Abstract
Past research has sought to improve the production of cyclopropane fatty acids by the oleaginous yeast Yarrowia lipolytica by heterologously expressing the E. coli fatty acid synthase gene and improving cultivation processes. Cyclopropane fatty acids display properties that hold promise for biofuel applications. The E. coli fatty acid synthase gene was introduced into several genetic backgrounds of the yeast Y. lipolytica to optimize lipid synthesis; the mean cyclopropane fatty acid productivity was 43 mg L−1 h−1 on glucose, and the production rate reached its maximum (3.06 g L−1) after 72 h of cultivation in a bioreactor. The best strain (JMY6851) overexpressed simultaneously the E. coli cyclopropane fatty acid synthase gene under a hybrid promoter (hp8d) and Y. lipolytica LRO1 gene. In fed-batch process using crude glycerol as carbon source, JMY6851 strain displayed high lipid accumulation (78% of dry cell weight) and high biomass production (56 g L−1). After 165 h of cultivation, cyclopropane fatty acids represented 22% of the lipids produced; cyclopropane fatty acid productivity (103.3 mg L−1 h−1) was maximal at 72.5 h of cultivation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The oleaginous yeast Yarrowia lipolytica can be used as a cell factory taking advantage of its ability to accumulate large amounts of lipids (> 40% of cell biomass), which are stored as triacylglycerols (TAGs) in lipid bodies [1,2,3]. It has been the focus of extensive research and has been modified to produce bioproducts (e.g., lipids, proteins, organic acids, polysaccharides, food additives, etc.) of great value for the biotechnology, pharmaceutical, and food industries [4,5,6]. It has many desirable traits, such as the ability to produce lipids from a wide variety of low-cost substrates (e.g., glycerol) [7, 8].
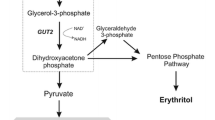
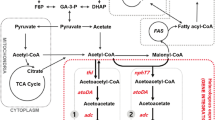
In Y. lipolytica, fatty acid (FA) biosynthesis and the storage of lipids as TAGs or sterol esters (SEs) are processes that have been well described. The TAG precursor glycerol-3-phosphate (G3P) is produced from dihydroxyacetone phosphate (DHAP) during glycolysis via the action of a glycerol-3-phosphate dehydrogenase (GPD1) [9,10,11]. Then, using G-3-P, diacylglycerol (DAG) can be produced. DAG is a crucial precursor in triglyceride synthesis, and it takes part in a series of enzymatic reactions that include the synthesis of lysophosphatidic acid (LPA) catalyzed by a Gly3P acyltransferase (SCT1). LPA acyltransferase (SLC1) adds an additional acyl group to LPA, generating phosphatidic acid (PA) and resulting in the release of DAG through the action of PA phosphohydrolase (PAP) [12]. DAG can also be produced via the phospholipid (PL) biosynthesis pathway. Choline phosphotransferase 1 (CPT1), which is a DAG choline phosphotransferase, transfers a phosphate group, releasing a DAG [13]. The final step in TAG biosynthesis is the addition of an acyl group to DAG, either via the action of a DAG acyltransferase with acyl-CoA serving as an acyl donor or via the action of a PL DAG acyltransferase (LRO1) with a phospholipid serving as an acyl donor [1, 12]. In Y. lipolytica, there are two DAG acyltransferases (DGAT), one is a member of the DGAT1 family (DGA2) and the other is a member of the DGAT2 family (DGA1).
Several genetic modifications were shown to push and pull lipid accumulation via metabolic engineering, as recently reviewed by Ledesma and Nicaud [14]. Among them is the deletion of the competitive pathways such as the degradation of FAs, which, in Y. lipolytica, takes place exclusively via β-oxidation in peroxisomes. The first step is catalyzed by six acyl-CoA oxidases (Aox1–6), which are encoded by the POX1–POX6 genes, respectively. A strain in which all six genes were deleted (pox1-6Δ) could not degrade FAs [10, 15]. Similarly, the deletion of genes involved in TAG remobilization (i.e., TGL3 and TGL4, which encode triacylglycerol lipases) results in strains that accumulate larger amounts of lipids [16, 17].
It has been demonstrated that overexpression of key genes could also boost lipid accumulation. The rate of DGAT activity can limit the rate of lipid accumulation, because DGAT plays an essential role in the quantitative and qualitative flux of FAs into storage TAGs [9, 18]. Overexpression of DGA1 or DGA2 resulted in a high level of lipid accumulation (70% of DCW) in recombinant Y. lipolytica [8, 18, 19]. Lipid accumulation was shown to be dependent on DGA1 and DGA2 gene copy number [19]. Another modification, such as the overexpression of GPD1, a gene that encodes a glycerol-3-phosphate dehydrogenase, also displays an enhanced ability to accumulate lipids due to the increase of the glycerol-3-phosphate pool (up to 65–75% of DCW) [10].
