Abstract
The whitefly Bemisia tabaci (Gennadius) has caused notable damage to vegetable and cotton crops in the eastern Mediterranean region since about 1994, and has become particularly problematic in southern Turkey beginning in 2000. The development of squash silverleaf symptoms in Cucurbita species and the unprecedented high population levels in the region suggested that the B biotype, notable for the latter phenotypes, had been introduced. To test this hypothesis and determine the host distribution of the suspect introduced B biotype and its associated natural enemies, B. tabaci immature instars and adults, and the associated natural enemies were collected from cultivated and uncultivated plant species. From the southern Turkey collections, B. tabaci was found to colonize 152 species from 43 plant families. Of the plant species upon which B. tabaci was found to reproduce, 152 of them were reported as hosts of B. tabaci in Turkey. Five species of predators and two species of parasitoids were identified as natural enemies of the B biotype of B. tabaci in southern Turkey. Using the mitochondrial cytochrome oxidase I gene all B. tabaci were identified as the B biotype of the B. tabaci complex, at 96–100% shared identity with reference B biotype sequences. Results indicate that this invasive biotype has displaced the local Turkey-cotton haplotype that was known to occur previously in southern Turkey.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bemisia tabaci (Gennadius) has been known to cause economic losses in cotton and vegetable crops in southern Turkey since 1966 (Kaygısız 1976; Ulusoy 2001). This whitefly species has become an increasingly important pest and vector of plant viruses in agricultural crops in tropical and subtropical regions throughout the world (Avidov and Harpaz 1969; Mound and Halsey 1978; Costa and Brown 1991; Costa et al. 1993; Gill 1992; Cock 1993; Brown 1994, 2000, 2001). The B biotype was first described in the southwestern U.S.A. from poinsettia (Costa and Brown 1991). It is considered a distinct biological type based on its unusually broad host range, high reproductive success, and ability to induce a silvering phenotype in leaves of colonized Cucurbita species (Costa and Brown 1991) and irregular ripening in tomato fruits (Schuster et al. 1990), both which are reminiscent of phytotoxic disorders. The further ability of the B biotype develop insecticide resistance to certain commonly used compounds, has made it particularly difficult to control, and enabled its rapid spread throughout the Americas (Costa et al. 1993; Brown et al. 1995; Kirk et al. 2000; Viscarret et al. 2003), Europe, Asia, and to Australia (Brown 2000).
Biological variants of B. tabaci cannot be distinguished using traditional taxonomic methods because they do not have distinguishing morphological characters (Mound and Halsey 1978; Gill 1992; Rosell et al. 1997). A distinctive esterase profile for the ‘A biotype’ indigenous to the Southwestern U.S.A., when compared to that for the invasive ‘B’ biotype provided the first evidence that the two variants were polymorphic (Costa and Brown 1991). The unique ability of the ‘B’ biotype to cause silvering in Cucurbita spp. has provided a reliable phenotype by which the ‘B’ biotype (and its closest relatives) can be distinguished from all New World B. tabaci, and most other Old World B. tabaci, because other bio/haplotypes do not express this phenotype (Costa et al. 1993; Bedford et al. 1994; Brown et al. 1995; Brown 2000).
A number of studies have revealed unexpected variation within B. tabaci (Costa et al. 1993; Brown et al. 1995, 2000; see refs in Brown 2001), leading to the proposal that B. tabaci is a sibling species complex (Brown et al. 1995). This has since spurred a great interest in the ecology, diversity, population genetics, and evolutionary history of the B. tabaci complex. Molecular markers, including the mitochondria (mt) 16S rRNA and mt cytochrome oxidase I (mtCOI) genes (Brown et al. 1995; Frohlich et al. 1999; Kirk et al. 2000; Viscarret et al. 2003), and the ITS1 (nuclear, non-coding) sequence (De Barro et al. 2000) have been used to document extensive genetic polymorphisms within the B. tabaci complex, worldwide. Analysis of the mtCOI for geographically representative B. tabaci has revealed that the mtCOI sequence is informative and predictive of extant phylogeographical relationships for the B. tabaci complex (Frohlich et al. 1999; Brown 2000; Brown and Idris 2005; Kirk et al. 2000; Viscarret et al. 2003).
