, samples were blocked and then stained with the primary antibodies to detect Type I collagen, aggrecan or Type II collagen (Abcam, Cambridge, MA), respectively. Secondary antibodies were conjugated to Alexa488 or Alexa568 (Molecular Probes). Nuclei were visualized with 200 μg/L DAPI (4′, 6-diamidino-2-phenylindole, dilactate) stain. Sections were mounted in Fluorescent Mounting Medium (DakoCytomation), and photographed using an Axioskop mot plus microscope (Zeiss, Thornwood, NY) with a digital camera (Axiocam) and Openlab software.
Live-Dead Assay
In order to assess cell viability, a live-dead assay was performed following standard protocols. Alginate beads encapsulated with hMSCs were soaked in Hank’s buffered saline solution (HBSS, Life Technologies) for 10 min to remove extra media and other active reagents. The beads were then incubated in 2 μM ethidium homodimer and 4 μM calcein AM (Life Technologies) combined with HBSS for 30 min in the dark. The bead was visualized via fluorescence microscopy (Axiovert 40 CFL with filter set 23; Zeiss, Thornwood, NY) using a digital camera (Diagnostic Instruments 11.2 Color Mosaic, Sterling Heights, MI).
Histochemical Staining
All samples were collected and fixed in 4% paraformaldehyde in 0.1 M sodium cacodylate buffer containing 10 mM CaCl2 (pH 7.4) for 4 h at room temperature and then transferred to 0.1 M sodium cacodylate buffer with 10 mM CaCl2 (pH 7.4) for 24 h at room temperature to re-crosslink alginate. Then samples were dehydrated through a series of ethanol washes followed by two Citrisolv (Fisher Scientific, Pittsburgh, PA) washes. The samples were embedded in Paraplast X-tra Paraffin (Fisher Scientific) and sectioned to 5 mm thick sections and placed on positively charged glass slides (Fisher Scientific). Prior to staining, sections were oven-dried at 37 °C for 2 h, deparaffinized in Citrisolv, and rehydrated in decreasing ethanol washes. Samples were stained using alcian blue (Poly Scientific, Bay Shore, NY) for 30 min, followed by standard washes. Samples were counterstained under nuclear fast red (Poly Scientific) for 5 min. After dehydration and clearance, the slides were mounted and imaged.
Statistical Analysis
Data from all the studies were analyzed using analysis of variance and Tukey’s multiple-comparison test. A significant level of 95% was chosen, and a p value less than 0.05 was considered to indicate a significant difference among samples.
Results
DNA quantification of hMSCs in dynamic culture and static culture was performed to determine the effect of the TPS bioreactor on cell proliferation. By day 2, the total DNA content per bead in the dynamic group increased 2.7 fold in the dynamic group compared to the static group. For all remaining time points, hMSCs proliferation in the dynamic group was statistically greater than that observed in static controls (Fig. 1a). Fluorescent images showing viability of the whole beads were obtained, and the majority of the cells appeared viable in both groups (Fig. 1b). The cell distribution in dynamic group on day 21 appeared more spatially homogenous than those in static groups. The Live/Dead data showing increased cell number under dynamic culture was consistent with the DNA quantification results.
hMSC proliferation and proteoglycan expression in TPS. a The DNA contents in static and dynamic culture based on DNA quantification from PicoGreen assay. The cell population remained higher in the dynamic group than static group throughout the study (n = 3). b Live-dead images of beads in static and TPS culture throughout the study. The merged images showed that the majority of the cells appeared viable in all groups on each time point. All scale bars represent 1000 μm. c Alcian blue staining of hMSCs in static and dynamic groups. Cell nuclei stained pink, and acidic GaG residues stained a light blue color (alginate bead stains dark blue). After day 14, the lighter blue extracellular matrix was observed in both groups with higher expression in the static group. All scale bars represent 200 μm.