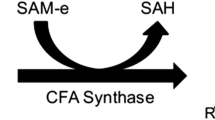
Y. lipolytica is considered to be an excellent biological platform for exploring how genetic engineering can yield both common and unusual lipids [1, 20, 21]. Among the unusual lipids, cyclopropane fatty acids (CFAs) display physicochemical properties of industrial interest (e.g., oxidative resistance, fluidity at low temperatures): they are excellent compounds for creating a variety of chemical products, cosmetics, paints, and lubricants [22,23,24], to name a few examples. CFAs result from post-synthetic modification of the phospholipid bilayer, which primarily occurs when cultures of E. coli or other bacterial species enter the stationary phase of growth [23, 25, 26]. The reaction is catalyzed by cyclopropane fatty-acyl-phospholipid synthase (CFAS), which transfers a methylene group from S-adenosyl-L-methionine to either hexadecenoyl or octadecenoyl, replacing the phospholipid’s double bond. Unsaturated cis-Δ9 and cis-Δ11 FAs are preferentially employed in cyclization. Palmitoleic (Δ9 C16: 1), oleic (Δ9 C18: 1), and vaccenic (Δ11 C18: 1) acids can serve as precursors in the synthesis of C17 CFA, C19 CFA, and lactobacillic acid, respectively [26]. Several genes encoding CFAS have been isolated from organisms such as Lactobacillus, E. coli, Brucella arbutus, Pseudomonas putida, Sinorhizobium meliloti, and Mycobacterium tuberculosis [27,28,29,30] as well as from plant species [31, 32]. Some have been overexpressed in various organisms, leading to an increase in CFAs production [28, 29, 31,32,33,34,35,36]. This research has shown that both C17:0 CFA and C19:0 CFA are produced upon the expression of the E. coli CFAS. CFAs level was dependent on the promoter strength used for CFAS expression [28, 30].
For gene overexpression, several promoters have been developed for Y. lipolytica. One of the strong constitutive promoters was developed by Novo using the TEF1 gene, which encodes the translation elongation factor-1α [37]. Later, hp4d promoter was discovered by Madzak et al. [38]; it contains four UAS1 tandem elements from the XPR2 promoter. Madzak and colleagues showed that promoter strength increased as the number of UAS1 tandem elements increased [38]. Dulermo et al. demonstrated that optimal expression levels were enzyme specific for various enzymes, whose genes were expressed under different promoters [39]. Markham and Alper use the strong hybrid promoter UAS1B16-TEF (contains 16 copies of UAS1 upstream of the TEF promoter) for the expression of ecCFAS). Using bioreactor fermentation, the researchers attained CFAs concentrations of 3 g L−1 and CFAs representation of 32% in the total lipid pool [30]. Czerwiec and coworkers optimized CFAs production using six promoters of different strength. The best production was obtained with the hp8d promoter (8 copies of UAS1), allowing to produce 2.33 g L−1 with CFAs representation of 45% of the total lipid content [28].
In this study, we aimed to optimize CFAs production in Y. lipolytica by introducing a copy of the CFAS gene from E. coli into Y. lipolytica strains with different genetic backgrounds. First, we characterized the strains’ CFAs productivity in a bioreactor. Then, CFAs production was analysed under various conditions: in flasks versus in a bioreactor and in the presence of different pure or crude carbon sources (i.e., glucose, glycerol, and sucrose). To increase CFAs yield, we also overexpressed the LRO1 gene. We additionally examined how CFAs productivity was affected by the use of a fed-batch strategy, where glucose and crude glycerol were employed as carbon sources. CFAs productivity was greatest (103.3 mg L−1 h−1) in the strain that expressed both the LRO1 gene and the EcCFAS gene and that was cultured for 72.5 h via a fed-batch process.
Materials and methods
Yeast strains and culture media
The Y. lipolytica strains used in this study were derived from the auxotrophic strain Po1d (MATA ura3-302 leu2-270 xpr2-322, U−L−) [40]. The auxotrophic strain JMY3122 (pox1-6∆, tgl4∆, Leu−, Ura−) [41] is unable to degrade FAs and remobilized TAGs. The auxotrophic obese strain JMY3820 (U−L−) was derived from the prototrophic obese strain JMY3501 (pox1-6Δ tgl4Δ pTEF-GPD1 pTEF-DGA2) [41, 42], which contains pox1-6Δ tgl4Δ and overexpresses YlDGA2 and YlGPD1, to push and pull TAG biosynthesis [18, 43]. Strains carrying these modifications will be referred to as “obese” in this study for simplification. All strains used in this study are described in Table 1. Two rich media—yeast extract peptone dextrose (YPD) medium and yeast extract peptone glycerol (YPG) medium—were used to preculture yeasts at 28 °C and 160 rpm. The media contained 10 g L−1 of yeast extract (Difco, Paris, France), 20 g L−1 of Bacto™ Peptone (Difco, Paris, France), and either 20 g L−1 of glucose (Merck, Fontenay-sous-Bois, France) or 20 g L−1 of glycerol (Sigma-Aldrich, St. Louis, USA), respectively. To select the transformants, minimal glucose (YNB) media were used; they were prepared as described previously [18]. The media and growth conditions used for E. coli have been described elsewhere [44].
In order to avoid confusion, we use the following nomenclature: for the gene we use CFA, for the enzyme we use CFAS and CFAs for cyclopropane fatty acids production.
Vectors
All the plasmids, inserts, and promoters used in this study are listed in Table 2. Plasmids were constructed as described in Dulermo and Nicaud [10] and Czerwiec et al. [28].
