Abstract
Rhabosciadium aucheri is an Iranian endemic herbaceous species that grows in the west, center, and south regions of Iran. In the present study, genetic variation of 70 individuals belonging to seven natural populations of four provinces was investigated using Random Amplified Polymorphic DNA (RAPD) markers. Ten out of twenty-two RAPD primers employed in this study, generated 110 highly amplified and reproducible loci and a mean of 11.1 bands per primer and 48.13% of polymorphism was obtained. According to our results, the primer OPA10 presented the highest effective number of alleles, Shannon’s index, and genetic diversity. The highest value of genetic identity (0.916) was determined between Hamadan, Nahavand and Hamadan, Alvand Mts. populations and the highest genetic distance (0.277) was observed between Hamadan, Asadabad and Kurdistan, Qorveh populations. Therefore, there is an obvious correlation between genetic diversity and geographical distribution. PCA was obtained based on RAPD molecular data and Neighbor Joining (NJ) dendrogram was provided successively. Similar results were attained employing UPGMA and Neighbor Joining dendrograms, supported by PCA ordination plot. Overall, almost moderate level of polymorphism was obtained by RAPD molecular markers at the population level that shows a reasonable amount of intraspecific variability. The reason might be due to the low level of gene flow between populations that could give rise to high genetic differentiation.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Apiaceae (Umbelliferae), is considered a large and important family of the flowering plants in the world [1,2,3,4,5]. Iran is one of the significant diversification centers of the family members [6]. Rhabdosciadium Boiss. (Apiaceae) encompasses only eight herbaceous species, that are distributed in Iran and Turkey exclusively [7,8,9,10]. Only three species i.e., R. strausii Hausskn. ex Bornm., R. petiolare Boiss. & Hausskn. ex Boiss. and R. aucheri Boiss., are known as endemic to Iran [3, 4]. Rhabdosciadium aucheri Boiss., is regarded as a rare and endemic species, which has limited distribution and found only in the west (Kurdistan, Kermanshah, Hamadan, and Lorestan provinces), center (Tehran, Arak, Esfahan, and Chaharmahal & Bakhtiari provinces) and south (Kohgiluye & Boyerahmad and Fars) regions of the country [3, 4]. This Iranian species differs from two other related ones morphologically by its flattened leaves with ovate-oblong segments, mericarps 10–15 mm and short-conical stilopodium embedded in corky pericarp [3, 4, 7]. Based on the current morphological analyses, the species has shown significant differences especially in quantitative morphological traits among seven natural populations [11]. Also, chemical components of the species have been evaluated properly [12, 13].
Due to the undeniable importance of genetic diversity in conservation management especially about rare and endemic species and consequently its effective role in survival of populations by species adaptation [14, 15], molecular markers have been asserted as remarkable tools in the evaluation of genetic diversity and conservation strategies in different types of plants [16,17,18,19]. Lately, genetic diversity and population structure of R. aucheri populations have been investigated employing ISSR molecular markers [11].
One of the DNA-based molecular markers, that has been extensively used by several authors, is random amplified polymorphic DNA (RAPD), which analyses a large number of bands in contrast with other molecular markers. Therefore, a more comprehensive assessment of the genome would be provided. Some other probable benefits of using such a marker are related to its cost, speed, and consequently its potential in distinguishing polymorphism. It successfully recognizes genetic diversity between populations [20]. Although RAPDs have some mentioned above advantages, their predominant allelic expression, as well as low level of reproducibility, are regarded as their undeniable drawbacks [21]. Polymorphism is regarded as a common phenomenon that occurs in nature, which is related to existing more than one divergent phenotype in the same population of a species. It is related to biodiversity, genetic variation, and adaptation. Evolutionary procedures could result in polymorphism, that could be modified by natural selection. The species adaptation to the environmental fluctuations relies on genetic variation [22].
In the present study, we aimed to investigate the genetic variability of the species employing RAPD molecular markers to estimate the level of diversity between specimens collected from different localities of Iran.