Alcian blue staining was used to demonstrate the produced ECM contained glycosaminoglycans (GAGs). In these images, the nuclei and cytoplasm of the cells appeared pink, while the GAGs appeared light blue (the alginate stains a dark blue color). Increased GAGs production in both groups was observed after day 14, apparent by increased levels of light blue stain surrounding the cell nuclei. From the results, we noted an overall increased presence of GAGs in the static group. Additionally, hMSCs proliferated into large cell clusters in the dynamic group towards later time points (Fig. 1c).
We also analyzed the gene expression of chondrogenic phenotype markers by the hMSCs in the dynamic and static groups at each time point. In general, three positive markers, aggrecan, type II collagen, and Sox 9 genes were expressed at higher levels in the static group when compared to the dynamic group (Fig. 2a). On day 7, a burst of aggrecan and Sox 9 expression was observed in the static group and then was maintained at a similar level throughout the study. However, chondrogenic markers expression was not significantly increased in the dynamic group. Type I collagen is a negative marker of chondrogenic phenotype. For both groups, the expression of type I collagen decreased on day 2, but increased to the original level during the later time points in the static group.
mRNA expression and protein expression of chondrogenic phenotype markers. a mRNA expression of positive chondrogenic markers including aggrecan, sox 9 and type II collagen. Type I collagen is a negative marker for chondrogenesis. For all genes, the expression level in static culture condition was higher than dynamic culture. *All samples were reported as average from technical triplicates plus standard deviation, although a few error bars were too small to visualize. All values were first normalized to GAPDH, then to the first time point. b Fluorescent immunostaining for chondrogenic phenotype markers. Type I collagen expression decreased during culture in both groups while aggrecan and type II collagen expression increases. Both aggrecan and type II collagen showed higher level in static group compared to dynamic group. All scale bars represent 50 μm.
Immunostaining for type II collagen, type I collagen, and aggrecan in both groups was performed to evaluate expression at the protein level. The staining of type I collagen in both groups decreased in apparent expression with time, indicating chondrogenic differentiation was maintained throughout the study (Fig. 2b). In agreement with the gene expression results, more intense type II collagen staining (red) was observed in static group on day 21 although both groups showed an increase of protein expression compared to the previous time point. Aggrecan expression (green) in both groups appeared similar for all time points (Fig. 2b).
The preliminary results brought us to the next set of experiments to investigate if the addition of compressive force could be beneficial to chondrogenic differentiation of hMSCs in the TPS bioreactor. The pressure changes while the roller moves across the tubular growth chamber and a peak pressure appears when the roller moves onto the target bead containing the pressure sensor (Figs. 3a, 3b). We recorded the peak pressure in each cycle and calculated the average peak gauge pressure at different conditions (Fig. 3b). The pressure caused by only dynamic flow was not detectable by the sensor. The addition of compressive force by the metal roller applied a peak pressure of 6813 ± 2195 Pa onto the bead and the encapsulated cells during movement.
Dynamic compression TPS bioreactor set-up and results. a Bioreactor assembly, the roller moves at a controlled speed provided by the rocker. The right picture shows the pressure detection experimental set up with a close look of the detailed sensor design. b Gauge pressure measured inside the alginate beads. The left figure shows the detected peak pressure at different conditions. Only the pressure change caused by compression was detectable. The right figure shows real time sensor response. A peak pressure appeared during each cycle of compression. c Live-dead images of beads in static and TPS culture throughout the study. The bead cultured in dynamic flow for 7 days was taken as a control. The merged images showed that the majority of cells appeared viable in all groups on each time point. All scale bars represent 300 μm.
A Live/Dead assay was used to investigate the hMSC viability under the combined mechanical stimuli including shear force and compressive force. The hMSCs cultured in the TPS bioreactor for 7 days in the presence of TGF-β3 were considered as a control group for the entire study. No significant difference of the cell viability was observed when applying additional compression to the encapsulated cells compared to the 7 days pre-culture control and the dynamic group (Fig. 3c).