Bioreactor and flask culture conditions
The strains GY1005, GY1070, and JMY5578 were cultivated in a 5-L bioreactor (TRYTONI, Pierre Guérin, Niort, France) with a final operating volume of 3.5 L at 28 °C. A pH level of 6 was maintained using KOH 3 M, an agitation speed of 1000 rpm, and the dissolved oxygen was set up at 20% saturation level. Culture media (initial volume: 3 L) were inoculated at 3% with cultures grown overnight in YPD in shake flasks. The media contained 15 g L−1 of yeast extract, 77 g L−1 of dextrose, 12 g L−1 of NH4Cl, 3 g L−1 of KH2PO4, and 1 mL L−1 of the defoamer EROL™ ANBIO 1397 K (PMC OUVRIE, France). After 25 h of cultivation, a fed-batch process using dextrose (990 g L−1) at a flow rate of 2 g L−1 h−1 was employed. The strain JMY5578 was cultivated in 500 mL flasks containing 100 mL of culture medium (55 g L−1 of dextrose, 5 g L−1 of yeast extract, 5 g L−1 of NH4Cl, and 3 g L−1 of KH2PO4). The strain JMY6851 was cultivated in rich medium using a fed-batch process. A 5-L bioreactor (Sartorius Stedim Biotech, Göttingen, Germany) was employed; it contained a medium (initial volume: 2 L) with 10 g L−1 of yeast extract, 20 g L−1 of peptone, and 5 g L−1 of glycerol. A fed-batch process using crude glycerol at a flow rate of 6 g h−1 was utilized as described elsewhere [6].
Cultivation of Yarrowia lipolytica using different carbon sources
Strain responses to different substrates were evaluated using the protocol previously described [45]. Briefly, the strain JMY5578 was grown in a synthetic medium containing 0.85 g L−1 of yeast nitrogen base without amino acids and ammonium sulfate (BD Difco, Franklin Lakes, USA), 50 mM of phosphate buffer (pH 6.8), and 4.6 g L−1 of ammonium chloride to which different carbon sources were added. More specifically, glucose (Sigma Aldritch), glycerol (Fisher Scientific), sucrose (Sigma Aldritch), glucose syrup (Roquette, 98% m/m), crude glycerol (SAS PIVERT, Rouen 2, 80% m/m), and molasses (Lesaffre, 54% m/v of sucrose) were used. The carbon sources were mixed with ammonium chloride to attain a molar carbon-to-nitrogen ratio of 35.
Quantification of dry cell weight
Biomass was quantified by measuring dry cell weight (DCW). Fifteen-mL samples of culture medium were centrifuged at 8000 rpm (772.6 g) for 10 min. Pellets were washed twice by resuspending them in one volume of distilled water to remove any traces of the medium. They were then dried overnight at 105 °C, and their DCW was determined. Biomass was also quantified by measuring sample OD at 600 nm.
Lipid extraction
Lipids were extracted using the modified Folch method [46]. First, samples were ground and homogenized using a high-throughput homogenizer (Precellys 24, Ozyme, France). They were then lyophilized using an Alpha 1–4 LD Plus freeze dryer (Martin Christ, Germany). Subsequently, 100 mg of the lyophilized biomass was placed in screw cap microtubes (3 replicates per sample); the tubes contained a ceramic bead (diameter: 4.9 mm). The tubes were agitated (3 × 30 s, 30 Hz). One mL of a chloroform/methanol mixture (2:1, v:v) was added. The tubes were again agitated (3 × 30 s, 30 Hz) and then centrifuged for 3 min at 6,000 g. The organic phase was transferred to a new tube, and the extraction process continued. Non-lipid components were removed by adding 2 mL of a KCl (1 M)/methanol solution [4:1 (v/v)] containing 0.034% of MgCl2; the resulting mixture was vortexed for 30 s and centrifuged for 2 min at 5000 g. Finally, the lower phase was collected and evaporated under a nitrogen stream at 50 °C. The recovered lipids were weighed and analyzed using gas chromatography–flame ionization detection (GC-FID). Lipid production dynamics were also quantified via the direct transmethylation of freeze-dried cells. Lipids were converted into their fatty acid methyl esters (FAMEs) using the method described by Browse et al. [47]; the FAMEs were analyzed using GC as described elsewhere [48].
Identification of fatty acid methyl esters
FAMEs were prepared from the extracted lipids using the method described by Merlier et al. 2018 [49]. They were then analyzed using gas chromatography–mass spectrometry (GC–MS). A Trace GC Ultra chromatograph (Thermo Fisher, France) equipped with a Restek Rt-2560 capillary column (100 m × 0.25 mm, 0.2 μm, SGE Analytical Science, UK) was employed. The temperature program was as follows: 170 °C for 2 min after injection, a 5-min hold at 200 °C (ramp rate of 3 °C/min), a 3-min hold at 210 °C (ramp rate of 0.5 °C/min), and finally a 2-min hold at 250 °C (ramp rate of 20 °C/min). The carrier gas was helium (flow rate of 2.5 mL/min). One-μL samples of FAMEs was introduced in split injection mode. Injector temperature was 230 °C. FAMEs were separated using the electron ionization (EI) method of MS. A quadrupole mass analyzer (QMA) was used; a magnetic field was applied to the QMA to enhance its performance. Compounds were identified based on their m/z ratios; reference spectra from the 2008 NIST database were used.