Molecular genetic (RAPDs) analysis of B. tabaci in Spain revealed that at least two indigenous B. tabaci haplotypes occur there. The S type is restricted to the Convolvulaceae, and the Q haplotype (Guirao et al. 1997), which is a member of a sister subclade to the B biotype, has a broad host range that also includes the Convolvulaceae. The Q biotype is a polyphagous biotype from the Mediterranean/North African region and was shown to occur sympatrically with the ‘B’ biotype in Spain in about 1994. This result has been corroborated by mtCOI data (Frohlich et al. 1999; Brown 2000), which suggests that the B biotype is not indigenous to Spain, and that it was recently introduced there from the Middle East/Africa, and further that the Q population is native to the Mediterranean region (Brown 2000; Brown and Idris 2005). The B biotype now occurs throughout much of the world where it has successfully established in arid, irrigated agricultural production areas, including mild climate regions of most continents (Fig. 1; Costa et al. 1993; Brown et al. 1995; Summers et al. 1995; Frohlich et al. 1999; Brown 2000; Viscarret et al. 2003).
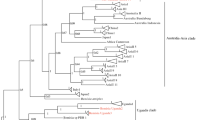
Phylogenetic tree (Clustal W) showing four (of seven) the main clades containing haplotypes of the Bemisia tabaci complex, and the affiliation of selected field populations from Turkey (Tu5-14) with the North Africa–Mediterranean–Middle East clade and specifically with the sister clade containing the B biotype (single line box). Also illustrated is that the B. tabaci native to Turkey, TC (double line box) is most closely related to the sister clade containing the Q biotype and close relatives. The outgroup (genus) is the greenhouse whitefly, Trialeurodes vaporariorum (West.)
During its invasive period, the B biotype also was introduced into Southern Turkey in about 2000 (Ulusoy et al. 2002). However, its distribution and the extent to which it could colonize potential hosts there is not known. Previously, an indigenous B. tabaci population had been reported from cotton in Turkey, designated TC (Turkey-cotton) (Bedford et al. 1994). A mtCOI analysis showed that the native TC population grouped in the subclade that also contains the Q type from Spain, and that the two are close relatives but that they diverge by about 3% based on a comparison of their mtCOI sequences (Brown 2001; Kirk et al. 2000; Viscarret et al. 2003).
This objective of this study was to: (i) determine the distribution of the invasive B biotype and other prospective sympatric B. tabaci populations, (ii) document the natural host range for B. tabaci, and (iii) identify the natural enemies associated with B. tabaci in cultivated and uncultivated hosts, in southern Turkey.
Materials and methods
Natural host range
Plant species growing within or around agriculture and non-agriculture fields in East Mediterranean region in Turkey were examined for colonization by B. tabaci. Collections were made in the southern Turkish provinces of Adana, Mersin, and Hatay during 2000–2002 (Fig. 2). Whitefly-infested leaves were detached from colonized plants, placed into individual plastic bags, and transported to the laboratory for species identification. Leaves were examined to locate the exuvia of the pupal stage (4th instar) using a binocular microscope. Whiteflies were identified to species using key taxonomic criteria of the pupae (Chang 1969; Gill 1992). Plant species for which two or more adults enclosed from exuvia were considered reproductive hosts of B. tabaci.
Natural enemies
Predators associated with whiteflies were collected with modified vacuum samplers in survey areas of B. tabaci in the East Mediterranean region of Turkey (Fig. 2). When immature predators were observed associated with whitefly nymphs, the insects were studied in the laboratory to determine if the suspect natural enemy consumed whitefly nymph. Immature predators were collected, together with the plant material infested by the respective prey nymphs. Predator larvae were reared on nymphs in plastic boxes. In field areas (Fig. 2) or in a plastic box in the laboratory observations were used to verify whether the natural enemy consumed nymphs or adult of B. tabaci. Also, all parasitoids associated with B. tabaci were identified following the collection of nymphs from leaves (Kirk et al. 2000). Leaves were placed in a plastic box in the laboratory and waiting for the adults to emerge 15 days according to Ryckewaert and Alauzet (2002).