Gene expression of chondrogenic phenotype markers by the hMSCs in the uncompressed dynamic group and compression groups was analyzed (Fig. 4a). To prevent unexpected cell apoptosis under mechanical loading, all hMSCs were culture in static environment for 7 days to allow for ECM production; thus gene expression values in this study were specifically normalized to day 7. Sox 9 and aggrecan gene expression in the compression group significantly increased 15 fold and 7 fold on day 14 compared to day 7 pre-culture, respectively. Both Sox 9 and aggrecan expression remained low throughout the study for the uncompressed dynamic group. On day 21, the aggrecan gene expression decreased in the compressed group compared to day 14. The expression of Sox 9 maintained similar levels as day 14 until the last time point in the compressed group. We observed a delay of type II collagen gene expression in the compressed group during the study such that on day 14, the expression was higher in the uncompressed dynamic group compared to compressed group. However, on day 21, while the uncompressed group experienced a significant decrease of type II collagen expression, hMSCs exposed to mechanical compression showed an increasing trend when compared to the previous time point. Sox 9, type II collagen, aggrecan and type I collagen proteins were detected by Western blot.
Chondrogenic phenotype marker expression in compressed group and uncompressed group during dynamic culture. a mRNA expression of positive chondrogenic markers including aggrecan, sox 9 and type II collagen. Type I collagen is a negative marker for chondrogenesis. For aggrecan and Sox9, the mRNA expression level in the compression condition was higher than the uncompressed condition. After day 21, Type II collagen gene expression in the compression group was higher than the uncompressed group. All gene expressions were first normalized based on GAPDH, then normalized to the first time point. b Alcian blue staining of hMSCs in compressed and uncompressed groups. Cell nuclei stained pink, and acidic GAG residues stained a light blue color (alginate bead stains dark blue). After day 14, the lighter blue extracellular matrix was observed in both groups with higher expression in the compressed group. The expression level remained similar through day 21. All scale bars represent 100 μm.
Alcian blue staining was conducted to see the difference in ECM production between the compressed and uncompressed groups. We saw significantly increased GAG formation in both groups after day 14 by visualizing increased light blue staining around the pink cell nuclei (Fig. 4b). When comparing the images from the two groups at each time point, a clear difference was observed. With additional compression, more GAGs were produced in the compressed group. In addition, cells were more frequently observed to aggregate into clusters and modify the surrounding alginate in the compressed group. The expression level of GAGs on day 21 was not significantly different than day 14 for both compressed and uncompressed groups.
Discussion
As previously demonstrated, encapsulated cells in the TPS bioreactor experience shear forces in addition to increased nutrient and oxygen flow.33,34 Many studies have demonstrated the positive effect of dynamic culture on hMSCs proliferation using other different types of bioreactors.5,11,38,39 Our novel bioreactor can provide both shear and compressive forces at the same time to better mimic the natural environment of articular cartilage. Another advantage of this system is that by incorporating a customized sensor, the pressure inside the beads can be directly measured as the compression is applied. In this study, we investigated hMSC proliferation during chondrogenesis and expression of hMSC chondrogenic markers in both static culture and dynamic culture in the TPS bioreactor. We also investigated the chondrogenic behavior of hMSCs under periodic compression.
Our first objective was to investigate the hMSCs proliferation during the chondrogenesis process in the dynamic culture provided by the TPS bioreactor. Our DNA quantification results showed that a greater cell population in the dynamic group was maintained throughout the study than in the static group. This result is consistent with previous work demonstrating the importance of dynamic culture and 3D scaffolds in cell proliferation.23,32 Here we have demonstrated the ability to develop a platform that increases the in vitro expansion of hMSCs with satisfactory cell viability. The TPS bioreactor can provide an avenue to address this critical issue of initial cell population to allow for stem cell implantation and the treatment of articular cartilage defects. However, it is important to evaluate if the proliferated hMSCs are phenotypically stable for future chondrogenesis.