Materials and methods
Plant material
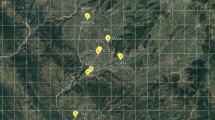
In this study, fresh leaves belonging to 70 individuals in seven geographical populations of R. aucheri, from four provinces of Iran, i.e., Hamadan, Lorestan, Kurdistan and Chaharmahal and Bakhtiari, were collected from July to August 2018 and dried in plastic bags using silica gel powder for further analyses. Details of sampling sites are provided (Table 1, Fig. 1).
The distribution map belonging to seven populations of Rhabdosciadium aucheri (Abbreviations: Pop 1: Hamadan, Asadabad population; Pop 2: Hamadan, Nahavand population; Pop 3: Hamadan, Alvand Mts. population; Pop 4: Lorestan, Noorabad population; Pop 5: Lorestan, Dorud population; Pop 6: Kurdistan, Qorveh population; Pop 7: Chaharmahal and Bakhtiari, Shahr-e-Kord, Sabzkouh population). Note: populations from different localities are also differentiated with different colors and shapes in the map
DNA extraction and RAPD assay
Total genomic DNA was extracted by plant genomic DNA extraction Kit (DNPTM Kit) from silica-dried leaves (approximately 0.5 g material per sample) using CTAB activated charcoal protocol [23, 24] and stored at −20 °C for further study. DNA quality was determined after electrophoresis on 1% agarose gel. Density and concentration of the extracted DNA were examined on a NanoDrop ™ 2000 (Thermo Scientific). Overall, 22 RAPD primers of Operon technology (Alameda, Canada) were applied in this research. These primers comprised OPA, OPB, OPC, OPH, OPI and OPM sets. Polymerase Chain Reaction (PCR) were carried out in a 25 μl volume containing 10 mM Tris-HCl buffer at pH 8; 50 mM KCl; 1.5 mM MgCl2; 0.2 mM of each dNTP (Bioron, Germany); 0.2 μM of a single primer; 20 ng genomic DNA and three U of Taq DNA polymerase (Bioron, Germany). PCR amplification was performed with Techne thermocycler (Germany) using the following program profile: 94 °C for 5 min, followed by 40 cycles of 1 min at 94 °C; 1 min at 52 °C and 2 min at 72 °C and a final extension step of 7–10 min at 72 °C. RAPD fragments were separated by electrophoresis, running on 1.0% (w/v) agarose gel, stained with ethidium bromide, and observed under UV transilluminator using a digital camera.
RAPD analyses
Each amplified fragment employing RAPD primers was treated as a binary character in this research. Therefore, it was scored “0” and “1” for its absence and presence respectively. Jaccard’s similarity coefficient [25], as well as Nei’s genetic distance [26], was detected among the studied populations and employed for grouping using Unweighted Paired Group with Arithmetic Average (UPGMA), Neighbor-Joining (NJ) clustering methods, and ordination based on principal component analysis (PCA) [27, 28]. The bootstrap values were applied to construct NJ tree using PAUP* ver. 4.0a168 software [29]. The PCA analysis was performed using DARwin ver. 5 [30] software. To estimate molecular differences between populations, AMOVA test was performed using GenALEX 6.5 [31].
