Abstract
Objectives
To develop and validate a machine learning based automated segmentation method that jointly analyzes the four contrasts provided by Dixon MRI technique for improved thigh composition segmentation accuracy.
Materials and methods
The automatic detection of body composition is formulized as a three-class classification issue. Each image voxel in the training dataset is assigned with a correct label. A voxel classifier is trained and subsequently used to predict unseen data. Morphological operations are finally applied to generate volumetric segmented images for different structures. We applied this algorithm on datasets of (1) four contrast images, (2) water and fat images, and (3) unsuppressed images acquired from 190 subjects.
Results
The proposed method using four contrasts achieved most accurate and robust segmentation compared to the use of combined fat and water images and the use of unsuppressed image, average Dice coefficients of 0.94 ± 0.03, 0.96 ± 0.03, 0.80 ± 0.03, and 0.97 ± 0.01 has been achieved to bone region, subcutaneous adipose tissue (SAT), inter-muscular adipose tissue (IMAT), and muscle respectively.
Conclusion
Our proposed method based on machine learning produces accurate tissue quantification and showed an effective use of large information provided by the four contrast images from Dixon MRI.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Research has shown that maintaining a proper balance in body fat and muscle composition is a key factor in good health [1]. Inappropriate amounts of fat can greatly increase the risks to cardiovascular disease, diabetes, and even cancers [2–4]. This problem has grown to such an extent that obesity now contributes significantly to the global burden of disease [5]. Conversely, sarcopenia, in which there is a general decline in skeletal muscle mass in relation to body fat, leads to frailty and functional impairment. Sarcopenia is usually an age-related process that is becoming increasingly prevalent in developed countries where populations are progressively ageing [6, 7]. However, it has been shown that loss of lean muscle mass alone does not correlate proportionately with declines in muscle strength and function; the quantity of intermuscular adipose tissue (IMAT) may instead be a better predictor [8].
MRI is an imaging technique that is capable of estimating the volume of body components. The main advantage of MRI over other techniques (such as DEXA) is its capacity for accurate depiction of regional body composition without any ionizing radiation. Recent advances in fat and water discrimination (e.g. Dixon sequence) using 3D multi-echo gradient recalled echo imaging have further improved the soft-tissue contrast and measurement accuracy of fat-infiltration within skeletal muscle attainable by MRI [9].
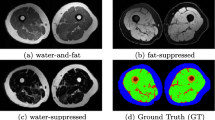
Accurate segmentation of multiple structures is the key to regional body composition study. Using fat and water discrimination technique in MRI is able to achieve more accurate and less variable segmentations [10, 11]. However, while Dixon technique delivers up to four contrasts in one measurement, i.e. in-phase, opposed-phase, water and fat images (Fig. 1), it remains an open question how these four contrasts can be efficiently used for the task of segmentation. Most studies have not used the combined image contrast space, but rather use a single fat or water image one at a time to segment the fat or muscle component separately. Thresholding of a fat image or fat fraction image is a popular choice for the segmentation of adipose tissue [10, 12–14]. Some authors have adapted the atlas/registration-based segmentation method by registering a water/fat image alone [15, 16].
Some studies used the combined contrast image space for body component segmentation. Makrogiannis et al. applied K-means clustering to the combined space of fat image and water image intensities to separate muscle and intermuscular adipose tissue (IMAT) in the thigh [11]. Wang et al. also employed K-means clustering to fat and water images to segment adipose tissue in the abdomen [17]. Joshi et al. selected fat image and water image combined space to perform registration-based segmentation [18]. However, since the fat and water images are secondarily derived through subtraction/addition, there may be artifacts introduced through mis-registration, motion, etc. As a result, including the raw echo images (in-phase and opposed-phase) should be helpful for segmentation. Kullberg et al. used fuzzy clustering in combined contrasts of in-phase, fat, and water images, but only for adipose tissue segmentation in the abdomen [19]. Valentinitsch et al. applied a multi-parametric clustering method in combined in-phase, fat, and water images to segment different structures in the calf and thigh [20]. However, their clustering method was applied several times with different combination of contrast images to segment one structure at a time and only in single slice data.