The second objective was to evaluate the effect of dynamic culture (shear force only) on hMSC chondrogenic differentiation. Our results revealed that in this study, static culture was more desirable for hMSCs chondrogenesis based on conclusions drawn from gene expression and histological data. For the mRNA expression, higher expression of chondrogenic markers including Sox 9, aggrecan and type II collagen in the static group were observed throughout the study. Similar to our results, Kock et al. reported reduced chondrogenesis in flow perfusion bioreactor with a rate of 1.22 mL/min.12 In fact, previous studies have shown the potentially negative effects of high oxygen level on chondrocyte metabolism.1,10 It has been indicated that the shear stress may play a negative role in regulating the mRNA expression for chondrogenesis with a shear stress range of 0.61–1.64 Pa.27 The shear stress that our TPS bioreactor provided is about 0.1 Pa according to a previously developed computational model,33 which is relatively lower than the in vivo loading. As such, it is not likely to be a predominant factor in guiding the chondrogenic differentiation in this study. In another aspect, our bioreactor provides not only flow induced shear stress, but also significantly increased mass transfer (e.g., nutrients and oxygen). Several groups have demonstrated that a hypoxic environment can induce the increase of collagen II expression and proteoglycan deposition significantly for MSCs undergoing chondrogenic differentiation compared to normoxic conditions.10 Therefore, although the mechanical stimuli could have played a role in enhancing chondrogenesis, the enhanced mass transfer might have potentially counteracted the positive effect, thus leading to the static culture being the preferred case for chondrogenesis in this study.
In articular cartilage tissue engineering, it has been demonstrated that dynamic compression and hydrostatic pressure are crucial to the healthy development of chondrocytes or chondrogenic differentiation and phenotype maintenance.25,28 One study demonstrated that hydrostatic pressure enhanced the cartilaginous matrix formation of MSCs during chondrogenesis.3 Previous studies have shown the application of compression on chondrocytes and progenitor cells would induce apoptosis at early time points.14,20,24 Particularly, Thorpe et al. pointed out that MSC seeded constructs should be first allowed to undergo chondrogenesis prior to transferring to a load-bearing environment.30 Therefore cells encapsulated in alginate beads were cultured in TPS with TGF-β3 for 7 days to form ECM, demonstrated by alcian blue staining, as a support against future mechanical loading.
Our third objective aimed to answer the question whether additional compression would enhance the chondrogenic differentiation of hMSCs in our TPS system. Our results showed that compared to hMSCs in dynamic culture in the TPS bioreactor, hMSCs exposed to additional periodic compression had higher expression levels of aggrecan and type II collagen, the two main positive chondrogenic markers, at later time points after 14 days. The expression of transcription factor Sox 9 was maintained at a higher level in the compressed group throughout the study. Interestingly, one previous study reported that little ECM gene expression was observed by mechanical loading introduced on day 8, while significant ECM gene expression and matrix synthesis were induced by mechanical loading introduced on day 16.21 In our study, the aggrecan gene expression peak correlated with the observed GAG content via alcian blue staining at day 14 in the compression group. Interestingly, type I collagen gene first increased in the compression study, but deceased at later time points, perhaps indicating a transition from hMSCs proliferation to differentiation. These results may suggest that a mechanotransduction process occurred during chondrogenesis, where compressive stimulation induced higher metabolic activity of the cells. During early time points, hMSCs are balancing the response to dynamic culture that slows the chondrogenesis and periodic compression that improves the chondrogenesis. However, at later time points, the differentiation effect caused by compression becomes predominant. Notably, the combined effects of growth factors and mechanical stimuli on chondrogenesis have not been well understood. Future studies to evaluate different combinations of growth factor loading and mechanical loading would be necessary to better address the complex consequence. For both shear only dynamic culture and dynamic compression culture, we observed a trend of increased chondrogenic marker expression towards the end of the study. A study with later time points to gain insight into the longer term differentiaion process and associated cell phenotype will be beneficial to understand the behavior of hMSCs undergoing chondrogenic differentiation in this complex culture system. Detailed effects such as hypertrophic phenotype prevention induced by the compression dynamic culture will be further evaluated in future studies. The small dimension and moderate mechanical loading in this study tried to minimize the possible heterogeneity, but further investigation into force distribution and cellular response will help us better understand the effects of this bioreactor system. Based on the above results, we suggest expanding 3D encapsulated hMSCs in dynamic culture for a short period for proliferation, and subsequently the cells can be transferred to static culture for more desirable differentiation. Furthermore, the addition of compression applied to the 3D cell encapsulated construct may help prevent dedifferentiation. Overall, we suggest that a combination of dynamic and static culture to proliferate phenotypically stable hMSCs might be preferred and necessary for future clinical cell treatment for articular cartilage defects with prominent outcome.