Results
Overall, 10 out of 22 RAPD primers produced 110 highly amplified and reproducible loci, and an average of 11.1 bands per primer was attained in this study (Table 2). Only 10 bands were common in all 7 studied populations and 100 amplicons presented polymorphism (Fig. 2). RAPD primers containing OPA 10 (85.52%), OPM 10 (75.72%) and OPH 07 (70.57%), generated the highest percentages of polymorphism, while some other primers, produced the lowest polymorphism percentages (almost 18–28%). In general, the average percentage of 48.13% polymorphism was calculated in this research (Table 2). Also, the highest effective number of alleles (1.359), Shannon’s Index (The popular diversity index that is commonly used to characterize species variation in a population) [32] (0.529) and genetic diversity (0.317) were dedicated to the primer OPA 10 (Table 2). As is observed from Table 2, maximum of 16 bands were produced by the primer OPA 09, while minimum of three bands was created by the primer OPI 12. Moreover, the maximum number of polymorphic fragments was produced by the primer OPA 09 with 17 specific bands, while the primers OPA 13 and OPA 05 were placed in the next ranks, with five and four specific fragments, respectively. However, the primers OPC 13, OPM 10, and OPB 05 created a unique polymorphic amplicon. Among the studied populations, Hamadan, Asadabad displayed the presence of polymorphic loci of 1100 bp of the primer OPA 13, 950 bp of the primer OPC 01 and 1200 bp of the primer OPB 05. However, Lorestan, Noorabad population revealed specific bands of 350 bp of the primer OPM 10, and Hamadan, Alvand Mts. population was the only one having a band of 650 bp of the primer OPA 05, a band of 450 bp of OPA-13, and 550 bp of the OPA 05 primers. Also, Lorestan, Dorud population showed the band of 150 bp of the primer OPA-04. Some specific loci were present in all except one population. For instance, both polymorphic bands of 850 bp of OPH-07 and 900 bp of OPA-09 primers were absent only in Hamadan, Nahavand population. Nei’s genetic identity and genetic distance was detected among the studied populations using molecular RAPD data (Table 3). The value of genetic identity differed from 0.757 between Hamadan, Asadabad (no. 1) and Kurdistan, Qorveh (no. 6) populations to 0.916 between Hamadan, Nahavand (no. 2) and Alvand Mts. (no. 3) populations. Moreover, the mean value of 0.85 was determined for genetic identity based on RAPD molecular data. Comparatively higher genetic distance (0.277) was found between Hamadan, Asadabad (no. 1) and Kurdistan Qorveh (no. 6) populations (Table 3). Also, similar results were obtained using UPGMA and Neighbor Joining (NJ) dendrograms, supported by PCA ordination plot (Figs. 3 and 4). Also, cophenetic correlation (the correlation of entities’ scores on the dimensions) (r) [33] values between the Neighbor Joining dendrogram and the dissimilarity matrix presented a satisfying degree of fit (r = 0.97). The results of NJ tree based on RAPD molecular data for the studied populations of R. aucheri are presented in Fig. 3. According to our results, three populations including Hamadan, Nahavand, Hamadan, Heidareh and Kurdistan (populations 2, 3 and 6) constituted unique major group among which, population 2 and most of the members of population 3 were placed in a closer distance. However, the bootstrap value for this grouping was not supported significantly. Moreover, all members of population 5 (with bootstrap value of 65) formed another group exclusively. However, most of the members belonging to population 4 (with bootstrap support of 58) were separately grouped. Also, all members of Chaharmahal and Bakhtiari, Shahr-e-Kord population (no. 7) formed a separate major cluster that was not supported significantly. Most of the individuals of the populations 1, 4, 5 and 7 constituted a considerable distance from the other mentioned populations. However, the exact position of the populations Hamadan, Asadabad (no. 1) and Shahr-e-kord, Sabzkouh (no. 7) were not supported properly (Fig. 3). The PCA plot based on RAPD molecular data confirmed the above results (Fig. 4). Both populations 5 and 7 placed far from each other in the space that corroborates their genetic distance. Moreover, the populations 1 and 3, as well as the populations 2 and 3, showed a level of affinities.