Quantification of glucose and glycerol levels
Rapid glucose levels in bioreactor experiments were determined using the dinitrosalicylic acid (DNS) method [50]. Glucose and glycerol quantifications were also performed using high-performance liquid chromatography (HPLC), as described in Larroude et al. [6].
Results and discussion
Metabolic pathway for producing cyclopropane fatty acids in Yarrowia lipolytica and a strategy for building of recombinant strains
First, the strain GY1005 (pTEF-EcCFA) was created by introducing the EcCFA gene expressed under the control of the strong constitutive promoter pTEF into the strain JMY3122. GY1005 contained the deletion of the TGL4 gene (tgl4∆) and deletion of the six POX genes (pox1-6∆). The strain GY1070 (obese pTEF-EcCFA) was derived from JMY3820, an obese strain (tgl4∆ pox1-6∆ GPD1 DGA2) previously described by Lazar et al. [41] and contained the E. coli CFAS expressed under the control of the TEF promoter. Strain JMY5578 contained the pTEF-EcCFA expression cassette and co-expressed the Y. lipolytica LRO1 gene with the goal of improving transfer of CFAs into TAG. The strain JMY6851 (obese hp8d-EcCFA pTEF-LRO1) was obtained optimizing CFAs production (via the expression of EcCFA under the strong promoter hp8d), and overexpressing LRO1. JMY3501 was the control obese strain (Table 1).
CFAs production dynamics in the strains GY1005 and GY1070
The abilities of the strains GY1005 and GY1070 to effectively produce CFAs were compared and contrasted. The two strains were cultivated for 136 h. Samples were taken every 24 h, and biomass, total lipid levels, and CFAs levels were measured (Fig. 1a, b, Table 3). After 72 h of cultivation, accumulated lipids accounted for 12% of DCW and 19 g.L−1 of biomass in GY1005 and 36% of DCW and 26 g.L−1 of biomass in GY1070 (Fig. 1b, Table 3).
At 136 h of cultivation, biomass levels were higher in GY1005 than in GY1070 (29.7 g L−1 vs. 23 g L−1, respectively). Levels of accumulated lipids were unchanged (12% of DCW) for GY1005; they were higher for GY1070 (48% of DCW). Total lipid levels were fourfold greater in GY1070 than in GY1005. The combined overexpression of DGA2 and GPD1 in GY1070 led to a total lipid yield that was 300% greater than that in GY1005. However, there was only a slight increase in CFAs production in the two strains. They reached their maximum at 136 h: they were 1.2 g L−1 (33.8% of total lipids) and 1.5 g L−1 (13.2% of total lipids) in GY1005 and GY1070, respectively. Similar results were obtained by Czerwiec and colleagues: their best CFA producer had lower lipid content [28]. In contrast, the highest levels of CFAs productivity were achieved within 72 h of cultivation: they were 10 mg L−1 h−1 and 17 mg L−1 h−1 for GY1005 and GY1070, respectively. Consequently, in subsequent experiments, Y. lipolytica strains were only cultivated for 72 h.
More detailed analysis of the lipid production profiles revealed that, over time, there was an increase in C17 CFA and C19 CFA accumulation and a concomitant decrease in the compounds’ precursors (C16:1 vs. C18:1 and C18:2, respectively; Supplementary data Table S1). In GY1005, CFAs represented 33.8 ± 1.1% of TFAs at 136 h of cultivation; this percentage was 13.2 ± 0.1% in GY1070. This result demonstrates that, in GY1070, increased lipid accumulation resulted in decreased CFAs content. It also suggests that, in this background, there are constraints on the expression of CFAS or of phospholipid DAG acyltransferase (LRO1).
Expression of LRO1 gene improves CFAs production
To further improve CFAs production, there were several additional genetic modifications that were possible, such as overexpressing the gene encoding phospholipase A2 (PLA2), to release FA-CFAs from PL-CFAs, or overexpressing the gene encoding phospholipid diacylglycerol acyltransferase (LRO1), to transfer the FA-CFA from PL-CFA and generate DAG or DAG-CFA. In addition, we recently reported that levels of CFAS can be limiting. Overexpression of the gene encoding CFAS under a strong promoter increased the relative representation of CFAs among TFAs; however, there was a concomitant decrease in total lipids [28].
Therefore, in an effort to enhance CFAs synthesis, the gene EcCFA was cloned under the control of the hybrid promoter (p4UAS1-TEF) [28] and used to transform the strain JMY3122. This transformant was used to generate the strain JMY5578, via the overexpression of the gene LRO1, which increased TAG synthesis by increasing the transfer of FAs from PLs to TAGs (Table 1). CFAs production dynamics (biomass, lipid levels, and CFAs levels) were compared for JMY5578 grown in flasks versus in a bioreactor (Fig. 2). A batch process was used in the flasks, and a fed-batch process was used in the bioreactor; glucose was the carbon source. Cultivation lasted 72 h. Additional production parameters are described in “Materials and methods”.
