Abstract
We studied avian haemosporidian parasites of the genera Plasmodium, Haemoproteus and Leucocytozoon in a riparian songbird community in Central California, USA, over a period of 2 years. We sequenced a well-characterized region of the mitochondrial cytochrome b gene to identify the prevalence and diversity of these parasites from 399 birds. Of the 39.8 % of birds infected with haemosporidian parasites, most (30.8 %) were infected with Plasmodium. We identified 35 lineages, including 13 from the Plasmodium genus, 12 from Haemoproteus, and 10 from Leucocytozoon, 14 of which were novel lineages. In addition, we provide the first report of haemosporidian infections in 13 host species. Plasmodium prevalence ranged widely among host species from 0.0 to 68.6 %. We identified 2 Plasmodium lineages that were generalists, infecting multiple species across several families. One Plasmodium species, P. homopolare, was found in 84 individual birds representing 9 host species from 5 families, but primarily from Emberizidae. This is the first avian haemosporidian study utilizing molecular methods in California, which increases our understanding of the diversity and prevalence of avian haemosporidia affecting Passeriformes in this region and beyond.
Zusammenfassung
Erste molekulare Studie zu Prävalenz und Diversität von Vogel-Hämosporidien einer Singvogelgemeinschaft in Zentral-Kalifornien
Wir untersuchten zwei Jahre lang Vogel-Hämosporidien der Gattungen Plasmodium, Haemoproteus und Leucocytozoon in einer Auwaldsingvogelgemeinschaft in Zentral-Kalifornien, USA. Mittels Sequenzierung einer gut charakterisierten Region des mitochondrialen Cytochrom-b-Gene haben wir die Prävalenz und Diversität dieser Malariaparasiten von 399 Vögeln ermittelt. Von den 39,8 % mit Hämosporidien infizierten Vögeln waren die meisten (30,8 %) mit Plasmodium befallen. Wir identifizierten 35 Linien, wovon 13 der Gattung Plasmodium angehörten, 12 der Gattung Haemoproteus und 10 der Gattung Leucocytozoon. 14 Linien waren bisher unbekannt. Erstmalig wurden 13 Arten auf ihren Hämosporidienbefall geprüft. Die Prävalenz mit Plasmodium betrug zwischen 0 und 68,6 %. Zwei der Plasmodium Linien waren Generalisten, die mehrere Vögel-Arten bzw. Familien infizierten. Einer dieser Generalisten, P. homopolare, wurde in 84 Individuen von neun Arten aus fünf Familien nachgewiesen, vornehmlich aber Emberizidae. Diese Studie ist die erste, die molekulare Methoden zum Nachweis von Hämosporidien in Vögeln in Kalifornien verwendete. Diese Ergebnisse erweitern unsere Kenntnis zu Diversität und Prävalenz von Vogel-Hämosporidiena bei Passeriformes in der Region und darüber hinaus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Historically, avian haemosporidian species of the genera Plasmodium, Haemoproteus and Leucocytozoon were identified based on morphological characteristics, as well as the hosts within which the parasites were found, often resulting in a “one-host–one-species” classification. More recently, molecular studies utilizing polymerase chain reaction (PCR) of the cytochrome b (cyt b) gene have revealed an astounding diversity of species (Ricklefs and Fallon 2002; Waldenström et al. 2002; Ricklefs et al. 2005; Clark et al. 2014), which are quite capable of host switching and infecting a wider range of host species than originally thought (Waldenström et al. 2002; Lauron et al. 2014; Ricklefs et al. 2014). Currently, there are approximately 250 morphologically described species of Plasmodium, Haemoproteus and Leucocytozoon (Valkiūnas et al. 2014), but recent molecular studies suggest there may be thousands of species within these three combined genera (Bensch et al. 2004; Outlaw and Ricklefs 2014).
These genera share a complex life cycle requiring a susceptible vertebrate host and a competent dipteran vector for reproduction (Garnham 1966; Valkiūnas 2005; Atkinson 2008). However, they differ in geographical and host range because they rely on different families of blood-feeding vectors for transmission (Valkiūnas 2005) and present varying degrees of prevalence and virulence among species and families of birds (Atkinson et al. 2001b; Beadell et al. 2004; Ricklefs et al. 2005; Valkiūnas 2005; Palinauskas et al. 2008; Zehtindjiev et al. 2008; Dimitrov et al. 2015). Avian haemosporidia have recently gained popularity as a model study system for the ecology and evolution of host–parasite relationships (Njabo et al. 2010; Knowles et al. 2011; Lachish et al. 2011; Loiseau et al. 2012; Carlson et al. 2013; Dodge et al. 2013). However, significant unresolved taxonomic issues limit the study of these complex multiple-parasite/multiple-host systems, including the absence of established species limits, unresolved intra-generic evolutionary relationships, and limited sampling of potential hosts and geographic ranges. Relatively few avian haemosporidian studies have been conducted in the western United States, and in particular California, where only a handful of microscopy studies have been conducted over the last three-quarters of a century (Wood and Wood 1937; Herms et al. 1939; Herman et al. 1954; Clark and Swinehart 1966; Super and van Riper 1995; Martinson et al. 2008). Here, we present the results of a descriptive, PCR-based study to quantify haemosporidian prevalence and diversity in a California riparian songbird community across all seasons over 2 years. Our study aims to increase the understanding of avian haemosporidian prevalence, diversity, evolutionary relationships, and host and geographical range. Our goals were to: (1) quantify the prevalence and diversity of haemosporidians at a highly biodiverse California study site; (2) identify variation in host-sharing among parasite lineages identified; (3) determine which parasites are locally transmitted; and (4) relate host–parasite associations in California to those of other geographic areas.