The adult predators and parasitoids were identified by Prof. Dr Nedim Uygun (Coccinellidae), Prof. Dr M. Rifat Ulusoy (Aphelinidae and Chrysopidae), and Prof. Dr Suat Kiyak (Anthocoridae, Lygaeidae, and Miridae).
Molecular identification of the B. tabaci complex
DNA lysis, polymerase chain reaction, and mtCOI sequences
Adults were removed from leaves of field-infested plants and placed in 95% alcohol. After their identification as B. tabaci, 3–4 adults from 68 field collections were homogenized in 30 ml lysis buffer containing 50 mM NaCl, 10 mM Tris–HCl, pH 8.0, 1 mM EDTA, and 0.5% Nonidet P-40. Total nucleic acids were incubated with Proteinase K at 56°C for 2 h, followed by boiling at 95°C for 10 min to inactivate Proteinase K. DNA lysis was stored at –20°C. Polymerase chain reaction (PCR) primers were synthesized in China or the U.S.A. by local, commercial laboratories, respectively (Frohlich et al. 1999): C1-J-2195 (5′-TTGATTT TTTGGTCATCCAGAAGT-3′) and L2-N-3014 (5′-TCCAATGCACTAATCTGC CATATT A-3′).
A fragment of the mtCOI sequence was amplified for three or more adult whiteflies per field collection, using PCR (Mullis and Faloona 1987). The PCR reaction (40 μl) contained 20 pM each primer, 0.25 mM dNTPs, 2.0 mM Mg2+, 1 μl DNA solution, 1.5 unit Taq polymerase (Master Taq, Eppendorf). PCR parameters were: predenaturation at 94°C for 5 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1.5 min, and extension at 72°C for 1.5 min, with a final extension at 72°C for 10 min. A 2-μl aliquot for each PCR product was separated by electrophoresis on a 1.0% agarose gel in Tris–acetate–EDTA buffer, pH 8.0, containing 10 mg ml-1 ethidium bromide (Sambrook et al. 1989). PCR products of the expected size (∼820–850 bp) were visualized under an ultraviolet light.
The DNA sequence was determined for each PCR product (n = using an ABI Model 377 DNA sequencer, available in the Genomics and Technology Center, The University of Arizona, Tucson, AZ, U.S.A.). The PCR product for each individual was sequenced bi-directionally using the same primers as were employed for mtCOI amplification. The mtCOI sequences (780 bases) were edited manually using a minimal overlap of 400 bases and a consensus sequence was obtained for each population.
Whitefly haplotype/biotype determination
Sequences (780 bases) were aligned and the percent nucleotide identity was estimated using the Clustal W algorithm (Thompson et al. 1994) available in the DNASTAR software (Lasergene, Madison, WI, U.S.A.). Reference mtCOI sequences were obtained from UA laboratory collection and/or from the GenBank database (Bedford et al. 1994; Frohlich et al. 1999; Brown 2000; Kirk et al. 2000; Viscarret et al. 2003; Berry et al. 2004), including the native ‘TC’ population from Turkey. Representative sequences have been deposited as accessions in the NCBI GenBank database. Nucleotide distances were calculated for aligned sequences using Clustal V (DNASTAR, Lasergene).
For tree construction, a single haplotype sequence was included for only selected populations because all field collections were identified as the same haplotype. This was considered appropriate in light of the extreme homogeneity of the field populations examined here (n = 68), and to illustrate their common affiliation with reference B. tabaci haplotypes. The outgroup (genus level) included in the alignment was the greenhouse whitefly Trialeurodes vaporariorum (Westwood) {AF342774}.
