Abstract
Migratory birds travel long distances and use diverse habitats, potentially exposing them to a broad range of microbes that could negatively affect their health and survival. Gut microbiota composition may be related to organismal health especially during periods of impaired immunity due to stress, by functioning as a reservoir for potential pathogens. We provide an insight into the composition of the gastrointestinal microbiota in migratory Red Knot (Calidris canutus) and Ruddy Turnstone (Arenaria interpres) staging in Delaware Bay, USA, by analyzing fecal bacterial communities of three individuals per species with 16S rRNA clone libraries. In the 313 bacterial sequences we analysed from Red Knots, we identified 19 bacterial classes across 29 genera, and from the 218 Ruddy Turnstone sequences, we identified 11 bacterial classes across 17 genera. In Red Knots and Ruddy Turnstones, 27 and 41 % of all sequences were closely related to Campylobacter spp., which include several human pathogens. Only 5 of the 46 genera, and 8 out of 124 operational taxonomic units were shared between species, suggesting that gut microbial community structure can be species-specific under environmentally similar conditions. Our study provides baseline information that can be used in future studies to better understand diversity and function of gut microbes, and can be expanded to investigate how gut microbiota of migratory birds affects their body condition, immune function, and demographic performance.
Zusammenfassung
Die Magen-Darmflora zweier Arten ziehender Küstenvögel während des Frühlingszuges im Rastgebiet Delaware Bay, USA
Zugvögel ziehen lange Strecken und nutzen verschiedenartige Lebensräume, wo sie einer großen Anzahl verschiedener Mikroben ausgesetzt sind, die sich negativ auf Gesundheit und Überleben auswirken könnten. Die Zusammensetzung der Darmflora könnte mit der Gesundheit zusammenhängen, da sie ein Reservoir möglicher Krankheitserreger bildet, besonders in Zeiten von geschwächter Immunabwehr durch Stress. Wir untersuchten die Zusammensetzung der mikrobiellen Biozönose in Magen und Darm aus Stuhlproben von jeweils drei Individuen des Knutts (Calidris canutus) und des Steinwälzers (Arenaria interpres) aus dem Rastgebiet Delaware Bay anhand von 16S rRNA Klon-Datenbanken. In den 313 Sequenzen bakterieller RNA, die wir für die Knutts analysierten, waren 19 Klassen und 29 Gattungen Bakterien vertreten. In den 218 Sequenzen der Steinwälzer fanden wir 11 Klassen und 17 Gattungen von Bakterien. In Knutts und Steinwälzern waren jeweils 27 und 41 % der Sequenzen nah verwandt mit Campylobacter spp., welche einige menschliche Krankheitserreger beinhalten. Nur fünf der 46 Gattungen und acht von 124 taxonomischen Einheiten waren beiden Vogelarten gemeinsam, was darauf hindeutet, dass die Magen-Darmflora auch unter ähnlichen ökologischen Bedingungen artspezifisch sein kann. Unsere Studie gibt eine Grundidee der Vielfalt und Funktion von Magen-Darmbakterien, auf die sich zukünftige Studien stützen können. Darauf aufbauend könnte untersucht werden, wie die Magen-Darmflora von Zugvögeln ihre Körperkondition, Immunabwehr und demographische Merkmale beeinflusst.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Migratory animals travel long distances and use diverse habitats, potentially exposing themselves to a broad range of pathogens with potential for the spread of infectious disease. However, the assumption that migrating animals affect the spread of pathogens has not been rigorously tested (Altizer et al. 2011). An important step toward testing the role of migratory animals as potential disease vectors is to ascertain what microorganisms these animals encounter and carry with them as their geographic locations change within their annual cycles.
Migrating shorebirds visit many locations with varying habitat types over their annual cycle, and some of these habitats are known hotspots for pathogen exposure (Krauss et al. 2010). Shorebird annual migrations span a large latitudinal range and total distances of over 5,000 km are not uncommon. To accomplish long-distance migrations, most shorebird species rely on staging sites between their origin and destination. Staging sites are visited annually with often high site fidelity because they provide an abundance of quality food, allowing for a rapid gain of body mass (Pfister et al. 1998). One such site, used extensively by ~half a million shorebirds during spring migration is Delaware Bay, USA (Clark and Niles 1993). This stopover site is one of the main spawning grounds for Horseshoe Crabs (Limulus polyphemus). Horseshoe Crabs deposit eggs at extremely high densities on Delaware Bay beaches in May, and these eggs serve as a key resource for staging shorebirds, providing a protein-rich food source that enables them to complete migration to their breeding grounds.