Methods
Study site
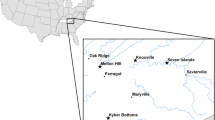
Sampling took place over 38 trapping days between April 2011 and January 2013, during the months of April, May, June, October and January. Trapping took place at China Creek Park, a 120-acre (48.5-ha) riparian habitat in the southern Central Valley of California, approximately 16 miles (25.75 km) east of Fresno (36°44′N, 119°29′W, 120 m above sea level). The site is located in the King’s River watershed of the Sierra Nevada mountain range, and includes a small stream and two ponds, as well as areas of oak grassland. China Creek Park was selected as a study site for researching parasite–vector–host questions related to avian malaria because of the known prevalence and diversity of mosquito species, and the suspected diversity of bird species, at this site. Results of the vector research, conducted by collaborators, is being published separately (Carlson et al. 2015).
Bird sampling
Birds were captured using mist nets. Upon capture, birds were identified to species, weighed, measured, and banded with a U.S. Fish and Wildlife Service numbered aluminum band. Birds were categorized as residents or migrants based on data from 2 websites managed by the Cornell Laboratory of Ornithology: All About Birds, which includes range maps, (http://www.allaboutbirds.org) and E-bird (http://ebird.org), which includes bird species sightings for Fresno County.
We obtained 25–50 µl of blood from each bird via brachial venipuncture, which was placed in lysis buffer (10 mM Tris–HCL pH 8.0, 100 mM EDTA, 2 % SDS) and stored at ambient temperature while in the field, and then preserved at −80 °C in the laboratory until further processing.
DNA extraction, PCR amplification, and sequencing
Parasite DNA was extracted from whole blood stored in lysis buffer following animal tissue protocols recommended for the Wizard SV Genomic DNA Purification kits (Promega, Madison, WI, USA). Success of each DNA extraction was verified with primers that amplify the brain-derived neurotrophic factor (BDNF) (Sehgal and Lovette 2003). Plasmodium and Haemoproteus spp. were detected by nested PCR protocols that amplify sections of the mitochondrial cyt b gene. The primers used for the first and second reactions were HaemNF/HaemNR2 and HaemF/HaemR2, respectively, and have been described previously (Waldenström et al. 2004; Bensch et al. 2009). Presence of Leucocytozoon spp. was determined by a nested PCR following the protocol described by Hellgren et al. (2004) with two modifications: the annealing temperature was adjusted to 54.5 °C and reactions were performed in Accupower® PCR Pre-Mix (Bioneer, Alameda, CA,, USA). All PCR reactions were carried out in 25 or 20 µl (in the case of Accupower®) reactions and were accompanied by negative (ddH2O) and positive controls (samples from infected birds previously confirmed by sequencing and microscopy) to control for any contamination and to confirm success of the PCR. The resulting PCR products were visualized on 1.8 % agarose gels to check for positive infections.
We purified PCR product from all positive samples and performed bi-directional sequencing as outlined in Jasper et al. (2014). Sequences were edited and aligned using Sequencher 4.8 (GeneCodes, Ann Arbor, MI, USA). We then identified sequences to genus by identifying their closest sequence matches in GenBank via the National Center for Biotechnology Information (NCBI) nucleotide BLAST search. Novel lineages were defined as a lineages that differed by 1 or more nucleotides (nt) from any lineage deposited in GenBank prior to this study. The 1-nt difference is significant because it has been established that “mtDNA lineages of parasites differing by as little as one nucleotide in the cyt b gene frequently exhibit distinctly different areas of transmission and range of host species” (Bensch et al. 2009), and because several studies have shown differences in virulence and vector competence between avian haemosporidian lineages differing by only 1 nt (Palinauskas et al. 2015; Žiegyte and Valkiunas 2014). Novel lineages found in a single individual (as opposed to multiple individuals) were confirmed by 2 replicate PCRs to ensure that the 1-nt difference(s) was not due to PCR error. All sequences were deposited in GenBank™ (Accession numbers KJ482708; KJ584585–KJ584606; KJ620777–KJ620788).
Phylogenetic analysis
Sequences were edited using Sequencher 4.8 (GeneCodes) and aligned using SEAVIEW software (Galtier et al. 1996). The appropriate model of sequence evolution was determined by the software MrModeltest (Nylander et al. 2004) to be GTR+Γ+I for species of Plasmodium and Haemoproteus, and GTR+Γ for species of Leucocytozoon. Maximum likelihood (ML) bootstrap analyses were performed using a heuristic search consisting of 100 replicates and the starting tree as a neighbor-joining tree. ML trees for Plasmodium and Haemoproteus were generated using RAxML (Stamatakis 2006), under the GTR+Γ+I model. A ML tree for Leucocytozoon was generated using RAxML under the GTR+Γ model. In all cases, a thorough ML search was performed along with 1000 rapid bootstrap inferences.
We based our ML phylogenetic analysis on 13 Plasmodium spp. lineages isolated from birds sampled at China Creek, as well as 11 reference sequences downloaded from GenBank. The final alignment included 447 nucleotides of the cyt b gene. Haemoproteus majoris served as outgroup. The Haemoproteus phylogeny included 12 lineages found at China Creek and 10 reference sequences downloaded from GenBank. The final alignment included 439 nucleotides of the cyt b gene. Leucocytozoon schoutedeni served as outgroup. The final alignment for the lineages in the Leucocytozoon genus was 449 nucleotides long and included 10 lineages of the cyt b gene from China Creek and 5 reference sequences from GenBank. Plasmodium relictum served as outgroup. Our analyses included only reference sequences from GenBank that have well-established positive morphological identifications (Valkiūnas et al. 2014).
Phylogenetic reconstruction was also done using Bayesian Inference in MrBayes v.3.1.2 (Ronquist and Huelsenbeck 2003) using the appropriate models of sequence evolution described above. Two Markov Chain Monte Carlo (MCMC) simulations were run simultaneously for 10 million generations with sampling every 200 generations, generating 50,000 trees. The first 12,500 trees were discarded from the sample as the “burn-in” period, accounting for 25 % of the trees. The remaining 37,500 trees were used to construct a majority rule consensus tree and to calculate the posterior probabilities of the individual clades. Because phylogenetic reconstructions using Bayesian Inference were consistent with that of ML, only the ML trees are presented here.