Results and discussion
Natural host range
Bemisia tabaci B biotype populations were collected from a total of 152 species from 43 plant families (Table 1). All 152 species were new records for Turkey. In a survey of uncultivated plants in the region, 69 species belonging to 20 plant families were utilized as reproductive hosts, and all hosts supported the complete life cycle of the B biotype. The Malvaceae harbored the greatest number of B. tabaci host species at 14, following by Leguminosae (13), Solanaceae (13), Compositae (10), and the Euphorbiaceae and Labiatae, at 9 each. Thus, the B biotype colonizes at least 152 species in 43 plant families in southern Turkey. Similarly, a great number of host species for the B. tabaci complex have been reported throughout the world, and it is known that the B biotype has an extensive host range (Brown et al. 1995). For example, 52 hosts of B. tabaci have been reported in Israel (Avidov and Harpaz 1969), 87 in Taiwan (Chang 1969), 172 in Egypt (Bedford et al. 1994), 115 in Sudan (Mound and Halsey 1978), and between 1988 and 1999 63 hosts were reported in Turkey for indigenous B. tabaci (Ulusoy 2001) which was prior to the introduction of the B biotype. Also, Mound and Halsey (1978) reported 315 hosts for B. tabaci, worldwide, and Cock (1993) listed more than 506 species, worldwide (does not discriminate biotypes or haplotypes).
Natural enemies
Five species of predators and two species parasitoids were associated with B. tabaci in southern Turkey (Table 2). The predators belonged to the Neuroptera: Chrysopidae, the Hemiptera: Anthocoridae, Lygaidae, and Miridae, the Coleoptera: Coccinellidae. Clitostethus arcuatus Rossi was the most common predator associated with B. tabaci examined in this study. The parasitoids belonged to the Hymenoptera: Aphelinidae. The most common parasitoids associated with the B. tabaci in southern Turkey were Eretmocerus mundus and Encarsia lutea. These results are in agreement with studies reported elsewhere showing that parasitic wasps, Eretmocerus and Encarsia, are among the most important natural enemies of B. tabaci (Gerling 1990; Kirk et al. 2000).
Molecular identification of B. tabaci biotype and displacement of the native TC population
The subset of B. tabaci collections analyzed from southern Turkey were identified as the ‘B’ biotype based on alignment (Clustal W) with well-studied reference haplotype or biotype sequences. The tree shown here resolves three major B. tabaci clades, representing the presumed extant geographical origin of represented haplotypes. The exception to this is the ‘B’ biotype, which occurs worldwide as a result of multiple, recent introductions, albeit its origin clearly is the Eastern Hemisphere (Costa et al. 1993; Brown 2000; Brown et al. 1995; Kirk et al. 2000).
The B. tabaci clades herein are delineated geographically as: (i) Americas and Caribbean (three sister clades), (ii) Mediterranean/North Africa/Middle East (two sister clades), (iii) Southeast Asia I, II, and Australia (three or more sister clades) (Fig. 1). Representatives from sub-Saharan Africa were not included because they are not relevant to the data set (see Brown 2000). The B biotype grouped within one sister clade within the large Mediterranean/North African clade, while the native Turkey TC reference population grouped with the second sister clade, which contains the Spanish Q biotype and its close relatives, e.g., haplotypes of uncharacterized Q- and TC-like populations from the region (Brown 2000).
The B biotype is not native to Turkey, but is a recent introduction (Brown 2001). A B. tabaci population collected from cotton in southern Turkey in 1985 and designated the TC (Bedford et al. 1994; Brown et al. 1995) is probably indigenous to the region. The TC/Q-like and B haplotype/biotype represent distinct lineages, or sister clades, within the large Mediterranean–North Africa/Middle East clade, at > 8% divergence (data not shown). No B. tabaci collections examined here were identified as either the local TC or the Q haplotype, the latter, which is known to be a pest/vector in vegetable crops in Spain/Sudan/Morocco (Fig. 1).