One consequence of large aggregations of migratory birds in Delaware Bay is that transmission of infectious agents could be increased through direct contact between birds and/or indirectly through ingestion of other birds’ feces. Delaware Bay is a hotspot for low-virulent avian influenza (AI) viruses, with highest prevalence in Ruddy Turnstones (Arenaria interpres) (Krauss et al. 2010), but increased exposure is possible for all species due to contact with AI viral particles shed in Ruddy Turnstone feces. Migratory birds can be physiologically stressed when arriving at staging areas and may be vulnerable to infection due to the possible immuno-suppressing effect of migratory movements (Owen and Moore 2006). In Delaware Bay, recent arrivals are still recovering protein lost during their journey, and have lower indices of several constitutive immune parameters than birds that had recovered lost protein and were storing fat (Buehler et al. 2010).
To date, the majority of studies investigating disease in shorebirds have focused on AI infections. However, outside of Delaware Bay, the prevalence of AI viruses in shorebirds is extremely low (Munster et al. 2007; Pearce et al. 2012). One aspect of shorebird biology that may have a potentially large influence on health is the gastro-intestinal microbiota community. Mammalian gut microbiota are known to play an important role in maintaining health through close interactions with the immune system and by facilitating nutrient uptake from symbiotic and commensal interactions between the host and non-pathogenic gut microbes (Neish 2009; Clemente et al. 2012; Hooper et al. 2001). While bacterial effect in nutrient uptake in avian hosts is unknown, lowering nutrient uptake due to imbalances in gut microbial composition of migratory birds might decrease body condition, which may result in failure to migrate, reduced reproductive performance, or even mortality. Perhaps equally relevant is the potential for the avian gut microbiota to provide competitive exclusion from pathogens as in other animals, while in other instances providing a reservoir for potential pathogens, especially during periods of impaired immunity due to stress.
Birds can alter their environment’s microbial community through deposition of feces, especially at congregational sites like Delaware Bay. High prevalence of the human pathogens Campylobacter jejuni and Escherichia coli has been shown in gulls (Larus spp.) (Camarda et al. 2006; Keller et al. 2011; Lu et al. 2011), but the impact of fecal deposition at high shorebird densities on individual animals’ gut microbiota has received little attention. One study has examined shorebird microbiota along the East-Atlantic Flyway in Portugal (Santos et al. 2012), but it is unlikely that these birds using Eurasian and African flyways are exposed to the same microbial environment as shorebirds using the Delaware Bay area.
To assess how shorebird fecal deposition impacts their health as well as their environment and associated organisms, it is essential to first identify what microbes shorebirds carry in their gastro-intestinal tract and whether gut microbiota communities vary between species. Therefore, the goal of our study is to describe the microbial community in shorebirds during one period in their annual cycle, namely the spring migratory staging period in Delaware Bay. Furthermore, to gain insight into inter-specific differences in gut microbiota composition, we compared gut microbiota in two migratory shorebird species, the Red Knot (Calidris canutus) and Ruddy Turnstone. Very little is known about the microbes that shorebirds carry, and about how this microbial community changes over time. This may present a problem if microbial turnover rate is slow, as it would be impossible to distinguish between microbiota acquired at the staging site and microbiota remaining in the gut from the geographical origin of migration. However, shorebirds were observed to defecate every 5 min on their non-breeding grounds (Rose and Nol 2010). The defecation rate may be higher during the staging period of migratory birds in Delaware Bay, due to the high food intake necessary to reach the required pre-migration body mass. In addition, a study on food and fluid retention time of different bird species showed a range from 1.9 to 17 h, with the Rock Ptarmigan (Lagopus muta), which is closest in size to shorebirds, retaining food and fluid for only between 1.9 and 9.9 h (Stevens and Hume 1998). Although ptarmigans and shorebirds differ substantially in phylogeny and diet, these retention time frames suggest that it is reasonable for us to assume that shorebirds present in Delaware Bay for a number of days will have replaced their intestinal content several times. Hence, micro-organisms that shorebirds acquired from food and environment at their wintering ground are unlikely to still be present in the bird’s small intestine even after a short stay in Delaware Bay.