Lineages were given names in accordance with the proposed lineage naming criteria discussed by participants at the 2013 Malaria and Related Haemosporidian Parasites of Wildlife Research Coordination Network meeting, Vilnius, Lithuania. Lineage names reflect a code for the host from which the parasite was first obtained (we used the Institute for Bird Populations’ standardized 4-letter alpha code by English name), the locality code (CA for California, in our case), the initials of the person that collected the sample, and a unique assigned code for the parasite lineage that would also reflect to which genus the parasite belongs (i.e., P for Plasmodium).
Genetic distance was calculated in PAUP using the uncorrected-p model of sequence evolution per Mr. Modeltest.
Statistical analysis
For the prevalence and parasitemia values, 95 % Bayesian credible intervals were calculated. Credible intervals were calculated using the inverse of the cumulative distribution function of the beta distribution in R (i.e., qbeta) (Bolker 2008) where the shape 1 parameter was specified as m + 1 and the shape 2 parameter was specified as n − m + 1, where m was the number of positive samples and n was the total sample size. Prevalence data (% of infected individuals in sample) throughout the rest of this paper are followed by a 95 % Bayesian credible interval in parentheses, which indicates a 95 % probability that the actual prevalence falls within the Bayesian credible interval. Fisher’s exact test was used to compare numbers of infected and uninfected individuals in different samples.
We defined our minimum sample size, n, for analysis as 20 based on the work of Jovani and Tella (2006), which showed that the standard error rapidly decreases as the sample size reaches 10–20 individuals, but does not decrease much further with increased sample sizes.
Results
Parasite prevalence
We captured 399 birds representing 46 species and 16 families. Twenty-three of those individuals were recaptured between 1 and 5 times during the study, and were only sampled if a minimum of 3 days had passed since their most recent capture. Because individuals are known to gain, as well as lose, infections over their lifetimes (Valkiūnas 2005), data from recaptured individuals were included in our analysis. Overall, 159 of 399, or 39.8 % (35.1–44.6 %) of captures were infected with Plasmodium, Haemoproteus, Leucocytozoon, or some combination of these genera. Plasmodium infections were found in 123 birds, or 30.8 %, (26.4–35.4 %). Only 22 birds, or 5.5 % (3.7–8.2 %), were infected with Haemoproteus, and 21 birds, or 5.3 % (3.5–7.9 %), were infected with Leucocytozoon (Table 1). Seven birds were found to harbor inter-generic (e.g., Plasmodium–Leucocytozoon) co-infections based on PCR positives; 2 individuals infected with Plasmodium displayed sequences with double-peaks, suggesting intra-generic co-infections.
Prevalence and 95 % Bayesian Credible Intervals were quantified for 7 host species for which we had a sample size of 20 or greater. Plasmodium prevalence ranged from 0.0 % (0.1–10.9 %) in Bushtits (Psaltriparus minimus) (n = 31) to 68.6 % (54.9–79.7 %) in Song Sparrows (Melospiza melodia) (n = 51). Of these 7 host species, only Lincoln’s Sparrows (Melospiza lincolnii) (n = 22) were infected with Haemoproteus (4.5 %; 1.1–21.9 %). Only House Wrens (Troglodytes aedes) (n = 22) and Lincoln’s Sparrows were infected with Leucocytozoon. The prevalence in House Wrens was 4.5 % (1.1–21.9 %), and in Lincoln’s Sparrows was 13.6 % (5.0–33.6 %). Because the sample sizes in these 7 species ranged from 20 to 68, we tested the strength of the correlation between sample size and Plasmodium prevalence and found that sample size was a moderate predictor of prevalence, with R 2 = 0.6064.
We sampled 165 migratory, and 234 resident, individuals. Plasmodium was significantly more prevalent in resident species (44.4 %; 38.2–50.9 %) compared to migrants (11.5 %; 7.5–17.3 %) (P < 0.001). In contrast, Haemoproteus was significantly more prevalent (P < 0.05) in migrants (12.1 %; 8.0–18.0 %) compared to residents (0.9 %; 0.3–3.0 %), as was Leucocytozoon: 10.9 % (7.0–16.6 %) for migrants and 1.3 % (0.5–3.7 %) for residents (P < 0.05).
Parasite diversity
We found 13 lineages of Plasmodium at China Creek infecting 18 bird species from 10 families. Two of the Plasmodium lineages appear to be generalists (Table 2), which we define as a lineage infecting multiple species across several families. One of these two generalists, recently described as P. homopolare (Walther et al. 2014), infected 84 birds representing 9 host species from 5 families. Four of the 9 species, and over 80 % of the individuals infected with this parasite, were in the Emberizidae family. Of the 31 % of birds at China Creek infected with Plasmodium, 68 % were infected with P. homopolare; in other words, 1 in 5 birds at China Creek was infected with P. homopolare. The other generalist, Plasmodium cathemerium (designated as lineage SPTO_CA_ELW_6P), was found in 6 bird species from 3 families. By querying the MalAvi database, we identified 4 host species not previously found to be infected by Plasmodium: Lincoln’s Sparrow, Brown-headed Cowbird (Molothrus ater), Lazuli Bunting (Passerina amoena) and Black-headed Grosbeak (Pheucticus melanocephalus). Two Plasmodium lineages found at China Creek, CLSW_CA_ELW_1P and HOWR_CA_ELW_2P, differed by 1 and 6 nt, respectively, from any lineage in GenBank, and could be new lineages or species.
Twelve lineages of Haemoproteus at China Creek affected 10 host species from 7 families. Sample sizes for Haemoproteus lineages ranged from 1 to 4 and generally infected 1 to 3 host species from 1 or 2 families; however, the small sample sizes of individual Haemoproteus lineages observed prevent us from inferring host range for these lineages. By querying the MalAvi database, we identified 3 host species not previously found to be infected by Haemoproteus: MacGillivray’s Warbler (Geothlypis tolmiei), Brown-headed Cowbird, and Black-headed Grosbeak. Seven of the 12 lineages were novel and differed by at least 1 nt from any lineage in GenBank.