The percent nucleotide sequence divergence between the B biotype field collections from Turkey was low at ∼0–3% (data not shown), indicating there is minimal nucleotide variation between individuals examined in this study. Thus, the genetic composition of the B biotype collected in Turkey is highly homogeneous, as has been observed elsewhere for this invasive whitefly species (Frohlich et al. 1999; Brown 2000). Our results also suggest that the B biotype has displaced the indigenous TC haplotype (Fig. 1; Q and relatives sister clade) in southern Turkey, the latter, which was known to be present there in 1985. The TC halpotype was probably the predominant one in Turkey previously, and until 1999, when the B biotype was introduced there.
These results are consistent with the hypothesis that the B biotype is an exotic, introduced whitefly (Ulusoy et al. 2002) of extreme genetic heterogeneity (Brown 2000), which has many of the qualities characteristic to invasive species, including the ability to displace indigenous populations (Costa et al. 1993; Brown et al. 1995; Viscarret et al. 2003). The invasive B biotype is known to predominate upon its introduction into a number of locales, and to successfully establish in arid, irrigated agricultural areas (Brown et al. 1995; Brown 2000). The B biotype was first discovered in the Americas in approximately 1986–1987 (Costa and Brown 1991), and in other locations in the Americas and American Tropics, where it is known to have displaced the local B. tabaci haplotype(s) in many locations.
In contrast, the Q biotype, which is indigenous to southern Spain, has not been displaced by the introduction of the B biotype, and in fact has regained its status as the predominant biotype there (reviewed in Brown 2000). Whether the TC haplotype has survived the establishment of the B biotype in southern Turkey remains to be seen.
References
Avidov Z, Harpaz I (1969) Plant pests of Israel. Israel Universities Press, Jerusalem, pp 76–84
Bedford ID, Markham PG, Brown JK, Rosell RC (1994) Geminivirus transmission and biological characterization of whitefly (Bemisia tabaci) biotypes from different world regions. Ann Appl Biol 125:311–325
Berry SD, Fondong VN, Rey C, Rogan D, Fauquet CM, Brown JK (2004) Molecular evidence for five distinct Bemisia tabaci (Homoptera: Aleyrodidae) geographic haplotypes associated with cassava plants in sub-Saharan Africa. Ann Entomol Soc Am 97(4):852–859
Brown JK (1994) Current status of Bemisia tabaci as a pest and virus vector in world agroecosystems. FAO Plant Prot Bull 42:3–32
Brown JK (2000) Molecular markers for the identification and global tracking of whitefly vector–begomovirus complexes. Virus Res 71:233–260
Brown JK (2001) The molecular epidemiology of begomoviruses. In: Khan JA, Dykstra J (eds) Trends in plant virology. The Haworth Press Inc., New York, pp 279–316, 537 pp
Brown JK, Idris AM (2005) Genetic differentiation of the whitefly Bemisia tabaci (Genn.) mitochondria COI and geographic congruence with the coat protein of the plant virus genus: Begomovirus. Ann Entomol Soc Am 98:827–837
Brown JK, Frohlich D, Rosell R (1995) The sweetpotato/silverleaf whiteflies: biotypes of Bemisia tabaci (Genn.), or a species complex? Annu Rev Entomol 40:511–534
Brown JK, Perring TM, Cooper AD, Bedford ID, Markham PG (2000) Genetic analysis of Bemisia (Homoptera: Aleyrodidae) populations by isoelectric focusing electrophoresis. Biochem Genet 38:13–25
Chang YC (1969) Host plant and morphological variations of Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae) in Taiwan. Plant Prot Bull Taiwan 11:23–32
Cock MJW (ed) (1993) Bemisia tabaci—an update 1986–1992 on the cotton whitefly with an annotated bibliography. CAB International Institute of Biological Control, Ascot, UK, 78 pp
Costa HS, Brown JK (1991) Variation in biological characteristics and in esterase patterns among populations of Bemisia tabaci (Genn.) and the association of one population with silverleaf symptom development. Entomol Exp Appl 61:211–219