We predict that, if gut microbiota composition is mainly determined by underlying genetics, we would expect large differences in communities between shorebird species. However, if environment is the main force shaping gut microbiota composition, we hypothesize that gut microbiota composition of Red Knots and Ruddy Turnstones will be similar as both species use the same environment and food source during their stay in Delaware Bay. To better understand the microbial composition of migratory shorebirds gut, we analyzed 16S rRNA clone libraries developed from fecal DNA extracts. To our knowledge, this study is the first project to identify both culturable and unculturable gastro-intestinal microbiota of different shorebird species in the western hemisphere from fecal matter.
Methods
Sample collection
Red Knots and Ruddy Turnstones were captured at Delaware Bay (38°55′3″N, 75°18′29″W) beaches using canon nets between 7 and 29 May, 2010. Red Knots on average stage in Delaware Bay for 8–12 days between 10 May and 10 June (Gillings et al. 2009). Data on staging behavior of Ruddy Turnstone is lacking, but as they rely on the same temporary food source as Red Knot and face the same time constraints due to the short high-arctic breeding window, we assume similar staging durations for them. Upon capture, birds were placed in individual cardboard boxes lined with sterile waxed paper for up to 5 min for collection of feces. Feces of cecal origin were avoided during samples collection to avoid comparison between cecal and small intestine microbiota. Fecal pellets were transferred to sterile 2-ml collection tubes pre-filled with 96 % ethanol and stored at 4 °C until further analyses.
Molecular methods
We selected three Red Knot and three Ruddy Turnstone fecal samples for analyses of their gastrointestinal microbiota. All birds selected were captured within 2 days of each other (15–17 May) and were of similar body mass (Red Knot: 122.0 g ± 5.9; Ruddy Turnstone: 117.8 g ± 3.2) to limit bias due to variation in staging time prior to sampling. Also, by selecting individuals with a body mass that was above the arrival mass of 111.1 g (Gillings et al. 2009), we attempted to sample healthy individuals as low body mass is often linked to poor body condition (Møller et al. 1998). We are aware that sampling unhealthy individuals would provide us with more knowledge on potential pathogens these birds can transmit to the population, but as it is difficult to determine health of birds in the field, and as we are presenting a first overview of shorebird gut microbiota in Delaware Bay, we decided to sample presumably healthy birds to establish accurate baseline information.
We regressed body mass of both species against structural size measurements (wing length, total head length, and culmen length) of all conspecifics caught within 2 days of 16 May, to explore the need for a size correction on body mass of the individuals sampled for this study. We found no relationship between any structural size measurement and body mass for either species (maximum r 2 = 0.07 for Red Knot wing length-body mass linear regression; other results not shown), and therefore used original body mass in our staging duration estimations.
Body mass of Red Knot upon arrival was estimated at 111.1 g by Gillings et al. (2009), and the average body mass of 122.0 g in our study was estimated to be equivalent to an approximately 3-day presence in Delaware Bay prior to sampling based on a weight gain of 4.05 ± 0.32 g day−1. No body mass-staging duration relationship has been published for Ruddy Turnstone, for which body mass at the earliest catch (12 March) in 2010 averaged 97.1 ± 13.0 g (Delaware Division of Fish and Wildlife, unpublished data ) and as food availability is similar for Red Knots and Ruddy Turnstone, we assume that a comparable daily weight gain is possible for Ruddy Turnstone resulting in a sampling time of approximately 5 days after arrival.
Before DNA extractions, fecal samples were spun using a microcentrifuge at 14,000 rpm for 10 min to harvest the biomass. The fecal pellets were washed three times with sterile water to remove ethanol residue (Wilson 1997). After ethanol removal, we dried samples at 60 °C for 40 min. Total DNA was extracted from fecal samples using the Mo Bio Power soil kit following manufacturer’s instructions (Mo Bio Laboratorie, Carlsbad, CA, USA). DNA extracts were used to generate community 16S rRNA gene PCR products using general bacterial primers 8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 785R (5′-ACTACCRGGGTATCTAATCC-3′) (Ley et al. 2008; Lu et al. 2008). PCR reactions contained a buffer solution (10 mM Tris–HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.01 % gelatin, and 160 μg/ml bovine serum albumin (Hagelberg 1994), 0.04 mM each deoxyribonucleotide triphosphate, 0.2 μM of each forward and reverse primer, 1/4 U Taq DNA polymerase (Invitrogen, Carlsbad, CA, USA), and fecal DNA as the template in a final reaction volume of 25 μl. The PCR cycling conditions were 2 min at 94 °C, followed by 30 cycles of 45 s at 94 °C, 45 s at 56 °C, and 90 s at 72 °C and a final primer extension step at 72 °C for 5 min. PCR products were cloned into TOPO TA Cloning Kit One Shot TOP10 Chemically Competent E. coli (Invitrogen) and plated on 1.5 % agar plates with 100 mg ampicillin L−1. Clones were screened with M13F (5′-TTTTCCCAGTCACGACGTTGTA-3′) and M13R (5′-CAGGAAACAGCTATGACC-3′) as described by Ryu et al. (2012a). Positive clones were determined using electrophoresis, and DNA was recovered from agarose gels by band excision and filtered pipette tip centrifugation (Dean and Greenwald 1995). Clones were then sequenced using Big Dye Terminator v.3.1 chemistry on an ABI 3100 automated sequencer (Applied Biosystems, Foster City, CA, USA).