Electrophoresis patterns of amplified DNA products obtained from Rhabdosciadium aucheri populations using OPA10 (1–7) and OPA 13 (8–14) RAPD primers (Abbreviation: 1,8: Hamadan, Asadabad; 2,9: Hamadan, Nahavand; 3,10: Hamadan, Alvand Mts.; 4,11: Lorestan, Noorabad; 5,12: Lorestan, Dorud; 6,13: Kurdistan, Qorveh; 7,14: Shahr-e-Kord, Sabzkouh; N: No DNA; L: DNA Ladder)
NJ tree belonging to Rhabdosciadium aucheri populations based on RAPD molecular data created by Jaccard’s similarity coefficients (Abbreviations: Pop 1: Hamadan, Asadabad population; Pop 2: Hamadan, Nahavand population; Pop 3: Hamadan, Alvand Mts. population; Pop 4: Lorestan, Noorabad population; Pop 5: Lorestan, Dorud population; Pop 6: Kurdistan, Qorveh population; Pop 7: Shahr-e-Kord, Sabzkouh population). Note: The numbers above each branch shows the bootstrap support. Individuals are marked in different colors with their related population numbers
Principle component analysis plot related to Rhabdosciadium aucheri populations based on RAPD molecular data (Abbreviations: Pop 1: Hamadan, Asadabad population; Pop 2: Hamadan, Nahavand population; Pop 3: Hamadan, Alvand Mts. population; Pop 4: Lorestan, Noorabad population; Pop 5: Lorestan, Dorud population; Pop 6: Kurdistan, Qorveh population; Pop 7: Shahr-e-Kord, Sabzkouh population)
Discussion
In the present study, RAPD markers were employed to evaluate genetic diversity among the R. aucheri (Apiaceae) populations. Regarding the importance of R. aucheri as an Iranian endemic and rare species, its conservation is necessary to exhibit the size of its natural populations and investigate the level of genetic diversity. As a whole, the study of population genetics could give rise to valuable data about genetic structure and genetic variation of the populations [18, 23]. RAPDs are still considered the most inexpensive and the fastest technique for attaining inconsistency at the level of DNA [22].
Limitations
There may be some possible limitations in this research. Due to some drawbacks of RAPDs, it had better to use Next Generation Sequencing (NGS) technique in order to find microsatellite regions and primer development. However, NGS is regarded as an expensive technique that might be used in the near future studies.
In this research, RAPD data revealed a relatively moderate level of polymorphism (48.13%) at the population level that indicates a reasonable amount of intraspecific variability. Most of the previous investigations on other members of the family, presented a high level of polymorphism [34,35,36,37]. The reason might be due to the low level of gene flow between populations that might be the cause of several factors such as isolation of the populations, seed dispersal, etc. Restricted gene flow could result in high genetic differentiation [36]. Moreover, an acceptable percentage of genetic variability has recently been reported among the studied populations of the R. aucheri using ISSR molecular markers [11]. According to Kazemeini et al. [11], Lorestan, Noorabad population (no. 4) showed the highest percentage of polymorphism. While, Hamadan, Asadabad population (no. 1) indicated the lowest value for polymorphism percentage followed by the lowest gene diversity and value for Shannon’s information index. The previous results demonstrated that the level of polymorphism reported by RAPD, ISSR and AFLP markers for the family members were lower than those obtained by SRAP primer combinations [35, 38]. According to Zahid et al. [39], 48% of polymorphic bands were obtained for fennel germplasm accessions employing 30 RAPD primers. These results conform to those attained in this research. Due to the middle level of polymorphism, R. aucheri might be endangered species that has a limited distribution in Iran and it must be conserved properly.
The population distribution might be influenced by the distance and also gene migration might probably happen among the adjacent populations [40,41,42]. Therefore, populations located at a close distance are more genetically similar to each other. The correlation between genetic diversity and geographical distribution has been confirmed in different medicinal species belonging to the family Apiaceae [34,35,36,37]. The results of the present study conform to those achieved by the mentioned above authors. For instance, according to NJ tree and PCA results, the populations no. 1 and no. 6 located far from each other geographically, showed the highest level of genetic distance. Also, the populations no. 2 and no. 3, placing in the close distance displayed higher value of genetic identity. Moreover, the results of the recent investigation based on ISSR molecular data were in line with those obtained in this research. The closest genetic similarity was observed between the populations of Hamadan, Nahavand and Hamadan, Heidareh (populations 2 and 3) while, the lowest value for genetic similarity was observed between Hamadan–Asadabad and Kurdistan populations (populations 1 and 6) [11]. However, these results contradict those achieved by Martins et al. [43], Solouki et al. [44] and Pezhmanmehr et al. [45], who reported there is no correlation between genetic distance and geographical distribution among their studied populations. However, the results could not be generalized to all populations. As mentioned above, the reason might be due to some environmental and ecological factors, that could act as an obstacle against gene flow among some adjacent populations. Therefore, the mixture of genetic and morphological divergence, restricted gene flow, and local adaptation could result in diversification of R. aucheri populations in Iran that might be useful in the conservation of this valuable endemic and rare species.