In this study, we present a novel machine learning based segmentation method that fully uses the combined space of all four contrasts generated by Dixon technique to automatically segment multiple 3D structures in the thigh. We also show that segmentations that are more accurate can be generated by incorporation of all four contrasts compared to using non-fat suppressed (i.e. in-phase) MRI only and using combined fat only and water only images (two contrasts) analyses.
Materials and methods
Data
MRI scans of the thighs of 190 healthy community dwelling older adults (age 50–99 years, average age: 67.85 ± 7.90 years, 58 men and 132 women) were acquired and analyzed as a part of a larger longitudinal study. In this study, we have randomly chosen 40 subjects for validating our proposed segmentation method. The research protocol was approved by the National Healthcare Group Domain Specific Review Board. Informed written consent was obtained from all subjects prior to all examinations.
All the MRI scans were acquired using a 3T MR scanner (Siemens Magnetom Trio, Germany). Subjects were lying on a 6-channel spine-array coil and covered by a 6-channel body matrix external phased array coil. A rapid survey scan was obtained to identify axial slice locations, using the proximal and distal ends of the femur as landmarks. Next, four contrast images (in-phase, opposed-phase, water-only, and fat-only) were acquired using a 2D modified Dixon (multi-echo VIBE and T2 correction) T1-weighted gradient echo pulse sequence for each subject. The in- and opposed-phase images are source echo images while the water- and fat-only images were automatically constructed from the source echo images. Repetition time (TR), the first echo time (TE1), the second echo time (TE2), flip angle (FA), field of view (FOV), and matrix were: TR = 5.27 ms, TE1 = 1.23 ms, TE2 = 2.45 ms, FA = 9, FOV = 440 × 440 mm2, and matrix = 320 × 320. Both left and right thighs were encompassed in the image with in-plane resolution of 0.69 × 0.69 mm2 and with 72 slices (slice thickness: 5 mm). There is no gap between slices.
Machine learning based segmentation scheme
The key concept of our proposed approach is to use fully the image intensity information provided by the four contrast images of Dixon MRI. The approach can be summarized into two broad steps: (1) a machine learning 3-class (fat, muscle, and background/bone) classifier is learned from training samples and subsequently used for voxel classification on target subjects. The classifier is based on a set of image features extracted from all four contrast images. (2) Morphological operations are performed to smooth and generate segmentation masks for subcutaneous adipose tissue (SAT), intermuscular adipose tissue (IMAT), muscle, and bone. The overall scheme of the proposed approach is illustrated in Fig. 2.
Flow chart of the proposed machine learning based segmentation method. After preprocessing, a training set is segmented to assign labels in fat, muscle, and background/cortical bone. The labeled image and features extracted from four contrast images of the training set is passed to extreme learning machine (ELM) for training and then subsequently used to predict the unseen target. Finally, we apply some morphological operations to generate bone region, subcutaneous fat (SAT), muscle, and intermuscular fat (IMAT) volumetric images
Preprocessing: MRI intensity inhomogeneity correction
As the proposed scheme is based on the intensity of MR images, the first step was to correct the intensity inhomogeneity in the images due to bias in magnetic field during scans. In this study, we used a popular automated bias correction algorithm called N4ITK [21]. This algorithm was used to estimate iteratively the multiplicative bias field from the in-phase image that maximizes the high frequency content of the tissue intensity distribution. Sample images of the original in-phase image, estimated bias field, and bias corrected in-phase image are shown in Fig. 3 to demonstrate the effect of bias correction.
Segmentation of training data set
In order to provide classification information for machine learning, a set of training subjects is randomly selected and interactively segmented using an active contour based segmentation method [22] available in “ITK-SNAP” [23]. Each voxel of a training subject is assigned a target category t such that:
Feature extraction
For each voxel in both training data and unseen data, a feature vector is generated in terms of its intensity and neighborhood mean, variance, and entropy in all four-contrast image domains. These features were calculated in 3D. In this study, a cube of 5 × 5 × 5 voxels was used to calculate the corresponding feature vector for the center voxel in the cube. For each voxel sample, 16 different image features were generated.