Ten Leucocytozoon lineages, with sample sizes ranging from 1 to 5, were found in 15 host species from 10 families. While seven Leucocytozoon lineages were found in only 1–2 individuals, the small sample sizes of individual Leucocytozoon lineages prevent us from inferring host range for these lineages. Three lineages infected 2 or more species from 2 or more families. A Leucocytozoon lineage sampled from 5 individuals infected 5 host species from 5 host families, suggesting this genera includes generalists, a characteristic more often associated with Plasmodium than with Leucocytozoon. By querying the MalAvi database, we identified 6 host species not previously found to be infected by Leucocytozoon: Western Scrub Jay (Aphelocoma californica), House Finch (Haemorhous mexicanus), Lincoln’s Sparrow, Black-headed Grosbeak, Ruby-crowned Kinglet (Regulus calendula), and Warbling Vireo (Vireo gilvus). Seven of the 10 lineages differed by at least 1 nt from any lineage in GenBank.
No host species, except the Warbling Vireo, was observed to be parasitized by more than 3 lineages of parasite within any of the haemosporidia genera. Warbling Vireos were infected with 5 Haemoproteus lineages ranging in genetic distance from one another from 0.2 to 3.9 %, and with a mean of 2.7 %. Nine of 11 (82 %) Warbling Vireos sampled were infected with Haemoproteus; this represents 40.9 % (9 of 22) of Haemoproteus infections observed in our sample. Only one bird species for which we had a sample size greater than 20, the Common Bushtit (Psaltriparus minimus; n = 31), was observed to not be infected by any of the 3 haemosporidia genera.
A significantly greater diversity of Haemoproteus and Leucocytozoon parasites was found in migrants compared to residents (P < 0.05). Eleven distinct lineages of Haemoproteus were found in migrants (n = 165), compared to 1 lineage in residents (n = 234). The single Haemoproteus lineage found in a resident, isolated from a single House Finch, was not found in any migrant bird species. Nine distinct lineages of Leucocytozoon were found in migrants, compared to 2 lineages in residents. One Leucocytozoon lineage was found in both migratory and resident species. Migrants and residents had an equally diverse assemblage of Plasmodium parasites, with 8 lineages found in each group, and 3 lineages shared between migrants and residents.
Phylogenetic analysis
Phylogenetic relationships of Plasmodium, Haemoproteus and Leucocytozoon parasite lineages found at China Creek are shown in Figs. 1, 2 and 3. Although our phylogenetic trees are well supported only at the outer nodes, they provide insight into the diversity of lineages found in California and their relationships to known morphospecies of the same genus.
Locally-transmitted parasites
Based on data from concurrent isolations from mosquitoes captured at China Creek, we found that 7 Plasmodium lineages were transmitted locally based on their presence in the local mosquito population (Carlson et al. 2015). One additional Plasmodium lineage (LISP-SOSP_CA_ELW_9P) was identified in hatch year (HY) birds. Because HY birds have not yet migrated from their breeding grounds, we know that they were infected locally at China Creek. We also found 3 lineages of Haemoproteus (FOSP_CA_ELW_3H; HOFI_CA_ELW_4H; LISP_CA_ELW_5H) and 2 of Leucocytozoon (FOSP_CA_ELW_10L; LISP_CA_ELW_9L) that were locally transmitted, based on their presence in HY birds. Although blackflies (Diptera: Simuliidae) were collected concurrently at China Creek, none were found to be infected with Leucocytozoon (Carlson, personal communication).
Discussion
Parasite prevalence
Relative prevalences of Plasmodium, Haemoproteus and Leucocytozoon differed at China Creek from the majority of previous California studies of similar sampling effort. Plasmodium (30.5 %) was much more prevalent than Haemoproteus (5.8 %) and Leucocytozoon (5.3 %) at China Creek, in contrast to previous California studies, which reported prevalences of Plasmodium ranging from less than 4.0 to 11.9 % (Herman et al. 1954; Clark and Swinehart 1966; Super and van Riper 1995; Martinson et al. 2008). One likely reason for this is that previous California studies relied solely on microscopy and sometimes from a single blood smear prepared from each bird examined. Numerous papers have compared the use of microscopy versus PCR in prevalence studies (Jarvi et al. 2002; Richard et al. 2002; Fallon et al. 2003; Hellgren et al. 2004; Ricklefs et al. 2005; Valkiūnas et al. 2006; Fallon and Ricklefs 2008; Garamszegi 2010; Valkiūnas et al. 2014). Most researchers can agree that, while comparable prevalence results can be obtained for microscopy and PCR for all three genera, successful identification of haemosporidians by microscopy is highly dependent on the quality of blood smears and the skill and experience of the observer (Valkiūnas et al. 2008; Valkiūnas et al. 2014).
Avian haemosporidian infection dynamics include a brief pre-patent period in which the parasites are found only in the host tissues, followed by a patent stage that begins with a short acute infection lasting on the order of weeks and characterized by high parasitemia, followed by an indefinite period of chronic infection characterized by low parasitemia (Garnham 1966; Valkiūnas 2005; Atkinson, 2008). Researchers are most likely to trap a bird during the chronic infection because this period is relatively long and the bird is more likely to be active than during the acute infection. In fact, the acute stage of Plasmodium is relatively short, and the parasitemia level during chronic infections relatively low, compared to other haemosporidians (Greiner et al. 1975; Valkiūnas 2005), making morphological detection especially challenging. In addition, some morphological studies from California did not indicate the time of year during which sampling occurred (Clark and Swinehart 1966; Super and van Riper 1995). This is significant because parasitemias can be light or absent during early spring and/or late fall migrations; in fact, Plasmodium spp. are unlikely to be found in hosts in early spring and late fall in the Holarctic region using microscopic examination of blood films (Valkiūnas 2005). Other possible reasons why our study detected a much higher prevalence of Plasmodium than previous California studies include the differences in geographical sampling area, host community structure, and vector communities at different study sites. It must be noted that the China Creek site is located among several ponds, which serve as breeding grounds for mosquitoes (Carlson et al. 2015), and this may account for the high prevalence of Plasmodium.