Levels of biomass, lipids, and CFAs over time for the strain JMY5578 (p4UAS1-TEF-CFA, pTEF-LRO1). The level of glucose (the carbon source) is also indicated. a Results from batch cultivation in flasks. b Results from fed-batch cultivation in the bioreactor. The bars represent the standard deviation calculated from the three technical replicates
The results show that there was a trade-off between the production of biomass, lipids, and CFAs in the two processes (Fig. 2). In the batch process in the flasks, the final levels of biomass, lipids, and CFAs were 9.1 g L−1, 1.8 g L−1, and 0.4 g L−1, respectively (Fig. 2a, Table S2). Thus, biomass yield and CFAs productivity were 0.22 g g−1 of glucose consumed and 6 mg L−1 h−1, respectively. In the fed-batch process in the bioreactor, the final levels of biomass, lipids, and CFAs were 58.3 g L−1, 8.9 g L−1, and 3.1 g L−1, respectively (Fig. 2b, Table S3). Biomass yield and CFAs productivity were therefore 0.46 g g−1 of glucose consumed and 40 mg L−1 h−1, respectively. Thus, compared to the batch process, the fed-batch process resulted in sixfold and sevenfold greater biomass yield and CFAs productivity, respectively.
Past research has shown that lipid accumulation is highly dependent on the carbon-to-nitrogen ratio (the C/N ratio) and is induced by nitrogen starvation [3, 40]. We thus took a closer look at how batch and fed-batch processes affected CFAs production in JMY5578.
In the batch bioprocess, the lipid content was 19.5 ± 0.4% and CFAs content was 0.44 g L−1. However, the levels of biomass and lipid-free biomass remained low (9.1 ± 0.7 g L−1 and 1.8 g L−1, respectively). In the fed-batch process, lipid content was slightly lower (15.2%) and lipid-free biomass was higher, resulting in lipid and CFAs levels of 8.9 g L−1 lipid and 3.06 g L−1, respectively. These results match those obtained in previous research focusing on how batch and fed-batch processes affect lipid production in Y. lipolytica [17].
In JMY5578, CFAs represented up to 34.5% of total lipids. Based on this figure and the CFAs level cited above for the GY1005 strain at 72 h of cultivation (29.5%), CFAs level was 17% greater in JMY5578 than in GY1005. An increase of 155% in CFAs production was also observed, from 1.2 to 3.06 g L−1 using JMY5578 strain. As previously shown by Beopoulos et al., another strain overexpressing the gene LRO1 had 40% greater ricinoleic acid production (measured in % of TFAs) than its parental strain [20].
CFAs production is affected by carbon source
When developing microbial lipid production processes, cost is an important consideration. Economically feasible methods must be developed. More specifically, it is important to expand Y. lipolytica’s ability to use a range of cheap and readily available substrates. The impacts of different carbon sources (glucose, glycerol and sucrose, which are common experimental substrates, as well as glucose syrup, crude glycerol and molasses, which are cheap commercially available substrates) on lipid content and CFAs content are shown in Fig. 3. These results indicate that the carbon source can greatly affect lipid and CFAs accumulation. Different carbon sources resulted in similar relative levels of CFAs (% of total lipids; Fig. 3a), but different levels of lipid content and, consequently, different absolute levels of CFAs. Among the common experimental substrates, pure glucose appeared to best enhance lipid accumulation, while sucrose seemed to have a limited effect (Fig. 3b). However, when glucose syrup was used, the lipid content decreased; when crude glycerol was employed, lipid content increased (2.8 fold). Crude glycerol also resulted in a CFAs accumulation level that was 2.4 times higher than the mean (Table S4). As well known, crude glycerol contains some impurities such as fatty acids methyl esters (oleic acid is the main one), salts, soap glycerides and metal ion. Xu et al. [51] have demonstrated that soap plays a role as a surfactant, which may provide some influences in the fermentation process. As has been previously shown with other metabolites, salts such as Fe, which is a cofactor of enzymes, may be involved in SAM synthesis, required for CFAs production [52, 53]. Looking at the results overall (Table S4), crude glycerol shows promise as a substrate for lipid production in Y. lipolytica. It is cheap, widely available, and a by-product in the manufacture of soaps and FAs.
Improvement of CFAs production by combining strain optimization and fermentation process.
To further improve lipid and CFAs production, we sought to boost PAP levels to increase the conversion of PL-CFA into DAG-CFA. However, while past research on PAP regulation has found that PAP expression increases in high-glucose media [54], the overexpression of PAP appears to have a limited effect on lipid accumulation (Nicaud et al., unpublished). To explore these ideas, we created the strain JMY6851, which arose from the introduction of the pTEF-LRO1 LEU2ex and the hp8d-CFAS URA3ex expression cassettes into the obese auxotrophic strain JMY3820 (Table 1). JMY6851 thus displayed the obese phenotype (overexpression of DGA2 and GPD1), the overexpression of the gene EcCFA under the strong hybrid promoter hp8d, and the overexpression of the gene LRO1 under the pTEF promoter, with a view to enhancing lipid turnover. Since crude glycerol resulted in the highest levels of lipid and CFAs production (see the previous section), JMY6851 was grown in a 5-L bioreactor using crude glycerol and a fed-batch process optimized for Y. lipolytica (see [6]). A rich medium was employed, and crude glycerol was added at a rate of 6 g h−1 after 6 h of cultivation.