Conclusions
To solve the current problems associated with clinical cell-based treatments for articular cartilage defects, in vitro cell culture techniques must be improved. We have developed a bioreactor system that can enhance the in vitro proliferation of hMSCs encapsulated in 3D hydrogel beads. We have also demonstrated that mechanical stimuli can alter the chondrogenic differentiation in different ways. In the first study, static culture was preferred for differentiation compared to dynamic culture. The second study showed that additional compressive force during dynamic culture can improve the chondrogenic differentiation of the hMSCs. The findings in this study suggest combination culture conditions show promise in producing clinically relevant amounts of tissue through extended in vitro culture.
References
Adesida, A. B., A. Mulet-Sierra, and N. M. Jomha. Hypoxia mediated isolation and expansion enhances the chondrogenic capacity of bone marrow mesenchymal stromal cells. Stem Cell Res. Ther. 3:9, 2012.
Angele, P., D. Schumann, M. Angele, B. Kinner, C. Englert, R. Hente, B. Fuchtmeier, M. Nerlich, C. Neumann, and R. Kujat. Cyclic, mechanical compression enhances chondrogenesis of mesenchymal progenitor cells in tissue engineering scaffolds. Biorheology 41:335–346, 2004.
Angele, P., J. U. Yoo, C. Smith, J. Mansour, K. J. Jepsen, M. Nerlich, and B. Johnstone. Cyclic hydrostatic pressure enhances the chondrogenic phenotype of human mesenchymal progenitor cells differentiated in vitro. J. Orthop. Res. 21:451–457, 2003.
Bae, H., and M. Yu. Miniature Fabry-Perot pressure sensor created by using UV-molding process with an optical fiber based mold. Opt. Express 20:14573–14583, 2012.
Chen, X., H. Xu, C. Wan, M. McCaigue, and G. Li. Bioreactor expansion of human adult bone marrow-derived mesenchymal stem cells. Stem Cells 24:2052–2059, 2006.
Felson, D. T., R. C. Lawrence, P. A. Dieppe, R. Hirsch, C. G. Helmick, J. M. Jordan, R. S. Kington, N. E. Lane, M. C. Nevitt, Y. Q. Zhang, M. Sowers, T. McAlindon, T. D. Spector, A. R. Poole, S. Z. Yanovski, G. Ateshian, L. Sharma, J. A. Buckwalter, K. D. Brandt, and J. F. Fries. Osteoarthritis: new insights. Part 1: The disease and its risk factors. Ann. Intern. Med. 133:635–646, 2000.
Gruber, H. E., and E. N. Hanley, Jr. Human disc cells in monolayer vs 3D culture: cell shape, division and matrix formation. BMC Musculoskelet. Disord. 1:1, 2000.
Gupta, P. K., A. K. Das, A. Chullikana, and A. S. Majumdar. Mesenchymal stem cells for cartilage repair in osteoarthritis. Stem Cell Res. Ther. 3:25, 2012.
Hunziker, E. B. Articular cartilage repair: basic science and clinical progress. A review of the current status and prospects. Osteoarthritis Cartilage 10:432–463, 2002.
Kanichai, M., D. Ferguson, P. J. Prendergast, and V. A. Campbell. Hypoxia promotes chondrogenesis in rat mesenchymal stem cells: a role for AKT and hypoxia-inducible factor (HIF)-1alpha. J. Cell. Physiol. 216:708–715, 2008.