Machine learning: extreme learning machine
Supervised machine learning algorithms aim at learning from labeled data for prediction on unseen data. In this work, we used a state-of-art machine learning algorithm called extreme learning machine (ELM) to train and test all voxel samples. Compared to other machine learning algorithms, ELM is simple, fast, and achieves high performance in terms of accuracy [24].
Generating segmented volumes of SAT, IMAT, muscle, and bone
After ELM prediction, each voxel in unseen data is assigned a target category, i.e. 0 = background/cortical bone, 1 = fat, 2 = muscle. The voxel labels are mapped back to the spatial 3D image domain. However, SAT, IMAT, and bone marrow are all labeled as fat tissue, and skin and muscle are labeled as muscle tissue due to similar image intensity. It is necessary to separate different types of fat tissue properly. In this step segmented volumes of SAT, IMAT, muscle, and bone are generated using morphological operations.
- Step 1::
-
Generating segmented volume for muscle
Although some skin and muscle tissue are both categorized as muscle tissue by ELM, they are separated by SAT, hence not connected. The segmented volume for muscle (S muscle) was extracted by selecting the connected component (object) labeled as 2 = muscle with the largest number of voxels (Fig. 4b). A mask (M muscle) was generated by morphological closing of S muscle and filling all the holes within S muscle (Fig. 4c).
Fig. 4 Illustration of generating segmented volumes of SAT, IMAT, muscle, and bone. a Labeled image after ELM prediction. Image (b–g) is shown by overlaying binary mask onto the in-phase image for better illustration. b Segmented volume of muscle, c mask M muscle, d segmented volume of bone region, e mask \(M^{\prime}_{\text{muscle}}\), f segmented volume of SAT, and g segmented volume of IMAT
- Step 2::
-
Generating segmented volume for bone region
We defined the bone region as the bone marrow and its surrounding cortical bone. The cortical bone was selected as the largest connected component labeled as 0 = background/cortical bone within mask M muscle region. The segmented volume for bone region S bone was then generated by filling holes within the cortical bone (Fig. 4d). S bone was removed from M muscle to include only voxels belonging to muscle and IMAT, resulting mask \(M^{\prime}_{\text{muscle}}\) (Fig. 4e).
- Step 3::
-
Separating SAT and IMAT
In this last step, for voxels labeled as 1 = fat, IMAT was defined as voxels within \(M^{\prime}_{\text{muscle}}\) region (Fig. 4f) and SAT is the voxels outside \(M_{\text{muscle}}\) (Fig. 4g).
Evaluation
Datasets from 40 randomly selected subjects were used for quantitative validation. The characteristics of the 40 subjects are listed in Table 1. Usually, more training data means more accurate segmentation can be expected, but it is also more time consuming. In order to show that our proposed method works well with limited training data, the 40 subjects were grouped into four groups randomly with 10 subjects in each group. We applied leave-one-out cross validation (LOOCV) in each group to evaluate the accuracy and variability of the proposed method. LOOCV means that, out of the 10 subjects, one subject is selected as the target and the other nine as training set. This is repeated for all 10 subjects so that each subject is served as the target exactly once, resulting 10 automatically segmented volumes of four structures (SAT, IMAT, muscle, and bone region). The automated segmented volumes were compared to the ground truth in terms of Dice similarity coefficient (DSC), which is used to quantify how well two segmentations, A and B overlapped with each other [25]. All the ground truth segmentations were prepared interactively using “ITK-SNAP” [23] and subsequently verified by an experienced radiologist (Dr. C. H. Tan).
The proposed method, based on in-phase image only and combined fat and water images, was also implemented to benchmark the accuracy of our proposed method based on four contrasts. To do this we used the features extracted from in-phase image only (one contrast) and fat and water images only (two contrasts) as input to our machine learning algorithm, left the other stages of the methodology unchanged, and repeated the above experiments.