At 72.5 h, the level of biomass was 54.6 g L−1; lipids represented 70% (m/m) of DCW, and 19.6% of those lipids were CFAs (Fig. 4). The lipid concentration was 38.2 g L−1, and CFAs productivity was 103.3 mg L−1 h−1. The CFAs concentration was 7.5 g L−1 (137.4 mgCFA g−1 DCW), the best achieved during the study. It was 2.5-fold greater than the CFAs level produced by JMY5578 in this study (see above) and by JMY6068 in a previous study [18]. Additional information on biomass, lipid, and CFAs production dynamics is available in the Supplementary data (Table S5).
Levels of biomass (filled circle, blue), lipid content (filled triangle, red), and relative CFAs content (filled square, green) over time for the strain JMY6851 (obese hp8d-CFA pTEF-LRO1) grown via fed-batch cultivation using rich crude glycerol [whose level is also indicated (filled diamond, purple)]
Conclusion
The oleaginous yeast Y. lipolytica has been considered since several years as the best chassis to accumulate neutral lipids at high level. In this study, CFAs synthesis was metabolically engineered in Y. lipolytica through the combined expression of genes whose products improve fatty acid biosynthesis (GPD1 and DGA2), enhance phospholipid remodeling (LRO1) and prevent lipid remobilization and degradation (tgl4∆ and pox1-6∆). Here, the overexpression of DGA2 and GPD1 in an obese strain (GY1070) increased not only the lipid content, but also has significantly contributed to increase CFAs production at 72 h of cultivation, GY1070’s CFAs level was 1.23 g L-1 when compared to 0.71 g L-1 in GY1005. Our engineering strategy led to a creation of a recombinant Y. lipolytica, in which CFAs productivity reached its maximum at 72 h for both GY1005 and GY1070 (10 and 17 mg L−1 h−1, respectively). Moreover, in the engineered strain JMY5578, the integration of LRO1 gene also facilitated the FA transfer from the sn2 of PLs to the DAG to produce the TAGs-CFA to achieve CFAs level of 34.5% in TFAs. at 72 h of cultivation. This represented a 2.5-fold increase in CFAs production. In this strain background, the carbon source generally did not affect the relative percentage of CFAs (% of total lipids).
In this work, we have also combined the metabolic engineering to a proper bioprocess strategy of culture of Y. lipolytica strains. The created JMY6851, in which the obese phenotype (overexpression of DGA2 and GPD1) was combined with the overexpression of EcCFA under the hybrid promoter hp8d and overexpression of LRO1, was cultivated in a bioreactor using a fed-batch process. Even if the carbon source generally did not affect the relative percentage of CFAs (% of total lipids), in our case, when crude glycerol was used, CFAs accumulation increased 2.4-fold probably because impurities in crude glycerol can promote CFAs accumulation, as has been previously seen with other metabolites [51,52,53]. At 72.5 h, in JMY6851, lipid accumulation (70%), the CFAs level (7.49 g L−1), and CFAs productivity (103.3 mg L-1 h-1) were significantly greater than in the other strains in this study and in previous studies.
This study clearly shows that a combination of metabolic engineering and the proper bioprocess strategy can achieve excellent levels of CFAs and CFAs productivity, compared to what has been seen in research to date [28, 30, 36]. However, the analysis of CFA in the different lipid fractions (PL and TAG) remain to be determined to identify the gene modification required for further improving CFA production.
References
Beopoulos A, Cescut J, Haddouche R et al (2009) Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res 48:375–387. https://doi.org/10.1016/j.plipres.2009.08.005
Beopoulos A, Nicaud JM (2012) Yeast : A new oil producer ? OCL-Ol Corps Gras Lipides 19:22–28. https://doi.org/10.1684/ocl.2012.0426
Ratledge C, Wynn J (2002) The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 50:1–51
Groenewald M, Boekhout T, Neuvéglise C et al (2014) Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit Rev Microbiol 40:187–206
Rywińska A, Juszczyk P, Wojtatowicz M et al (2013) Glycerol as a promising substrate for Yarrowia lipolytica biotechnological applications. Biomass Bioenergy 48:148–166. https://doi.org/10.1016/j.biombioe.2012.11.021
Larroude M, Celinska E, Back A et al (2018) A synthetic biology approach to transform Yarrowia lipolytica into a competitive biotechnological producer of β-carotene. Biotechnol Bioeng. https://doi.org/10.1002/bit.26473
Ledesma-Amaro R, Nicaud JM (2016) Metabolic engineering for expanding the substrate range of Yarrowia lipolytica. Trends Biotechnol. https://doi.org/10.1016/j.tibtech.2016.04.010
Lazar Z, Neuvéglise C, Rossignol T et al (2017) Characterization of hexose transporters in Yarrowia lipolytica reveals new groups of sugar porters involved in yeast growth. Fungal Genet Biol. https://doi.org/10.1016/j.fgb.2017.01.001