King, J. A., and W. M. Miller. Bioreactor development for stem cell expansion and controlled differentiation. Curr. Opin. Chem. Biol. 11:394–398, 2007.
Kock, L. M., J. Malda, W. J. Dhert, K. Ito, and D. Gawlitta. Flow-perfusion interferes with chondrogenic and hypertrophic matrix production by mesenchymal stem cells. J. Biomech. 47:2122–2129, 2014.
Kock, L., C. C. van Donkelaar, and K. Ito. Tissue engineering of functional articular cartilage: the current status. Cell Tissue Res. 347:613–627, 2012.
Kong, D., T. Zheng, M. Zhang, D. Wang, S. Du, X. Li, J. Fang, and X. Cao. Static mechanical stress induces apoptosis in rat endplate chondrocytes through MAPK and mitochondria-dependent caspase activation signaling pathways. PLoS One 8:e69403, 2013.
Kupcsik, L., M. J. Stoddart, Z. Li, L. M. Benneker, and M. Alini. Improving chondrogenesis: potential and limitations of SOX9 gene transfer and mechanical stimulation for cartilage tissue engineering. Tissue Eng. A 16:1845–1855, 2010.
Larsson, T., R. M. Aspden, and D. Heinegard. Effects of mechanical load on cartilage matrix biosynthesis in vitro. Matrix 11:388–394, 1991.
Lee, K. B., J. H. Hui, I. C. Song, L. Ardany, and E. H. Lee. Injectable mesenchymal stem cell therapy for large cartilage defects—a porcine model. Stem Cells 25:2964–2971, 2007.
Li, W. J., R. Tuli, C. Okafor, A. Derfoul, K. G. Danielson, D. J. Hall, and R. S. Tuan. A three-dimensional nanofibrous scaffold for cartilage tissue engineering using human mesenchymal stem cells. Biomaterials 26:599–609, 2005.
Longobardi, L., L. O’Rear, S. Aakula, B. Johnstone, K. Shimer, A. Chytil, W. A. Horton, H. L. Moses, and A. Spagnoli. Effect of IGF-I in the chondrogenesis of bone marrow mesenchymal stem cells in the presence or absence of TGF-beta signaling. J. Bone Miner. Res. 21:626–636, 2006.
Lucchinetti, E., C. S. Adams, W. E. Horton, Jr, and P. A. Torzilli. Cartilage viability after repetitive loading: a preliminary report. Osteoarthritis Cartilage 10:71–81, 2002.
Mouw, J. K., J. T. Connelly, C. G. Wilson, K. E. Michael, and M. E. Levenston. Dynamic compression regulates the expression and synthesis of chondrocyte-specific matrix molecules in bone marrow stromal cells. Stem Cells 25:655–663, 2007.
Noel, D., F. Djouad, and C. Jorgense. Regenerative medicine through mesenchymal stem cells for bone and cartilage repair. Curr. Opin. Investig. Drugs 3:1000–1004, 2002.
Noriega, S., T. Mamedov, J. A. Turner, and A. Subramanian. Intermittent applications of continuous ultrasound on the viability, proliferation, morphology, and matrix production of chondrocytes in 3D matrices. Tissue Eng. 13:611–618, 2007.
Pelaez, D., C.-Y. Charles Huang, and H. S. Cheung. Cyclic compression maintains viability and induces chondrogenesis of human mesenchymal stem cells in fibrin gel scaffolds. Stem Cells Dev. 18:93–102, 2009.
Quinn, T. M., A. J. Grodzinsky, M. D. Buschmann, Y. J. Kim, and E. B. Hunziker. Mechanical compression alters proteoglycan deposition and matrix deformation around individual cells in cartilage explants. J. Cell Sci. 111:573–583, 1998.
Schnabel, M., S. Marlovits, G. Eckhoff, I. Fichtel, L. Gotzen, V. Vecsei, and J. Schlegel. Dedifferentiation-associated changes in morphology and gene expression in primary human articular chondrocytes in cell culture. Osteoarthritis Cartilage 10:62–70, 2002.