Results
All the segmentations were performed using Windows XP, on an Intel Xeon Processor (dual core, 3.00 GHz) with 9 GB RAM. The proposed method was implemented using MATLAB version 7.1.1 (The Mathworks, Inc, Natick, MA, USA) [26]. The MATLAB implementations used in this study were not particularly optimized for reduced computational cost and memory usage. The typical execution time for training of nine datasets was less than 2 s. Segmentation time was less than 45 s per unseen data.
Typical segmented images of the proposed segmentation method based on four contrasts are shown in Fig. 5. The proposed method robustly classified all the structures (SAT, IMAT, muscle, and bone region) in thigh images, even in extreme cases where a very thin SAT layer or severe fat infiltration in muscle exists (Fig. 6).
DSC values of comparing volumes segmented by the proposed method and the ground truth are presented in Table 2. The proposed method based on four contrasts achieved good segmentation accuracy and outperforms the method based on non-fat suppressed image and the method based on fat and water images, average DSCbone = 0.94 ± 0.03, DSCSAT = 0.96 ± 0.03, DSCIMAT = 0.80 ± 0.03, DSCMuscle = 0.97 ± 0.01. Friedman statistical test [27] is known for detecting differences in treatments across multiple test samples. The test was carried out using SATA 14 for Windows [28]. Results showed that there are significant differences (p < 0.0001) between the three schemes tested in terms of segmentation accuracy. Student’s t tests were performed and further showed significant improvement for the proposed scheme based on four contrasts over the scheme based on fat and water images (p = 0.0115 for bone region, p = 0.0001 for SAT, p < 0.0001 for IMAT, and p = 0.0025 for muscle). There were also significant differences between the scheme using fat and water images and using unsuppressed image only (student’s t test: p < 0.0001 for bone region, SAT, IMAT, and muscle).
Examples of segmented images using the proposed segmentation scheme based on different contrast images are shown in Fig. 7. Segmentation scheme based on four contrasts correctly segmented all the structures while the scheme based fat and water images failed to segment the bone region and underestimated IMAT and SAT. The scheme based on unsuppressed image only did not manage to segment muscle and fat content accurately.
Discussion
Our proposed segmentation method is shown to be accurate in segmenting 3D thigh MR images compared to ground truth segmentations. Implementing the machine learning technique for segmentation has shown to be easy and efficient in using all four contrasts generated by Dixon sequence. An advantage with the proposed method is that it segments multiple structures (bone region, SAT, IMAT, and muscle) in one shot, which offers great convenience to study relationship of different structures.
One concern over the machine learning technique is the variability faced with different training samples. In this study we used the LOOCV to show that our proposed method is consistently accurate using different sets of training data to segment different targets. The standard deviation of DSC is very low across four groups of total 40 validations, less than 0.03 for all components. The validation showed the high reproducibility of the proposed method.
A key element of this work is the incorporation of all four contrasts generated by Dixon sequence for a combined analysis. Makrogiannis et al. [11] have shown that the use of combined fat and water images analysis improves the accuracy of tissue decomposition compared to the use of non-fat suppressed images only. Results from our experiments corroborate with their study. In addition, our results suggest that the use of four contrast images further improves the segmentation accuracy of different structures compared to the use of fat and water images, especially in small structure IMAT. This could be contributed by the fact that more contrast information compensates for the noise and intensity non-uniformity of a single or dual set of MRI images, making the algorithm more robust in classification of different tissues.
Previous studies have shown that automatic muscle and fat segmentation using clustering method can be challenging in subjects with very thin SAT layer or very severe fat infiltration muscles [20]. However, such segmentation errors did not appear in our extreme subjects using the proposed segmentation scheme with four contrasts. This could be due to that we employed the machine learning method for the segmentation. Machine learning techniques (e.g. artificial neural networks) has been shown to outperform fuzzy clustering method in non-homogenous regions such as abnormal brain with edema, tumor, etc. [29]. In the cases with very thin SAT or large IMAT area, SAT and muscle region become more non-homogenous. As a result, machine learning technique would work better than clustering method in terms of classifying different tissues.