Sequences with >500-bp high-quality base calls were pooled per species and used in further analyses. Sequences were aligned and checked for chimeras using the Bellerophon program (http://comp-bio.anu.edu.au/bellerophon/bellerophon.pl; Huber et al. 2004). Base pair window was set at 300 bp and a Huber–Hugenholtz correction was applied to correct for potential partial length sequences in the alignment. 16S rRNA gene sequences were then compared to known bacterial 16S rRNA gene sequences in the GenBank (NR) database using National Center for Biotechnology Information (NCBI), BLAST (www.ncbi.nlm.nih.gov/BLAST/), and through the Greengenes program (DeSantis et al. 2006). We visually checked the ten closest matches to our sequences to ensure that identification of sequences was similar and reliable. Sequences were then classified to class or genus level using RDP Classifier with a 95 % confidence level (http://rdp.cme.msu.edu/classifier; Wang et al. 2007).
Nucleotide sequence accession numbers
Cloned 16S rRNA gene sequences obtained in this study were deposited in the GenBank database under the following accession numbers: KC478310–KC478351.
Results and discussion
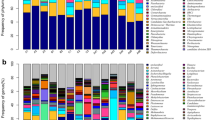
In our exploratory study, we investigated the composition of gut microbiota in two migratory shorebird species staging in Delaware Bay, through sequencing of bacterial 16S rRNA genes from fecal samples. A total of 531 clone sequences were analyzed (n = 313 for Red Knot; n = 218 for Ruddy Turnstone) and classified into 41 bacterial genera belonging to 18 classes (Table 1). Red Knots exhibited higher bacterial diversity, with sequences representing 29 known bacterial genera versus 17 in Ruddy Turnstones. An additional 16 unknown genera were identified to class level including two Bacilli classes (Enterococcaceales and Lactobacillales orders). The dominant bacterial classes were Bacilli in Ruddy Turnstone (37.8 %) and Epsilonproteobacteria (40.6 %) in Red Knot. The most common bacterial genus was Campylobacter comprising 40.6 and 26.8 % of all clones in Red Knot and Ruddy Turnstone, respectively. In Ruddy Turnstones, a large fraction of clones was attributed to unclassified Lactobacillales. All unclassified Lactobacillales clones were nearly identical to available sequences of uncultured bacteria with approximately 93 % sequence identity.
Only five genera were present in both Red Knot and Ruddy Turnstone. This large level of interspecific variation suggests that there are host-specific factors that influence the gut microbiota community structure of these species, rather than gut microbiota being solely impacted by environmental conditions which were similar for both shorebird species. However, because the number of sampled individuals in this study is quite small, it is possible that there could be large inter-individual differences causing intra-specific variation even within Red Knots or Ruddy Turnstones. Future studies with larger samples sizes and sampling over longer timescales will help resolve this possibility.
On an operational taxonomic unit (OTU) level (>97 % sequence similarity), Red Knot and Ruddy Turnstone sequences were assigned to 72 and 27 unique and 8 shared OTUs, respectively (Fig. 1). Despite the small overlap, 55.3 % of all sequences analyzed belonged to the 8 shared OTUs, suggesting that the most common OTUs are shared while OTUs unique to Red Knot and Ruddy Turnstone are relatively rare. Shared OTUs included members of the genera Enterococcus, Campylobacter, Corynebacterium, and Thalassobius, and could be matched to species level with 88–100 % similarities to Genbank sequences. Enterococci are a known member of the commensal flora community in humans and have been identified in several migratory and non-migratory bird species (Janiga et al. 2007; Silva et al. 2012; Ryu et al. 2013).