The high degree of variability (0.0–68.6 %) in Plasmodium prevalence among resident China Creek bird species may reflect differential susceptibilities among avian families and possibly species, which has been shown in previous studies (Valkiūnas 2005). Interestingly, Lincoln’s Sparrows had a relatively low prevalence of 13.6 %, and are in the same family as Song Sparrows and Spotted Towhees, which had much higher prevalences of 68.6 and 66.7 %, respectively. Lower prevalence in infection in the Lincoln’s Sparrow may be due to the fact that the species is a wintering migrant in China Creek, whereas Song Sparrow and Spotted Towhee are year-round residents and more likely exposed to local spring and summertime vector transmission of, particularly, P. homopolare, which was the most prevalent Plasmodium species in the China Creek songbird community. Of all Plasmodium infections in Song Sparrows and Spotted Towhees, 82.9 and 83.3 %, respectively, were P. homopolare.
One species, the Common Bushtit (n = 31), was not infected with any of the 3 genera of haemosporidia addressed in this study. Given that 58 % (18/31) of Common Bushtits were sampled during late spring and early summer, when mosquito abundance is highest, we suspect our negative finding indicates this host species has low susceptibility to Plasmodium infections. Haemoproteus and Leucocytozoon infections were relatively rare at China Creek, so these parasite genera in infected individuals may have been missed by random chance. Previous California studies found Haemoproteus in Common Bushtits (Martinson et al. 2008; Super and van Riper 1995), suggesting the limited sample size or lack of competent vector at China Creek, rather than an innate or acquired immunity, are likely reasons we did not observe Haemoproteus in this species. As for the lack of observed Plasmodium infections in this species, it is possible that PCR failed to detect an infected Common Bushtit because PCR only detects haemosporidian infections present in the peripheral blood. Because chronic infections can persist in individuals through periods in which the parasites recede into tissues, both PCR and microscopy can fail to detect infected hosts. For that reason, some researchers recommend antibody screening, which can detect chronic infections that were once present in the blood (Atkinson et al. 2001a, b; Jarvi et al. 2002). However, a query of the MalAvi database (Bensch et al. 2009) showed no record of infection of this host species by Plasmodium, Haemoproteus or Leucocytozoon.
We found that Plasmodium was significantly more prevalent in resident birds than in migrants, but that the opposite was true for Haemoproteus and Leucocytozoon. One reason could be that migratory birds infected with Plasmodium lacked the fitness required to reach China Creek; however, this is difficult to gauge as studies have shown both high variability in the effect of Plasmodium species on individual hosts (Zehtindjiev et al. 2008; Cornelius et al. 2014) as well as differences among haemosporidian genera in terms of their development in, and effects on, hosts (Valkiūnas 2005). The majority of migratory species sampled in our study are present at China Creek in late spring and summer, at the height of vector abundance. A combination of vector competence (Carlson et al. 2015) and parasite diversity probably account for local transmission effects.
Parasite diversity
As expected, for reasons explained earlier in this Discussion, parasite diversity at China Creek differed from that identified in earlier California studies of similar effort and scope (i.e., at least 300 passerines and near-passerines sampled), particularly for Plasmodium. We found 13 lineages of Plasmodium, 12 of Haemoproteus and 10 of Leucocytozoon, whereas Herman et al. (1954), who only studied Plasmodium, identified 8 lineages; Super and van Riper (1995) identified 1 Plasmodium lineage, 10 Haemoproteus lineages, and 7 Leucocytozoon lineages; and, Martinson et al. (2008) identified 2 Plasmodium lineages, 5 Haemoproteus lineages and 2 Leucocytozoon lineages. Diversity of Plasmodium parasites found at China Creek was comparable to a 4-year study in Missouri, USA, which utilized PCR to identify 8 lineages of Plasmodium and 12 of Haemoproteus, but found no Leucocytozoon (Ricklefs et al. 2005). Some PCR-based, multi-year community-level studies have been published from the Neotropics and Asia (Ishtiaq et al. 2007; Svensson-Coelho et al. 2013; Fecchio et al. 2013); however, comparisons are difficult because the prevalence and diversity of the three haemosporidian genera vary geographically and by host family (Valkiūnas 2005).
The 2 most common lineages found at China Creek were of the genus Plasmodium and were generalists that infected multiple host species across multiple families. P. homopolare (Walther et al. 2014) infected 84 individuals representing 9 different species from 5 different families. The second most abundant lineage, Plasmodium cathemerium (SPTO_CA_ELW_6P), infected 18 individuals representing 6 species from 3 families. This is in contrast to a study in which the 2 most common lineages (one each from Plasmodium and Haemoproteus) in a passerine bird community were each nearly restricted to a single host species (Ricklefs et al. 2005). However, the authors point out that these lineages are exceptions within their dataset and that increased sampling of individuals within a host species positively correlates with the number of infections of the lineage sampled. With the exception of the 2 generalist Plasmodium lineages and 2 generalist Leucocytozoon lineage found at China Creek, all other lineages from our study site infected 1–3 host species representing 1 to 2 families. Had we increased our sampling effort and identified additional infections from these lineages, we may have seen less host-specificity at the species level.
Because our study utilized only PCR, and not microscopy, there are several caveats associated with this methodology that could bias our lineage diversity results. First, the development of Plasmodium spp. and related haemosporidians can be abortive in resistant or partly resistant hosts, resulting in no development of gametocytes (Valkiūnas et al. 2011; Cannell et al. 2013). This can happen when (1) sporozoites are inoculated by a vector into the blood but are incapable of cellular invasion, so remain in the circulation for some time, or (2) remnants of tissue meronts, the development of which was aborted before the gametocyte stage, are produced and remain in hosts, serving as a PCR template. In both cases, the bird would be scored as infected by PCR when, in fact, it might not be a competent host of the parasite. Therefore, conclusions regarding the full range of avian hosts would require microscopic examination of blood films in addition to comparison of molecular results. Second, PCR masks co-infections, which are common in wild birds, and the presence of double peaks on sequence chromatograms cannot be relied upon to indicate the presence of co-infections (Pérez-Tris and Bensch 2005). In such cases, secondary light infections can be preferentially amplified (Valkiūnas et al. 2006), which can only be confirmed via microscopy. This means that certain very common lineages that occur with undetected (by PCR) co-infections could be the result of preferential PCR amplification and may reflect high detection, rather than high prevalence. Conversely, apparently uncommon lineages that occur with co-infections could be higher than detected in our study. The use of microscopy and the development of primers that can amplify specific lineages would shed light on this issue.