Costa HS, Brown JK, Sivasupramaniam S, Bird J (1993) Regional distribution, insecticide resistance, and reciprocal crosses between the ‘A’ and ‘B’ biotypes of Bemisia tabaci. Insect Sci Appl 14:127–138
De Barro PJ, Driver F, Trueman JWH, Curran J (2000) Phylogenetic relationships of world populations of Bemisia tabaci (Gennadius) using ribosomal ITS1. Mol Phylogenet Evol 16:29–36
Frohlich DR, Torres-Jerez I, Bedford ID, Markham PG, Brown JK (1999) A phylogeographic analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol 8:1593–1602
Gerling D (1990) Natural enemies of whiteflies: predators and parasitoids. In: D Gerling (ed) Whiteflies: their bionomics, pest status and management. Intercept Ltd, Andover, UK, pp 147–185
Gill RJ (1992) A review of the sweetpotato whitefly in California. Pan-Pac Entomol 68:144–152
Guirao P, Beitia F, Cenis JL (1997) Biotype determination of Spanish populations of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull Entomol Res 87:587–593
Kaygısız H (1976) Investigations on the morphological characters, biology, damage, host plants, distribution, and control of the cotton whitefly (Bemisia tabaci Genn. in the Mediterranean region of Turkey [in Turkish]. Tarım ve Orman Bakanlıı Zir. Müc. Ara. Ens. Müd. ve Zir. Kar. Gen. Müd. Adana Bölge Zir. Müc. Ara. Ens. Müd. Yayınları Aratırma Serisi No: 45, 53 pp
Kirk AA, Lacey LA, Brown JK, Ciomperlik MA, Goolsby JA, Vacek DC, Wendel LE, Napompeth B (2000) Variation within the Bemisia tabaci s.l. species complex (Hemiptera: Aleyrodidae) and its natural enemies leading to successful biological control of Bemisia biotype B in the USA. Bull Entomol Res 90:317–327
Mound LA, Halsey SH (1978) Whitefly on the World. A systematic catalogue of the Aleyrodidae (Homoptera) with host plant and natural enemy data. British Museum (Natural History) and Chichester. Wiley, London, UK, 340 pp
Mullis KR, Faloona FA (1987) Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol 155:335–350
Rosell RC, Bedford ID, Frohlich DR, Gill RJ, Brown JK, Markham PG (1997) Analysis of morphological variation in distinct populations of Bemisia tabaci (Homoptera: Aleyrodidae). Ann Entomol Soc Am 90:575–589
Ryckewaert P, Alauzet C (2002) The natural enemies of Bemisia argentifolii in Martinique. BioControl 47:115–126
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Schuster DJ, Mueller TF, Kring JB, Price JF (1990) Relationship of sweet potato whitefly to a new tomato fruit disorder in Florida. Hortic Sci 25:1618–1620
Summers CG, Elam P, Newton AS Jr (1995) Colonization of ornamental landscape plants by Bemisia argentifolii Bellows and Perring (Homoptera: Aleyrodidae). Pan-Pac Entomol 71:190–198
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment and sequencing weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Ulusoy MR (2001) Whitefly fauna of Turkey [in Turkish]. Baki Kitapevi Yayınları, Adana, 98 pp ISBN 975-7024-14-7
Ulusoy MR, Brown JK, Bayhan E (2002) The ‘B’ biotype of Bemisia tabaci now established in Turkey. European Studies Network on European Whiteflies Newsletter, May 2002, No. 13
Viscarret MM, Torres-Jerez I, Agostini de Manero E, López SN, Botto EE, Brown JK (2003) Mitochondrial DNA evidence for a distinct clade of New World Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae) from Argentina and Bolivia, and presence of the Old World B biotype in Argentina. Ann Entomol Soc Am 96:65–72
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Jürgen Gross
Rights and permissions
About this article
Cite this article
Bayhan, E., Ulusoy, M.R. & Brown, J.K. Host range, distribution, and natural enemies of Bemisia tabaci ‘B biotype’ (Hemiptera: Aleyrodidae) in Turkey. J Pest Sci 79, 233–240 (2006). https://doi.org/10.1007/s10340-006-0139-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-006-0139-4