Venn diagram for the shared and non-shared OTUs between Red Knot (Calidris canutus) and Ruddy Turnstone (Arenaria interpres). Number of unique and shared OTUs (bold) and percentage of total sequences are shown within the diagram (>97 % sequence similarity between our sequences and sequences available in Genbank)
The identification of sequences closely related to Campylobacter spp. in fecal samples of both Red Knots and Ruddy Turnstones is interesting since members of this genus are recognized as human pathogens. However, further in depth studies focusing on identification of these Campylobacter spp. to species level should be conducted to determine if the Campylobacter populations are pathogenic to bird hosts.
Actinobacteria were among the numerically dominant sequences in Red Knots (19.1 %), but were absent in Ruddy Turnstones. Members of the class of Actinobacteria are among the most common bacteria found in terrestrial and fresh marine environments. They have also been found to inhabit animals, as shown in a recent study investigating the ceca microbiota of wild and captive Western Capercaillie (Tetrao urogallus; Wienemann et al. 2011). In addition, Actinobacteria have been isolated from different gull species (Larus spp.; Lu et al. 2008; Ryu et al. 2012a), Canada Goose (Branta canadensis; Lu et al. 2009), New Zealand Mallard Duck (Anas platyrhynchos) feces (Murphy et al. 2005), and shorebird cloacal communities (Santos et al. 2012). Although the majority of Actinobacteria have not been identified as harmful, the class includes the Corynebacterium genus which includes several species harmful to mammals and birds (Potti et al. 2002; Hoelzle et al. 2013). Nearly all Actinobacteria found in our study were identified as belonging to this genus. Although in low abundance, Corynebacterium spp. have also been isolated from Black-winged Stilts (Himantopus himantopus) (Santos et al. 2012).
Approximately 38 % of clones of Ruddy Turnstone feces in our study were identified as belonging to the genera in the class Bacilli, whereas Red Knot clones only contained 9.5 % Bacilli, despite similarities in diet and habitat in Delaware Bay. Several studies have reported high prevalence of Bacilli in avian species such as Canada Goose (Lu et al. 2009), gull (Larus spp.; Lu et al. 2008; Ryu et al. 2012a), Sandhill Crane (Grus canadensis) and Snow Goose (Chen caerulescens; Ryu et al. 2012b) and three species of shorebird: Black-winged Stilt, Icelandic Black-tailed Godwit (Limosa limosa) and Common Redshank (Tringa totanus) (Santos et al. 2012). The class Bacilli includes the order Lactobacillales, which is believed to make up a large part of the human gastro-intestinal microbiota (Guarner and Malagelada 2003). Two known genera detected were Catellicoccus and Streptococcus, but the majority of Bacilli sequences found in Ruddy Turnstone could not be identified beyond the Lactobacillales order level. Interestingly, Catellicoccus-like sequences were identified in both bird species. Catellicoccus spp. have only been detected in large quantities in gulls (Lu et al. 2008; Ryu et al. 2012a). Ingestion of Catellicoccus originating from gull feces by shorebirds is possible in Delaware Bay because large numbers of gulls (predominantly Greater Black-backed Gull Larus marinus, Laughing Gull L. atricilla, and European Herring Gull L. argentatus) forage in mixed flocks with Red Knots and Ruddy Turnstones, but, as Catellicoccus was also isolated from Common Redshank in Portugal (Santos et al. 2012), microbes from this genus could also represent a regular component of shorebird gut microbiota.
One other study has examined gut microbiota in shorebirds (Santos et al. 2012). Compared to that study, our study detected a high proportion of Campylobacter ssp. clones for both Red Knot (41 %) and Ruddy Turnstone (26 %). Santos et al. (2012) detected <9 % Campylobacter spp. in their samples of three shorebird species (Common Redshank, Black-winged Stilt, and Black-tailed Godwit) staging in the Tagus estuary, Portugal. Moreover, the overall gut microbial diversity of shorebird species studied by Santos et al. (2012) was lower than in Red Knot and Ruddy Turnstone with a total of 9 classes and 25 genera, although this may be due to the fact that we analyzed close to 10 times more sequences than the previous study. Another difference between these studies was the type of sample studied, that is, cloacal swabs used by Santos et al. (2012) versus full fecal pellets used in our study. A total of 10 bacterial classes identified in our study were not found by Santos et al. (2012): Acidobacteria, Chloroflexi, Erysipelotrichi, Flavobacteria, Fusobacteria, Mollicutes, Nitrospira, Planctomycetacia, Sphingobacteria, and Delta-proteobacteria. Samples from these classes comprised 14 bacterial genera, but together made up only 8 % of total clones sequenced, suggesting low abundance. However, of the seven classes which shorebird species from both studies had in common, only six genera overlapped, which indicates large global variation in gut microbiota diversity in shorebird species.