We notice that a smaller DSC for the segmentation of IMAT was obtained. This is likely due to the small size and irregular shape of the IMAT structure. The voxel effects [30] become very significant in each step of the scheme and in the calculation of DSC. Any error in the procedures results in a relatively significant mismatch.
A limitation of this study is that a different preliminary segmentation of training set needs to be carried out for each MRI protocol. Nonetheless, once a small set of training data has been segmented for the machine learning algorithms, the scheme becomes fast, automatic and can be applied to any unseen data, making it suitable for large scale use in routine clinical practice.
Conclusion
This paper presents an accurate machine learning based segmentation method for thigh MRI images with Dixon technique. The method becomes fully automatic after initial segmentation of training samples. The proposed method incorporates combined image space of all four contrasts provided by Dixon sequence, improving the accuracy of tissue classification.
References
Wells JCK, Fewtrell MS (2006) Measuring body composition. Arch Dis Child 91(7):612–617
Do Lee C, Blair SN, Jackson AS (1999) Cardiorespiratory fitness, body composition, and all-cause and cardiovascular disease mortality in men. Am J Clin Nutr 69(3):373–380
Park SW, Goodpaster BH, Strotmeyer ES, de Rekeneire N, Harris TB, Schwartz AV, Tylavsky FA, Newman AB (2006) Decreased muscle strength and quality in older adults with type 2 diabetes. The health, aging, and body composition study. Diabetes 55(6):1813–1818
Collaboration PS (2009) Body-mass index and cause-specific mortality in 900,000 adults: collaborative analyses of 57 prospective studies. Lancet 373(9669):1083–1096
Finucane MM, Stevens GA, Cowan MJ, Danaei G, Lin JK, Paciorek CJ, Singh GM, Gutierrez HR, Lu Y, Bahalim AN (2011) National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9· 1 million participants. Lancet 377(9765):557–567
Lim JP, Leung BP, Ding YY, Tay L, Ismail NH, Yeo A, Yew S, Chong MS (2015) Monocyte chemoattractant protein-1: a proinflammatory cytokine elevated in sarcopenic obesity. Clin Interv Aging 10:605–609
Wang C, Bai L (2012) Sarcopenia in the elderly: basic and clinical issues. Geriatr Gerontol Int 12(3):388–396
Addison O, Marcus RL, LaStayo PC, Ryan AS (2014) Intermuscular fat: a review of the consequences and causes. Int J Endocrinol
Ma J (2008) Dixon techniques for water and fat imaging. J Magn Reson Imaging 28(3):543–558
Armao D, Guyon JP, Firat Z, Brown MA, Semelka RC (2006) Accurate quantification of visceral adipose tissue (VAT) using water-saturation MRI and computer segmentation: preliminary results. J Magn Reson Imaging 23(5):736–741
Makrogiannis S, Serai S, Fishbein KW, Schreiber C, Ferrucci L, Spencer RG (2012) Automated quantification of muscle and fat in the thigh from water-, fat-, and nonsuppressed MR images. J Magn Reson Imaging 35(5):1152–1161
Kullberg J, Johansson L, Ahlström H, Courivaud F, Koken P, Eggers H, Börnert P (2009) Automated assessment of whole-body adipose tissue depots from continuously moving bed MRI: a feasibility study. J Magn Reson Imaging 30(1):185–193
Sadananthan SA, Prakash B, Leow MKS, Khoo CM, Chou H, Venkataraman K, Khoo EY, Lee YS, Gluckman PD, Tai ES (2015) Automated segmentation of visceral and subcutaneous (deep and superficial) adipose tissues in normal and overweight men. J Magn Reson Imaging 41(4):924–934
Wald D, Teucher B, Dinkel J, Kaaks R, Delorme S, Boeing H, Seidensaal K, Meinzer HP, Heimann T (2012) Automatic quantification of subcutaneous and visceral adipose tissue from whole-body magnetic resonance images suitable for large cohort studies. J Magn Reson Imaging 36(6):1421–1434