Smith R. L., D. R. Carter and D. J. Schurman. Pressure and shear differentially alter human articular chondrocyte metabolism: a review. Clin. Orthop. Relat. Res. S89–S95, 2004.
Takahashi, I., G. H. Nuckolls, K. Takahashi, O. Tanaka, I. Semba, R. Dashner, L. Shum, and H. C. Slavkin. Compressive force promotes sox9, type II collagen and aggrecan and inhibits IL-1beta expression resulting in chondrogenesis in mouse embryonic limb bud mesenchymal cells. J. Cell Sci. 111(Pt 14):2067–2076, 1998.
Temenoff, J. S., and A. G. Mikos. Review: tissue engineering for regeneration of articular cartilage. Biomaterials 21:431–440, 2000.
Thorpe, S. D., C. T. Buckley, T. Vinardell, F. J. O’Brien, V. A. Campbell, and D. J. Kelly. The response of bone marrow-derived mesenchymal stem cells to dynamic compression following TGF-beta3 induced chondrogenic differentiation. Ann. Biomed. Eng. 38:2896–2909, 2010.
Vinatier, C., D. Mrugala, C. Jorgensen, J. Guicheux, and D. Noel. Cartilage engineering: a crucial combination of cells, biomaterials and biofactors. Trends Biotechnol. 27:307–314, 2009.
Xie, Y., P. Hardouin, Z. Zhu, T. Tang, K. Dai, and J. Lu. Three-dimensional flow perfusion culture system for stem cell proliferation inside the critical-size beta-tricalcium phosphate scaffold. Tissue Eng. 12:3535–3543, 2006.
Yeatts, A. B., and J. P. Fisher. Tubular perfusion system for the long-term dynamic culture of human mesenchymal stem cells. Tissue Eng. C Methods 17:337–348, 2011.
Yeatts, A. B., C. N. Gordon, and J. P. Fisher. Formation of an aggregated alginate construct in a tubular perfusion system. Tissue Eng. C Methods 17:1171–1178, 2011.
Yoon, D. M., S. Curtiss, A. H. Reddi, and J. P. Fisher. Addition of hyaluronic acid to alginate embedded chondrocytes interferes with insulin-like growth factor-1 signaling in vitro and in vivo. Tissue Eng. A 15:3449–3459, 2009.
Yoon, D. M., E. C. Hawkins, S. Francke-Carroll, and J. P. Fisher. Effect of construct properties on encapsulated chondrocyte expression of insulin-like growth factor-1. Biomaterials 28:299–306, 2007.
Yu, L., K. M. Ferlin, B. N. Nguyen, and J. P. Fisher. Tubular perfusion system for chondrocyte culture and superficial zone protein expression. J. Biomed. Mater. Res. A 103:1864–1874, 2015.
Zhang, Z. Y., S. H. Teoh, W. S. Chong, T. T. Foo, Y. C. Chng, M. Choolani, and J. Chan. A biaxial rotating bioreactor for the culture of fetal mesenchymal stem cells for bone tissue engineering. Biomaterials 30:2694–2704, 2009.
Zhao, F., P. Pathi, W. Grayson, Q. Xing, B. R. Locke, and T. Ma. Effects of oxygen transport on 3-d human mesenchymal stem cell metabolic activity in perfusion and static cultures: experiments and mathematical model. Biotechnol. Prog. 21:1269–1280, 2005.
Acknowledgments
This study was funded by the National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health (R01 AR061460) as well as by the National Science Foundation (CBET 1264517). This work was also funded by the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (Grant number: 230303). The authors thank Feng Gao from Cornell University for his help on data processing and Dr. Hannah B. Baker for reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Eric Darling oversaw the review of this article.
Ting Guo and Li Yu have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Guo, T., Yu, L., Lim, C.G. et al. Effect of Dynamic Culture and Periodic Compression on Human Mesenchymal Stem Cell Proliferation and Chondrogenesis. Ann Biomed Eng 44, 2103–2113 (2016). https://doi.org/10.1007/s10439-015-1510-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-015-1510-5