Beopoulos A, Nicaud JM, Gaillardin C (2011) An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl Microbiol Biotechnol 90:1193–1206. https://doi.org/10.1007/s00253-011-3212-8
Dulermo T, Nicaud JM (2011) Involvement of the G3P shuttle and Β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng 13:482–491. https://doi.org/10.1016/j.ymben.2011.05.002
Albertyn J, Hohmann S, Thevelein JM, Prior BA (1994) GPD1, which encodes Glycerol-3-Phosphate Dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol 14:4135–4144
Beopoulos A, Mrozova Z, Thevenieau F et al (2008) Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Env Microbiol 74:7779–7789. https://doi.org/10.1128/AEM.01412-08
Morash SC, Mcmaster CR, Hjelmstad RH et al (1994) Studies employing Saccharomyces cerevisiae CPT1 and EPT1 null mutants implicate the CPT1 gene in coordinate regulation of phospholipid biosynthesis. J Biol Chem 269:28769–28776
Ledesma Amaro R, Nicaud JM (2016) Yarrowia lipolytica as a biotechnological chassis to produce usual and unusual fatty acids. Prog Lipid Res 61:40–50. https://doi.org/10.1016/j.plipres.2015.12.001
Wang HJ, Le Dall MT, Waché Y et al (1999) Evaluation of acyl coenzyme A oxidase (Aox) isozyme function in the n-alkane-assimilating yeast Yarrowia lipolytica. J Bacteriol 181:5140–5148
Dulermo T, Tréton B, Beopoulos A et al (2013) Characterization of the two intracellular lipases of Y. lipolytica encoded by TGL3 and TGL4 genes: New insights into the role of intracellular lipases and lipid body organisation. Biochim Biophys Acta 1831:1486–1495. https://doi.org/10.1016/j.bbalip.2013.07.001
Friedlander J, Tsakraklides V, Kamineni A et al (2016) Engineering of a high lipid producing Yarrowia lipolytica strain. Biotechnol Biofuels 9:1–12. https://doi.org/10.1186/s13068-016-0492-3
Gajdos P, Nicaud JM, Rossignol T, Certik M (2015) Single cell oil production on molasses by Yarrowia lipolytica strains overexpressing DGA2 in multicopy. Appl Microbiol Biotechnol 99:8065–8074. https://doi.org/10.1007/s00253-015-6733-8
Beopoulos A, Haddouche R, Kabran P et al (2012) Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl Microbiol Biotechnol 93:1523–1537. https://doi.org/10.1007/s00253-011-3506-x
Beopoulos A, Verbeke J, Bordes F et al (2014) Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 98:251–262. https://doi.org/10.1007/s00253-013-5295-x
Ledesma-Amaro R, Dulermo R, Niehus X, Nicaud J-M (2016) Combining metabolic engineering and process optimization to improve production and secretion of fatty acids. Metabol Eng. https://doi.org/10.1016/j.ymben.2016.06.004
Svensson L, Hansson U, Gronowitz S, Klingstedt T (1997) The relationship between the structure of monoalkyl branched saturated triacylglycerols and some physical properties. Lipids 32:661–666. https://doi.org/10.1007/s11745-997-0084-2
Cronan JE, Reed R, Taylor FR, Jacksont MB (1979) Properties and biosynthesis of cyclopropane fatty acids in Escherichia coli. J Bacteriol 138:118–121
Duhot P, Gontier E, Thomas D, et al. (1999) Procedé de production d’acides gras ramifiés au moyen de plantes genétiquement modifiées. Patent PCT/FR 9802116. Europe, Canada, Etats Unis, Japon, WOFR9802116
Cronan JE, Nunn WD, Batchlor JG (1974) Studies on the biosynthesis of cyclopropane fatty acids in Escherichia coli. Biochim biophys acta 348:63–75
Grogan DW, Cronan JE (1997) Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol Rev MMBR 61:429–441
Montanari C, Sado Kamdem SL, Serrazanetti DI et al (2010) Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol 27:493–502. https://doi.org/10.1016/j.fm.2009.12.003
Czerwiec Q, Idrissi Taghki A, Imatoukene N et al (2019) Optimisation of cyclopropane fatty acids production in Yarrowia lipolytica. Yeast 36:143–151. https://doi.org/10.1002/yea.3379
Grogan DW, Cronan JE (1984) Cloning and manipulation of the Escherichia coli cyclopropane fatty acid synthase gene : physiological aspects of enzyme overproduction. J Bacteriol 158:286–295
Markham KA, Alper HS (2018) Engineering Yarrowia lipolytica for the production of cyclopropanated fatty acids. J Ind Microbiol Biotechnol. https://doi.org/10.1007/s10295-018-2067-8
Bao X, Katz S, Pollard M, Ohlrogge J (2002) Carbocyclic fatty acids in plants: biochemical and molecular genetic characterization of cyclopropane fatty acid synthesis of Sterculia foetida. Proc Natl Acad Sci USA 99:7172–7177. https://doi.org/10.1073/pnas.092152999
Bao X, Thelen JJ, Bonaventure G, Ohlrogge JB (2003) Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. J Biol Chem 278:12846–12853. https://doi.org/10.1074/jbc.M212464200
Zhao Y, Hindorff L, Chuang A et al (2003) Expression of a cloned cyclopropane fatty acid synthase gene reduces solvent formation in Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol 69:2831–2841. https://doi.org/10.1128/AEM.69.5.2831