Leinhard OD, Johansson A, Rydell J, Smedby Ö, NystrÖm F, Lundberg P, Borga M (2008) Quantitative abdominal fat estimation using MRI. In: Pattern recognition, 2008. ICPR 2008. 19th International Conference on, 2008. IEEE, pp 1–4
Karlsson A, Rosander J, Romu T, Tallberg J, Grönqvist A, Borga M, Dahlqvist Leinhard O (2015) Automatic and quantitative assessment of regional muscle volume by multi-atlas segmentation using whole-body water–fat MRI. J Magn Reson Imaging 41(6):1558–1569
Wang D, Shi L, Chu WC, Hu M, Tomlinson B, Huang W-H, Wang T, Heng PA, Yeung DK, Ahuja AT (2015) Fully automatic and nonparametric quantification of adipose tissue in fat–water separation MR imaging. Med Biol Eng Comput 53(11):1247–1254
Joshi AA, Hu HH, Leahy RM, Goran MI, Nayak KS (2013) Automatic intra-subject registration-based segmentation of abdominal fat from water–fat MRI. J Magn Reson Imaging 37(2):423–430
Kullberg J, Karlsson AK, Stokland E, Svensson PA, Dahlgren J (2010) Adipose tissue distribution in children: automated quantification using water and fat MRI. J Magn Reson Imaging 32(1):204–210
Valentinitsch A, Karampinos DC, Alizai H, Subburaj K, Kumar D, Link MT, Majumdar S (2013) Automated unsupervised multi-parametric classification of adipose tissue depots in skeletal muscle. J Magn Reson Imaging 37(4):917–927
Tustison NJ, Avants BB, Cook P, Zheng Y, Egan A, Yushkevich P, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29(6):1310–1320
Kass M, Witkin A, Terzopoulos D (1988) Snakes: active contour models. Int J Comput Vision 1(4):321–331
Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31(3):1116–1128
Huang G-B, Zhu Q-Y, Siew C-K (2006) Extreme learning machine: theory and applications. Neurocomputing 70(1):489–501
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
MATLAB (2010) Version 7.11.1 edn. The MathWorks Inc., Natick, Massachusetts
Conover WJ (1998) Practical nonparametric statistics
StataCorp (2015) Stata statistical software: release 14. StataCorp LP, College Station
Hall LO, Bensaid AM, Clarke LP, Velthuizen RP, Silbiger MS, Bezdek JC (1992) A comparison of neural network and fuzzy clustering techniques in segmenting magnetic resonance images of the brain. IEEE Trans Neural Netw 3(5):672–682
Rajon DA, Jokisch DW, Patton PW, Shah AP, Watchman CJ, Bolch WE (2002) Voxel effects within digital images of trabecular bone and their consequences on chord-length distribution measurements. Phys Med Biol 47(10):1741
Acknowledgments
This study was funded by Lee Foundation Grant 2013 (GERILABS—Longitudinal Assessment of Biomarkers for characterisation of early Sarcopenia and predicting frailty and functional decline in community-dwelling Asian older adults Study).
We would like to acknowledge Assistant Professor Poh Chueh Loo from Nanyang Technological University for his valuable advices in segmentation techniques. We would also like to extend our thanks to Mr. Samuel Neo, Medical Social Worker, Department of Continuing and Community Care, Tan Tock Seng Hospital, for his assistance in recruitment through the various Senior Activity Centres (SAC). We extend our appreciation to the following SACs (Wesley SAC, Care Corner SAC, Xin Yuan Community Service, Potong Pasir Wellness Centre, Tung Ling Community Services (Marine Parade and Bukit Timah), Viriya Community Services-My Centre@Moulmein, House of Joy) and the study participants who have graciously consented to participation in the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Rights and permissions
About this article
Cite this article
Yang, Y.X., Chong, M.S., Tay, L. et al. Automated assessment of thigh composition using machine learning for Dixon magnetic resonance images. Magn Reson Mater Phy 29, 723–731 (2016). https://doi.org/10.1007/s10334-016-0547-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-016-0547-2