In our study, we pooled bacterial 16S rRNA sequences derived from fecal matter of three individuals per species. As the main purpose of this exploratory study was to gain a first insight into shorebird gut microbiota, we feel that number of sequences analyzed was of higher importance than sampling a broad range of individuals. The latter would have resulted in fewer sequences per individual, reducing the overall sequencing depth. The use of next-generation sequencing (NGS) methods can circumvent the depth at which we can study the relatively less abundant member of the microbial community. However, the read length associated with these methods does not allow for the identification of members at lower taxonomic levels. Additionally, the error rates of associated with NGS can falsely introduce unique members, making it more difficult to perform microbial diversity analyses (Kozich et al. 2013).
It should also be noted that we studied the fecal samples rather than intestinal contents, as collecting latter samples can only be performed by sacrificing birds, which was not possible due to the protected status and current population declines of Turnstones and Knots. Although microbial structure associated with the intestine and feces may differ, we assume fecal microbiota as a proxy for intestinal microbiota due to its role as fecal storage area. Thus, we assume that comparison of fecal samples between bird species can provide a general picture of the bacterial composition of the intestinal microbiota. By avoiding cecal material, we were able to focus on the intestinal samples, rather than including bacteria that are selected in the cecum (Lu et al. 2003). Additionally, fecal samples are often used to isolate bacterial pathogens as well as intestinal parasites. In this study, we sampled only once and we made the assumption that micro-organisms acquired by shorebirds from food and environment at their wintering ground are unlikely to still be present in the bird’s gastro-intestinal tract even after a short stay in Delaware Bay. Future studies should increase the level of sampling and collect fecal and intestinal samples to further validate our findings with respect to microbial turnover rates and potential fecal/intestinal microbiota discrepancies in shorebirds.
The use of DNA-based methods for detecting bacteria in environmental samples cannot discriminate between viable and non-viable bacteria (Hirsh et al. 2010). Therefore, some of the bacterial species detected may not play an important role within the shorebird gut microbiome, as they may have been killed during digestion. However, the fact that some of these bacterial groups have been detected in other birds via cultured-based methods (Lu et al. 2003) suggests that many of these bacteria are capable of withstanding the prevailing conditions in the avian gut. Moreover, some of the bacterial groups have been detected in many different birds using molecular techniques suggesting that these bacteria are not transitory but rather persist and perhaps thrive in the avian gut (Lu et al. 2009; Ryu et al. 2012a, b). Future studies using RNA as the target molecule in PCR and cloning-based approaches (Revetta et al. 2011) may further confirm the presence of such bacteria, as well as provide information on the relative level of activity of different groups inhabiting migratory birds gut.
Our results have potential implications for determining the importance of fecal deposition by migratory birds on human and avian health in the Delaware Bay during stopover periods. Also, the differences we observed in the gut microbiota community of two ecologically similar species inhabiting the same environment will be valuable in helping to identify signature sequences that can be used in the development of methods to identify each bird species’ importance in fecal pollution (Ryu et al. 2012b).
Conclusion
Our study is the first to describe gut microbiota composition of Red Knots and Ruddy Turnstones using one of the most important staging sites in the western hemisphere, Delaware Bay, USA. Gut microbiota are known to play a large role in maintaining health in organisms and therefore an insight into gut microbiota of shorebirds is a necessary first step to the identification and prevention of spread of potentially harmful pathogens. We found that Red Knots harbor a higher gut microbial diversity than Ruddy Turnstones, and that Red Knots and Ruddy Turnstones differ from each other in gut microbial profiles, despite their shared habitat and food source during the staging period in Delaware Bay. Our study provides baseline information that can be used in future studies to better understand diversity and function of gut microbes, and can be expanded to investigate how gut microbiota of migratory birds affects their body condition, immune function, and demographic performance.