Yu X-H, Rawat R, Shanklin J (2011) Characterization and analysis of the cotton cyclopropane fatty acid synthase family and their contribution to cyclopropane fatty acid synthesis. Plant Biol 11:97. https://doi.org/10.1186/1471-2229-11-97
Yu X, Prakash RR, Sweet M, Shanklin J (2014) Coexpressing Escherichia coli cyclopropane synthase with Sterculia foetida lysophosphatidic acid acyltransferase enhances cyclopropane fatty acid accumulation. Plant Physiol 164:455–465. https://doi.org/10.1104/pp.113.230953
Machida S, Shiraiwa Y, Suzuki I (2016) Construction of a cyanobacterium synthesizing cyclopropane fatty acids. Biochim Biophys Acta 1861:980–987. https://doi.org/10.1016/j.bbalip.2016.05.012
Muller Sven ST (1998) Comparison of expression systems in the yeasts Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast 14:1267–1283
Madzak C, Blanchin-Roland S, Cordero Otero RR, Gaillardin C (1999) Functional analysis of upstream regulating regions from the Yarrowia lipolytica XPR2 promoter. Microbiology 145:75–87
Dulermo R, Brunel F, Dulermo T et al (2017) Using a vector pool containing variable-strength promoters to optimize protein production in Yarrowia lipolytica. Microb Cell Fact 16:31. https://doi.org/10.1186/s12934-017-0647-3
Barth G, Gaillardin C (1996) Yarrowia lipolytica. In: Wolf K (ed) Non conventional yeasts in biotechnology. Springer, Berlin, pp 314–388
Lazar Z, Dulermo T, Neuvéglise C et al (2014) Hexokinase a limiting factor in lipid production from fructose in Yarrowia lipolytica. Metab Eng 26:89–99. https://doi.org/10.1016/j.ymben.2014.09.008
Sagnak R, Cochot S, Molina-Jouve C et al (2018) Modulation of the glycerol phosphate availability led to concomitant reduction in the citric acid excretion and increase in lipid content and yield in Yarrowia lipolytica. J Biotechnol 265:40–45. https://doi.org/10.1016/j.jbiotec.2017.11.001
Dulermo R, Gamboa-Meléndez H, Dulermo T et al (2014) The fatty acid transport protein Fat1p is involved in the export of fatty acids from lipid bodies in Yarrowia lipolytica. Yeast Res 14:883–896. https://doi.org/10.1111/1567-1364.12177
Sambrook J, Fritsch E, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, New York
Back A, Rossignol T, Krier F et al (2016) High-throughput fermentation screening for the yeast Yarrowia lipolytica with real-time monitoring of biomass and lipid production. Microb Cell Fact 15:147. https://doi.org/10.1186/s12934-016-0546-z
Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226:497–509
Browse J, Mccourt PJ, Somerville CR (1986) Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem 152:141–145
Park YK, Dulermo T, Amaro RL, Nicaud JM (2018) Biotechnology for biofuels optimization of odd chain fatty acid production by Yarrowia lipolytica. Biotechnol Biofuels 11:1–12. https://doi.org/10.1186/s13068-018-1154-4
Merlier F, Imatoukene N, Octave S et al (2018) Spectrometry method for separation and characterization of 3-hydroxymethyl pyridine ester of fatty acids at low levels. J Chromatogr A 1575:72–79. https://doi.org/10.1016/j.chroma.2018.09.010
Sumner J, Noback C (1924) The estimation of sugar in diabetic urine, using dinitrosalicylic acid. J Biol Chem 62:290–297
Xu J, Zhao X, Wang W et al (2012) Microbial conversion of biodiesel byproduct glycerol to triacylglycerols by oleaginous yeast Rhodosporidium toruloides and the individual effect of some impurities on lipid production. Biochem Eng J 65:30–36. https://doi.org/10.1016/j.bej.2012.04.003
Chatzifragkou A, Papanikolaou S (2012) Effect of impurities in biodiesel-derived waste glycerol on the performance and feasibility of biotechnological processes. Appl Microbiol Biotechnol 95:13–27. https://doi.org/10.1007/s00253-012-4111-3
Samul D, Leja K, Grajek W (2014) Impurities of crude glycerol and their effect on metabolite production. Ann Microbiol 64:891–898. https://doi.org/10.1007/s13213-013-0767-x
Hardman D, McFalls D, Fakas S (2017) Characterization of phosphatidic acid phosphatase activity in the oleaginous yeast Yarrowia lipolytica and its role in lipid biosynthesis. Yeast 34:83–91. https://doi.org/10.1002/yea.3216
Acknowledgements
This work was performed in collaboration with SAS PIVERT (www.institut-pivert.com), which is an institute for energy transition (Institut pour la Transition Energétique [ITE] PIVERT)). SAS PIVERT received funding from the French government’s Investments for the Future programme (Investissements d’Avenir). This study was also supported by Investments for the Future funding (ANR-001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no financial or commercial conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Imatoukene, N., Back, A., Nonus, M. et al. Fermentation process for producing CFAs using Yarrowia lipolytica. J Ind Microbiol Biotechnol 47, 403–412 (2020). https://doi.org/10.1007/s10295-020-02276-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-020-02276-6