References
Downie SR, Katz-Downie DS, Watson MF (2000) A phylogeny of flowering plant family Apiaceae based on chloroplast DNA rpl 16 and rpoC1 intron sequence: towards a Supergeneric classification of subfamily Apioideae. Am J Bot 87(2):273–292. https://doi.org/10.2307/2656915
Duran A, Doğan B, Duman H, Martin E, Öztürk M, Çetin Ö (2010) Taxonomic studies on the genus Rhabdosciadium (Apiaceae) with particular reference to Turkish species and their relationships with some closely related genera. Biologia 65(3):451–458. https://doi.org/10.2478/s11756-010-0032-8
Hedge IC, Lamond JM (1972) Rhabdosciadium Boiss. In: Davis PH (ed) Flora of Turkey and the East Aegean Islands 4. Edinburgh University Press, Edinburgh, p 311
Hedge IC, Lamond JM (1987) Tribus Scandiceae. In: Rechinger KH (ed) Umbelliferae, Flora Iranica 162. Akademische Druck und Verlagsanstalt, Graz, pp 83–110
Lamamra M, Laouer H, Amira S, Orhan IE, Senol FS, Demirci B, Akkal S (2017) Chemical composition and cholinesterase inhibitory activity of different parts of Daucus aristidis Coss. Essential oils from two locations in Algeria. Rec Nat Prod 11(2):147–156
Amiri MS, Joharchi MR (2016) Ethnobotanical knowledge of Apiaceae family in Iran: a review. Avicenna J Phytomed 6(6):621–635. https://doi.org/10.22038/AJP.2016.6696
Akalın E, Akpulat A (2012) Rhabdosciadium urusakii sp. nov. (Apiaceae) from East Anatolia (Turkey). Nord J Bot 30(5):560–564. https://doi.org/10.1111/j.1756-1051.2012.01475.x
Firat M, Güzel Y (2019) Contributions to the taxonomy of the Irano-Turanian genus Rhabdosciadium (Apiaceae): nomenclatural notes, carpology, molecular phylogeny and the description of a new species from Bitlis (Turkey). Phytotaxa 395(3):179–198. https://doi.org/10.11646/phytotaxa.395.3.3
Lyskov D, Kljuykov E, Güner ED, Samigullin T (2017) Molecular phylogeny of the genus Rhabdosciadium (Apiaceae) with description of a new species R. anatolyi from Hakkâri province, eastern Turkey. Phytotaxa 331(2):253–258. https://doi.org/10.1111/j.1756-1051.2012.01475.x
Pimenov MG, Leonov MV (1993) The genera of the Umbelliferae. Royal Botanic Garden, Kew
Kazemeini F, Asri Y, Mostafavi G, Kalvandi R, Mehregan I (2020) Assessment of genetic diversity, population structure and morphological analyses in an Iranian endemic species Rhabdosciadium aucheri Boiss. (Apiaceae) using ISSR markers. Biologia. https://doi.org/10.2478/s11756-020-00637-1
Eghtesadi F, Mehrdad M, Sonboli A (2006) Essential oil composition of Rhabdosciadium aucheri from Iran. Chem Nat Compd 42(4):489–490. https://doi.org/10.1007/s10600-006-0189-0
Kalvandi R, Mirza M, Safikhani K, Naderi M (2013) Essential oil composition of Rhabdosciadium aucheri Boiss. From Iran. Iran J Med Aromat Plants 29(3):605–610 (In Persian with an abstract in English)
Cires E, Sanna M, Vázquez VM, Fernández Prieto JA (2018) Genetics and conservation of rare and endemic plants: the case of Genista sanabrensis (Fabaceae) in the Iberian Peninsula. Mediterr Bot 39(2):77–87. https://doi.org/10.5209/MBOT.60078
Tomasello S, Álvarez I, Vargas P, Oberprieler C (2015) Is the extremely rare Iberian endemic plant species Castrilanthemum debeauxii (Compositae, Anthemideae) a ‘living fossil’? Evidence from a multi-locus species tree reconstruction. Mol Phylogenet Evol 82:118–130. https://doi.org/10.1016/j.ympev.2014.09.007