Fourteen lineages across the 3 avian haemosporidian genera differed by at least 1 nt from any lineage in GenBank, and could be new lineages or species. Currently, there is no single definition of a species in terms of genetic difference for haemosporidian parasites (Perkins 2014). Ideally, studies of wild birds utilize PCR and microscopy, in order to maximize detection of parasites and identification to morphospecies. However, when morphological analysis is lacking, nt differences of as little as 1 base pair have been used to delineate lineages (Bensch et al. 2000, 2004; Hellgren 2005) and have been found to have important impacts, for example, on virulence (Palinauskas et al. 2015) and vector competence (Žiegytė and Valkiūnas 2014).
The Haemoproteus lineage diversity seen in Warbling vireos (5 lineages in 9 infected individuals) was unique to this species. Based on the close phylogenetic groupings of the lineages found in Warbling Vireos (Fig. 2), their relatively small genetic distances, and their presence in a common host species, it seems likely that these 5 parasite lineages represent polymorphisms of 1 or 2 Haemoproteus species. The other 4 Haemoproteus lineages found at China Creek differ from Pa. vireonis by 3 to 10 nt. Because these lineages infected only Warbling Vireos, and no other host species in our sample, it is tempting to infer that these lineages specialize on Vireonidae. In fact, one Haemoproteus lineage affecting Warbling Vireos in our sample is a 100 % match to the morphospecies Parahaemoproteus vireonis, found in the Red-eyed Vireo (Vireo olivaceus) in Colombia (Gonzalez et al. 2015); however, it is also a 100 % match to a Haemoproteus species found in the Blue-backed Manaquin (Chiroxiphia pareola) in Peru, a species in the Pipridae family (Parahaemoproteus is one of two sub-genera of Haemoproteus; Bennett et al. 1965).
We expected to find a higher diversity of parasites in migratory birds than in resident birds at our study site, since migratory birds encounter parasites in multiple ecosystems each year, whereas residents only encounter parasites in one ecosystem (Møller and Erriyzøe 1998), and possibly parasites carried by migrants. We found this to be the case for Haemoproteus and Leucocytozoon, but not Plasmodium, infections. Competent vectors for Haemoproteus and Leucocytozoon may be lacking at China Creek, resulting in a limited number of resident birds infected with these genera.
Phylogenetic analysis
A high diversity of haemosporidian parasites is shown in our trees (Figs. 1, 2, 3).
In addition, the figures allow us to infer how poorly parasite and host family relationships phylogenies correspond. In Fig. 1, for example, several parasite lineages infecting Cliff Swallows are paraphyletic, as are 2 lineages infecting House Wrens. Similar patterns can be seen in Figs. 2 and 3. This has also been shown in previous work, and likely reflects these parasites’ capacity for host-switching (Ricklefs and Fallon 2002; Ricklefs et al. 2004, 2014; Lauron et al. 2014).
Overall, this study reveals high prevalence and diversity of Plasmodium lineages, and high diversity of Haemoproteus and Leucocytozoon lineages, in California riparian songbirds. By examining a wide range of host species, we were able to report on the parasite–host community structure at China Creek Park, including the prevalence and diversity of lineages across host species. Many of the lineages identified have not been reported previously in California and/or in certain of the host species in our study. These new data should contribute to resolution of the taxonomic relationships of avian haemosporidia, as well as their relationships to hosts and geographical range, and provide a basis for further evolutionary and ecological study of avian haemosporidian in California riparian songbird communities.
References
Atkinson CT (2008) Haemoproteus, Avian malaria. In: Atkinson CT, Thomas NJ, Hunter DB (eds) Parasitic diseases of wild birds. Wiley, Iowa, pp 13–53
Atkinson CT, Dusek RJ, Lease JK (2001a) Serological responses and immunity to superinfection with avian malaria in experimentally-infected Hawaii Amakihi. J Wildl Dis 37:20–27
Atkinson CT, Lease JK, Drake BM, Shema NP (2001b) Pathogenicity, serological responses, and diagnosis of experimental and natural malarial infection in native Hawaiian thrushes. Condor 103:209–218
Beadell JS, Gering E, Austin J, Dumbacher JP, Peirce MA, Pratt TK, Atkinson CT, Fleischer RC (2004) Prevalence and differential host-specificity of two avian blood parasite genera in the Australo–Papuan region. Mol Ecol 3:3829–3844
Bennett GF, Garnham PCC, Fallis AM (1965) On the status of the genera Leucocytozoon Siemann, 1898 and Haemoproteus Kruse, 1890 (Haemosporidia: Leucocytozoidae and Haemoproteidae). Ibidem 43:927–932
Bensch S, Stjernman M, Hasselquist D et al (2000) Host specificity in avian blood parasites: a study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc R Soc Lond B 267:1583–1589
Bensch S, Perez-Tris J, Waldenstrom J, Hellgren D (2004) Linkage between nuclear and mitochondrial DNA sequences in avian malaria parasites: multiple cases of cryptic speciation. Evolution 58:1617–1621
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol 9:1353–1358
Bolker BM (2008) Ecological models and data in R. Princeton University Press, Princeton
Cannell BL, Krasnec KV, Campbell K, Jones HI, Miller RD, Stephens N (2013) The pathology and pathogenicity of a novel Haemoproteus spp. infection in wild little penguins (Eudyptula minor). Vet Parasitol 197(1–2):74–84. doi:10.1016/j.vetpar.2013.04.025