References
Altizer S, Bartel R, Han BA (2011) Animal migration and infectious disease risk. Science 331:296–302
Buehler DM, Tieleman BI, Piersma T (2010) Indices of immune function are lower in Red Knots (Calidris canutus) recovering protein than in those storing fat during stopover in Delaware Bay. Auk 127:394–401
Camarda A, Circella E, Pennelli D, Madio A, Bruni G, Lagrasta V, Marzano G, Mallia E, Campagnari E (2006) Wild birds as biological indicators of environmental pollution: biotyping and antimicrobial resistance patterns of Escherichia coli isolated from Audouin’s Gulls (Larus audouinii) living in the Bay of Gallipoli (Italy). Italian J Anim Sci 5:287–290
Clark KE, Niles LJ (1993) Abundance and distribution of migrant shorebirds in Delaware Bay. Condor 95:694–705
Clemente JC, Ursell LK, Parfrey LW, Knight R (2012) The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270
Dean A, Greenwald J (1995) Use of filtered pipet tips to elute DNA from agarose gels. Biotechniques 18:980
DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Env Microbiol 72:5065–5072
Gillings S, Atkinson PW, Baker AJ, Bennett KA, Clark NA, Cole KB, González PM, Kalasz KS, Minton CDT, Niles LJ, Porter RC, de Lima Serrano I, Sitters HP, Woods JL (2009) Staging behavior in Red Knot (Calidris Canutus) in Delaware Bay: implications for monitoring mass and population size. Auk 126:54–63
Guarner F, Malagelada JR (2003) Gut flora in health and disease. Lancet 361:512–519
Hagelberg E (1994) Mitochondrial DNA from ancient bones. In: Herrmann B, Hummel S (eds) Ancient DNA. Springer, New York, pp 195–204
Hirsh PR, Mauchline TH, Clark IM (2010) Culture-independent molecular techniques for soil microbial ecology. Soil Biol Biochem 42:878–887
Hoelzle LE, Scherrer T, Muntwyler J, Wittenbrink MM, Philipp W, Hoelzle K (2013) Differences in the antigen structures of Corynebacterium pseudotuberculosis and the induced humoral immune response in sheep and goats. Vet Microbiol 164:359–365
Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI (2001) Molecular analysis of commensal host–microbial relationships in the intestine. Science 291:881–884
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319
Janiga M, Sedlarova A, Rigg R, Novotna M (2007) Patterns of prevalence among bacterial communities of Alpine Accentors (Prunella collaris) in the Tatra Mountains. J Ornithol 148:135–143
Keller JI, Shriver WG, Waldenström J, Griekspoor P, Olsen B (2011) Prevalence of Campylobacter in wild birds of the mid-Atlantic region, USA. J Wildl Dis 47:750–754
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79(17):5112–5120
Krauss S, Stallknecht DE, Negovetich NJ, Niles LJ, Webby RJ, Webster RG (2010) Coincident Ruddy Turnstone migration and Horseshoe Crab spawning creates an ecological ‘hot spot’ for influenza viruses. Proc R Soc B 277:3373–3379
Ley R, Hamady M, Lozupone C, Turnbaugh P, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, Gordon JI (2008) Evolution of mammals and their gut microbes. Science 320:1647–1651
Lu J, Idris U, Harmon B, Hofacre C, Maurer JJ, Lee MD (2003) Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl Environ Microbiol 69:6816–6824
Lu J, Santo Domingo JW, Lamendella R, Edge TA, Hill S (2008) Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl Environ Microbiol 74:3969–3976
Lu J, Santo Domingo JW, Hill S, Edge TA (2009) Microbial diversity and host-specific sequences of Canada goose feces. Appl Environ Microbiol 75:5919–5926
Lu J, Ryu H, Domingo JWS, Griffith JF, Ashbolt N (2011) Molecular detection of Campylobacter spp. in California Gull (Larus californicus) excreta. Appl Environ Microbiol 77:5034–5039
Møller AP, Christe P, Erritzøe J, Mavarez J (1998) Condition, disease and immune defence. Oikos 83:301–306
Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus ADME, Fouchier RAM (2007) Spatial, temporal, and species variation in prevalence of Influenza A viruses in wild migratory birds. PLoS Pathog 3:e61. doi:10.1371/journal.ppat.0030061
Murphy J, Devane ML, Robson B, Gilpin BJ (2005) Genotypic characterization of bacteria cultured from duck faeces. J Appl Microbiol 99:301–309