Tabaripour R, Sheidai M, Talebi SM, Noormohammadi Z (2018) Genetic divergence and speciation within Ziziphora capitate (Lamiaceae): molecular and micromorphological evidences. Biodiversitas 19:747–755. https://doi.org/10.13057/biodiv/d190250
Savolainen V, Chase MW (2003) A decade of progress in plant molecular phylogenetics. Trends Genet 9(12):717–724. https://doi.org/10.1016/j.tig.2003.10.003
Sheidai M, Ziaee S, Farahani F, Talebi SM, Noormohammadi Z, Hasheminejad-Ahangarani Farahani Y (2014) Intra-specific genetic and morphological diversity in Linum album (Linaceae). Biologia 69, 32(1):–39. https://doi.org/10.2478/s11756-013-0281-4
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13(5):1143–1155. https://doi.org/10.1111/j.1365-294X.2004.02141.x
Rajora O, Rahman M (2003) Microsatellite DNA and RAPD fingerprinting, identification and genetic relationships of hybrid poplar (Populus x canadensis) cultivars. Theor Appl Genet 106:470–477. https://doi.org/10.1007/s00122-002-1082-2
Szmidt AE, Wang XR, Lu MZ (1996) Empirical assessment of allozyme and RAPD variation in Pinus sylvestris (L.) using haploid tissue analysis. Hered 76:412–420
Dwivedi S, Singh S, Chauhan UK, Tiwari MK (2018) Inter and interspecific genetic diversity (RAPD) among three most frequent species of macrofungi (Ganoderma lucidum, Leucoagricus sp. and Lentinus sp.) of tropical forest of Central India. J Genet Eng Biotechnol 16(1):133–141. https://doi.org/10.1016/j.jgeb.2017.11.008
Esfandani-Bozchaloyi S, Sheidai M, Hassanzadeh-Kalalegh M (2019) Comparison of DNA extraction methods from Geranium (Geraniaceae). Acta Bot Hung 61(3–4):251–266. https://doi.org/10.1556/034.61.2019.3-4.3
Yang L, Fu S, Khan MA, Zeng W, Fu J (2013) Molecular cloning and development of RAPD-SCAR markers for Dimocarpus longan variety authentication. SpringerPlus 2:501. https://doi.org/10.1186/2193-1801-2-501
Jaccard P (1908) Nouvelles recherches Sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Nei M (1972) Genetic distance between populations. Am Nat 106:283–291 http://www.jstor.org/stable/2459777
Podani J (2000) Introduction to the exploration of multivariate data [English translation]. Backhuys Publication, Leiden
Weising K, Nybom H, Pfenninger M, Wolff K, Kahl G (2005) DNA fingerprinting in plants: principles, methods, and applications. CRC press, Boca Raton
Swofford DL (2002) PAUP*: phylogenetic analyses using parsimony (*and other methods), version 4.0b10. Sinauer, Sunderland
Perrier X, Jacquemoud-Collet J (2006) DARwin software. Available from: http://darwin.cirad.fr/
Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech 27:379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x
Sokal RR, Rohlf FJ (1962) The comparison of dendrograms by objective methods. Taxon 11:33–40. https://doi.org/10.2307/121720
Gaudeul M, Till-Bottraud I, Barjon F, Manel S (2004) Genetic diversity and differentiation in Eryngium alpinum L. (Apiaceae): comparison of AFLP and microsatellite markers. Hered 92(6):508–518. https://doi.org/10.1038/sj.hdy.6800443
Maghsoudi-Kelardashti H, Rahimmalek M, Torabi M (2015) Genetic diversity in Iranian fennel (Foeniculum vulgare Mill.) populations based on sequence related amplified polymorphism (SRAP) markers. J Agr Sci Tech 17:1789–1803