Carlson JS, Martinez-Gomez JE, Valkiūnas G, Loiseau C, Bell DA, Sehgal R (2013) Diversity and phylogenetic relationships of haemosporidian parasites in birds of Socorro Island, Mexico, and their role in the re-introduction of the Socorro Dove (Zenaida graysoni). J Parasitol 99:270–276
Carlson JS, Walther EL, Trout Fryxell R, Staley S, Tell LA, Seghal RNM, Barker CM, Cornel AJ (2015) Identifying avian malaria vectors: sampling methods influence outcomes. Parasite Vector 8(1):1–16
Clark GW, Swinehart B (1966) Blood protozoa of passerine birds of the Sacramento (Calif.) region. Bull Wildl Dis Assoc 2:53–54
Clark NJ, Clegg SM, Lima MR (2014) A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): new insights from molecular data. Int J Parasitol 44(5):329–338
Cornelius EA, Davis AK, Altizer SA (2014) How important are haemoparasites to migratory songbirds? Evaluating physiological measures and infection status in three neotropical migrants during stopover. Physiol Biochem Zool 87:719–728
Dimitrov D, Palinauskas V, Iezhova TA, Bernotienė R, Ilgūnas M, Bukauskaitė D, Zehtindjiev P et al (2015) Plasmodium spp.: an experimental study on vertebrate host susceptibility to avian malaria. Exp Parasitol 148:1–16
Dodge M, Guers SL, Sekercioğlu C, Sehgal RNM (2013) North American transmission of haemosporidian parasites in the Swainson’s Thrush (Catharus ustulatus), a migratory songbird. J Parasitol 99:548–553
Fallon SM, Ricklefs RE (2008) Parasitemia in PCR-detected Plasmodium and Haemoproteus infection in birds. J Avian Biol 39:514–522
Fallon SM, Bermingham E, Ricklefs RE (2003) Island and taxon effects in parasitism revisited: avian malaria in the Less Antilles. Evolution 57:606–615
Fecchio A, Lima MR, Svensson-Coelho M, Marini MA, Ricklefs RE (2013) Structure and organization of an avian haemosporidian assemblage in a Neotropical savannah in Brazil. Parasitology 140:181–192
Galtier N, Gouy M, Gautier C (1996) SEAVIEWandPHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12:543–548
Garamszegi LZ (2010) The sensitivity of microscopy and PCR-based detection methods affecting estimates of prevalence of blood parasites in birds. J Parasitol 96(6):1197–1203
Garnham PCC (1966) Malaria parasites and other Haemosporidia. Blackwell, Oxford
Gonzalez AD, Lotta IA, Garcia LF, Moncada LI, Matta NE (2015) Avian haemosporidians from Neotropical highlands: Evidence from morphological and molecular data. Parasitol Int 64(4):48–59
Greiner EC, Bennett GF, White EM, Coombs RF (1975) Distribution of the avian hematozoa of North America. Can J Zool 53:1762–178
Hellgren O (2005) The occurrence of haemosporidian parasites in the Fennoscandian bluethroat (Luscinia svecica) population. J Ornithol 146:55–60
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. J Parasitol 90:797–802
Herman CM, Reeves WC, McClure HE, French EM, Hammon W (1954) Studies on avian malaria in vectors and hosts of encephalitis in Kern County, CA: infections in avian hosts. Am J Trop Med Hyg 3:676–695
Herms WB, Kadner CG, Galindo P, Armstrong DF (1939) Blood parasites of California birds. J Parasitol 25(6):511–512
Ishtiaq F, Gering E, Rappole JH, Rahmani AR, Jhala YV, Dove CJ, Milensky C, Olson SL, Peirce MA, Fleischer RC (2007) Prevalence and diversity of avian hematozoan parasites in Asia: a regional survey. J Wild Dis 43(3):382–398
Jarvi SI, Schultz JJ, Atkinson CT (2002) PCR diagnostics underestimate the prevalence of avian malaria (Plasmodium relictum) in experimentally-infected passerines. J Parasitol 88:153–188
Jasper WC, Linksvayer TA, Atallah J, Friedman D, Chiu JC, Johnson BR (2014) Large scale coding sequence change underlies the evolution of post-developmental novelty in honey bees. Mol Biol Evol 10.1093/molbev/msu292
Jovani R, Tella JL (2006) Parasite prevalence and sample size: misconceptions and solutions. Trends Parasitol 22:214–218
Knowles SCL, Wood MJ, Alves R, Wilkin TA, Bensch S, Sheldon BC (2011) Molecular epidemiology of malaria prevalence and parasitemias in a wild bird population. Mol Ecol 20:1062–1076
Lachish S, Knowles SCL, Alves R, Wood MJ, Sheldon BC (2011) Infection dynamics of endemic malaria in a wild bird population: parasite species-dependent drivers of spatial and temporal variation in transmission rates. J Anim Ecol 80:1207–1216
Lauron EJ, Loiseau C, Bowie RCK, Spicer G, Smith TB, Melo M, Sehgal RNM (2014) Coevolutionary patterns and diversification of avian malaria parasites in African sunbirds (Family Nectariniidae). Parasitology. doi:10.1017/S0031182014001681
Loiseau C, Harrigan RJ, Cornel AJ, Guers SL, Dodge M, Marzec T, Carlson JS, Seppi B, Sehgal RNM (2012) First evidence and predictions of Plasmodium transmission in Alaskan bird populations. PLoS ONE 7:e44729
Martinson ES, Blumberg BJ, Eisen RJ, Schall JJ (2008) Avian haemosporidian parasites from northern California oak woodland and chaparral habitats. J Wildl Dis 44:260–268
Møller AP, Erriyzøe J (1998) Host immune defence and migration in birds. Evol Ecol 12:945–953
Njabo KY, Cornel AJ, Bonneaud C, Toffelmier E, Sehgal RNM, Valkiūnas G, Russell AF, Smith TB (2010) Nonspecific patterns of vector, host and avian malaria parasite associations in a central African rainforest. Mol Ecol 20:1049–1060
Nylander JAA, Ronquist JP, Huelsenbeck JP, Nieves-Aldrey JL (2004) Bayesian phylogenetic analysis of combined data. Syst Biol 53:47–67
Outlaw DC, Ricklefs RE (2014) Species limits in avian malaria parasites (Haemosporida): how to move forward in the molecular era. Parasitology 141(10):1223–1232
Palinauskas V, Valkiūnas G, Bolshakov CV, Bensch S (2008) Plasmodium relictum (lineage P-SGS1): effects on experimentally infected passerine birds. Exp Parasitol 120:372–380