Neish AS (2009) Microbes in gastrointestinal health and disease. Gastroenterology 136:65–80
Owen JC, Moore FR (2006) Seasonal differences in immunological condition of three species of thrushes. Condor 108:389–398
Pearce JM, Ruthrauff DR, Hall JS (2012) Paired serologic and polymerase chain reaction analyses of Avian Influenza prevalence in Alaskan shorebirds. J Wildl Dis 48:812–814
Pfister C, Kasprzyk MJ, Harrington BA (1998) Body-fat levels and annual return in migrating Semipalmated Sandpipers. Auk 115:904–915
Potti J, Moreno J, Yorio P, Briones V, Garcia-Borboroglu P, Villar S, Ballesteros C (2002) Bacteria divert resources from growth for magellanic penguin chicks. Ecol Lett 5:704–714
Revetta RP, Matlib RS, Santo Domingo JW (2011) 16S rRNA gene sequence analysis of drinking water using RNA and DNA extracts as targets for clone library development. Curr Microbiol 63:50–59
Rose M, Nol E (2010) Foraging behaviour of non-breeding Semipalmated Plovers. Waterbirds 33:59–69
Ryu H, Griffith J, Hill S, Edge T, Toledo-Hernandez C, Gonzalez-Nieves J, Toranzos G, Santo Domingo J (2012a) Comparison of gull-feces specific assays targeting 16S rRNA gene of Catellicoccus marimammalium and Streptococcus spp. Appl Environ Microbiol 78:1909–1916
Ryu H, Lu J, Vogel J, Chávez-Ramírez F, Ashbolt N, Santo Domingo J (2012b) Development and evaluation of a qPCR assay targeting fecal pollution of sandhill cranes (Grus canadensis). Appl Environ Microbiol 78:4338–4345
Ryu H, Henson M, Elk M, Toledo-Hernandez C, Griffith J, Blackwood D, Noble R, Gourmelon M, Glassmeyer S, Santo Domingo J (2013) Development of quantitative PCR assays targeting 16S rRNA gene of Enterococcus spp. and their application to the identification of Enterococcus species in environmental samples. Appl Environ Microbiol 79:196–204
Santos SS, Pardal S, Proença DN, Lopes RJ, Ramos JA, Mendes L, Morais PV (2012) Diversity of cloacal microbial community in migratory shorebirds that use the Tagus estuary as stopover habitat and their potential to harbor and disperse pathogenic microorganisms. FEMS Microbiol Ecol 82:63–74
Silva N, Igrejas G, Goncalves A, Poeta P (2012) Commensal gut bacteria: distribution of Enterococcus species and prevalence of Escherichia coli phylogenetic groups in animals and humans in Portugal. Ann Microbiol 62:449–459
Stevens C, Hume I (1998) Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol Rev 78:393–427
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Wienemann T, Schmitt-Wagner D, Meuser K, Segelbacher G, Schink B, Brune A, Berthold P (2011) The bacterial microbiota in the ceca of Capercaillie (Tetrao urogallus) differs between wild and captive birds. Syst Appl Microbiol 34:542–551
Wilson IG (1997) Inhibition and facilitation of nucleic acid amplification. Appl Environ Microbiol 63:3741–3751
Acknowledgments
We would like to thank Nigel A. Clark, Jacquie A. Clark (British Trust for Ornithology) and Kevin S. Kalasz (US Fish and Wildlife Services) for granting us permission to sample shorebirds and for providing logistical assistance in Delaware Bay. We would also like to thank Yvonne I. Verkuil for valuable help during both field and laboratory work. Lastly, we thank three anonymous reviewers for their insightful comments which notably improved our manuscript. This study was funded by grants from the Natural Science and Engineering Council of Canada [PDF-373488-2009] and the Netherlands Organisation for Scientific Research (Rubicon 825.09.0190) to D.M.B. and by the EPA Office of Research and Development. All sampling procedures were in compliance with animal welfare laws of the USA and all permits necessary for shipping and handling of biological samples were obtained.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. C. Klasing.
Rights and permissions
About this article
Cite this article
Grond, K., Ryu, H., Baker, A.J. et al. Gastro-intestinal microbiota of two migratory shorebird species during spring migration staging in Delaware Bay, USA. J Ornithol 155, 969–977 (2014). https://doi.org/10.1007/s10336-014-1083-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10336-014-1083-3