Masoumi SM, Kahrizi D, Rostami-Ahmadvandi H, Soorni J, Kiani S, Mostafaie A, Yari K (2012) Genetic diversity study of some medicinal plant accessions belong to Apiaceae family based on seed storage proteins patterns. Mol Biol Rep 39(12):10361–10365
Qiu TX, Hong DY, Fu CX, Cameron KM (2004) Genetic variation in the endangered and endemic species Changium smyrnioides (Apiaceae). Biochem Syst Ecol 329(6):583–596. https://doi.org/10.1016/j.bse.2003.08.004
Bahmani K, Izadi-Darbandi A, Ashraf-Jafari A, Sadat-Noori S, Farajpour M (2012) Assessment of genetic diversity in Iranian fennel using ISSR markers. J Sci 4(9):79–84. https://doi.org/10.5539/jas.v4n9p79
Zahid N, Abbasi N, Hafiz I, Ahmad Z (2009) Genetic diversity of indigenose fennel (Foeniculum vulgare Mill.) germplasm in Pakistan an assessed by RAPD markers. Pak J Bot 41:1759–1767
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influence of gene flow and drift on the distribution of genetic variability. Evolution 53:1898–1914. https://doi.org/10.1111/j.1558-5646.1999.tb04571.x
Medrano M, Herrera CM (2008) Geographical structuring of genetic diversity across the whole distribution range of Narcissus longispathus, a habitat-specialist, Mediterranean narrow endemic. Ann Bot 102:183–194. https://doi.org/10.1093/aob/mcn086
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279. https://doi.org/10.2307/2410134
Martins SR, Vences FJ, Sáenz de Miera LE, Barroso MR, Carnide V (2006) RAPD analysis of genetic diversity among and within Portuguese landraces of common white bean (Phaseolus vulgaris L.). Sci Hortic 108(2):133–142
Solouki M, Mehdikhani H, Zeinali H, Emamjomeh AA (2008) Study of genetic diversity in chamomile (Matricaria chamomilla) based on morphological traits and molecular markers. Sci Hortic 117(3):281–287. https://doi.org/10.1016/j.scienta.2008.03.029
Pezhmanmehr M, Hassani ME, Jahansooz F, Najafi AA, Sefidkon F, Mardi M, Pirseiedi M (2009) Assessment of genetic diversity in some Iranian populations of Bunium persicum using RAPD and AFLP markers. Iran J Biotechnol 7(2):93–100
Acknowledgements
The authors gratefully acknowledge the financial supports of the Islamic Azad University, Science and Research Branch (Tehran, Iran). The authors also wish to thank all laboratory staffs for providing equipment to accomplish this research and Mr. A. Sadeghi for improving the quality of English language of the article.
Funding
This work was supported by the Islamic Azad University, Science and Research Branch, Tehran, Iran.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author declare that, there is no conflict of interest.
Ethical approval
On behalf of all authors, the corresponding author declare that, permission to collect an endemic and rare species was not required in Iran.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kazemeini, F., Asri, Y., Mostafavi, G. et al. RAPD-based evaluation of genetic diversity among populations of the Iranian endemic species Rhabdosciadium aucheri Boiss. (Apiaceae). Mol Biol Rep 47, 9345–9352 (2020). https://doi.org/10.1007/s11033-020-06028-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-06028-8