Palinauskas V, Žiegyte R, Ilgūnas M, Iezhova TA, Bernotiene R, Bolshakov C, Valkiūnas G (2015) Description of the first cryptic avian malaria parasite, Plasmodium homocircumflexum n. sp., with experimental data on its virulence and development in avian hosts and mosquitoes. Int J Parasitol 45:51–62
Pérez-Tris J, Bensch S (2005) Dispersal increases local transmission of avian malarial parasites. Ecol Lett 8:838–845
Perkins SL (2014) Malaria’s many mates: past, present and future of the systematics of the order haemosporidia. J Parasitol 100:11–25
Richard FA, Sehgal RNM, Jones HI, Smith TB (2002) A comparative analysis of PCR-based detection methods for avian malaria. J Parasitol 88:819–822
Ricklefs RE, Fallon SM (2002) Diversification and host switching in avian malaria parasites. Proc R Soc Lond B 269:885–892
Ricklefs RE, Fallon SM, Bermingham E (2004) Evolutionary relationships, cospeciation, and host switching in avian malaria parasites. Syst Biol 52:111–119
Ricklefs RE, Swanson BL, Fallon SM, Martinez-Abraín A, Scheuerlein A, Gray J, Latta SC (2005) Community relationships of avian malaria parasites in Southern Missouri. Ecol Monogr 75:543–559
Ricklefs RE, Outlaw DC, Svensson-Coelho M, Medeiros MCI, Ellis VA, Latta S (2014) Species formation by host shifting in avian malaria parasites. Proc Natl Acad Sci USA 111:14816–14821
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Sehgal RNM, Lovette IJ (2003) Molecular evolution of three avian neurotrophin genes: implications for proregion functional constraints. J Mol Evol 57:335–342
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Super PE, van Riper C (1995) A comparison of avian hematozoan epizootiology in two California coastal scrub communities. J Wildl Dis 31:447–461
Svensson-Coelho M, Blake JG, Loiselle BA, Penrose AS, Parker PG, Ricklefs RE (2013) Diversity, prevalence, and host specificity of avian Plasmodium and Haemoproteus in a western Amazon assemblage. Ornithol Monogr 76:1–47
Valkiūnas G (2005) Avian malaria parasites and other haemosporidia. CRC, Boca Raton
Valkiūnas G, Bensch S, Iezhova TA, Križanauskienė A, Hellgren O, Bolshakov CV (2006) Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. J Parasitol 92(2):418–422
Valkiūnas G, Iezhova TA, Križanauskienė A, Palinauskas V, Sehgal RNM, Bensch S (2008) A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J Parasitol 94:1395–1401
Valkiūnas G, Ashford RW, Bensch S, Killick-Kendrick R, Perkins S (2011) A cautionary note concerning Plasmodium in apes. Trends Parasitol 27(6):231–232
Valkiūnas G, Palinauskas V, Ilgūnas M, Bukauskaitė D, Dimitrov D, Bernotienė R, Zehtindjiev P, Ilieva M, Iezhova TA (2014) Molecular characterization of five widespread avian haemosporidian parasites (Haemosporida), with perspectives on the PCR-based detection of haemosporidians in wildlife. Parasitol Res 113:2251–2263
Waldenström J, Bensch S, Kiboi S, Hasselquist D, Ottosson U (2002) Cross-species infection of blood parasites between resident and migratory songbirds in Africa. Mol Ecol 11:1545–1554
Waldenström J, Bensch S, Hasselquist D, Östman Ö (2004) A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J Parasitol 90:191–194
Walther EL, Valkiūnas G, González AD, Matta NE, Ricklefs RE, Cornel A, Sehgal RNM (2014) Description, molecular characterization, and patterns of distribution of a widespread New World avian malaria parasite (Haemosporida: Plasmodiidae), Plasmodium (Novyella) homopolare sp. nov. Parasitol Res 113:3319–3332
Wood FD, Wood SF (1937) Occurrence of haematozoa in some California birds and mammals. J Parasitol 23:197–201
Zehtindjiev P, Ilieva M, Westerdahl H, Hansson B, Valkiūnas G, Bensch S (2008) Dynamics of parasitemias of malaria parasites in a naturally and experimentally infected migratory songbird, the great reed warbler, Acrocephalus arundinaceus. Exp Parasitol 119:99–110
Žiegytė R, Valkiūnas G (2014) Recent advances in vector studies of avian haemosporidian parasites. Ekologija 60(4):73–83
Acknowledgments
This work was supported by a grant from the San Francisco State University Arthur Nelson Scholarship and the San Francisco State University Instructionally Related Activities Grant. The authors wish to thank the County of Fresno and the Consolidated Mosquito Abatement District for access to the field site and the Kearney Agricultural Research and Extension (KARE) Center in Parlier, California, for providing lodging. We thank Annette Chan (SFSU staff) for microscopy assistance, Greg Spicer and Andrea Swei (SFSU faculty) for phylogenetics and R statistical software expertise, respectively; Elvin Lauron for laboratory assistance; and, Tija Altergott, Holly Archer, Doug Bell, Molly Dodge, Sierra Flynn, Ariana LaPorte, Leonard Liu, Claire Loiseau, Tim Marzec, Allison Nelson, Katherine Purcell, Elaine Vo and Laura Wilkinson for assistance with data collection in the field.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted. The bird sampling methodology was approved by the Institutional Animal Care and Use Committee (IACUC) and was performed under permits supplied by the United States Geological Survey Bird Banding Laboratory and a Scientific Collecting Permit issued by the California Natural Resources Agency, Department of Fish and Game.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by K. C. Klasing.
Rights and permissions
About this article
Cite this article
Walther, E.L., Carlson, J.S., Cornel, A. et al. First molecular study of prevalence and diversity of avian haemosporidia in a Central California songbird community. J Ornithol 157, 549–564 (2016). https://doi.org/10.1007/s10336-015-1301-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-015-1301-7