Abstract
Yarrowia lipolytica is a promising platform for single cell oil production. It is well-known for its metabolism oriented toward utilization of hydrophobic substrates and accumulation of storage lipids. Multiple copies of DGA2 under constitutive promoter were introduced into the Q4 strain, a quadruple mutant deleted for the four acyltransferases (Δdga1, Δdga2, Δlro1, and Δare1) to improve lipid accumulation. The Q4-DGA2 x3 strain contains three copies of DGA2. Further increase in accumulation was accomplished by blocking the β-oxidation pathway through MFE1 gene deletion yielding Q4-Δmfe DGA2 x3. In order to use molasses as a substrate for single cell oil production, sucrose utilization was established by expressing the Saccharomyces cerevisiae SUC2 gene yielding Q4-SUC2 DGA2 x3 and Q4-Δmfe SUC2 DGA2 x3. During cultivation on sucrose medium with a carbon to nitrogen ratio of 80, both strains accumulated more than 40 % of lipids, which was a 2-fold increase in lipid storage. Q4-Δmfe SUC2 DGA2 x3 accumulated more lipids than Q4-SUC2 DGA2 x3 (49 vs. 43 %) but yielded less biomass (13.7 vs. 15 g/L). When grown on 8 % (v/v) molasses, both strains accumulated more than 30 % of lipids after 3 days, while biomass yield was higher in Q4-SUC2 DGA2 x3 (16.4 vs. 14.4 g/L). Further addition of molasses at 72 h resulted in higher biomass yield, 26.6 g/L for Q4-SUC2 DGA2 x3, without modification of lipid content. This work presents genetically modified strains of Y. lipolytica as suitable tools for direct conversion of industrial molasses into value added products based on single cell oils.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yarrowia lipolytica is a non-pathogenic ascomycetous yeast. Its natural habitats are lipid and protein-rich environments. Y. lipolytica is a yeast of great biotechnological potential. It has been used as a producer of extracellular enzymes with proteolytic and lipolytic activities (Barth and Gaillardin 1996; Fickers et al. 2011) but also other industrial and therapeutical enzymes and proteins (Madzak et al. 2004; Gasmi et al. 2012). Due to its natural ability to utilize various hydrophobic compounds, it was considered as suitable microorganism for bioremediation of contaminated soils and treatment of oil-polluted water (Bankar et al. 2009). Another potential application of Y. lipolytica is organic acid production such as citric, isocitric, α-ketoglutaric, pyruvic, succinic, and acetic acids (Groenewald et al. 2014). Y. lipolytica has the ability to accumulate over 40 % of lipids in biomass mainly in the form of triacylglycerols (TAGs) stored in lipid bodies (LBs). It is therefore considered as an oleaginous microorganism (Papanikolaou and Aggelis 2002) suitable for single cell oil (SCO) production. One of bottlenecks for TAG production in Y. lipolytica is the last step of TAG synthesis occurring at the diacylglycerol (DAG) to TAG step catalyzed by acyl-CoA:diacylglycerol acyltransferase (DGAT). Besides the DGAT2 enzyme named Dga1p, Y. lipolytica harbors also a DGAT1 enzyme named Dga2p (Beopoulos et al. 2012). It also possesses a phospholipid:diacylglycerol acyltransferase Lro1p and a single acyl-CoA:sterol acyltransferase Are1p. Deletion of the four acyltransferases (Δdga1, Δdga2, Δlro1, and Δare1) results in a strain (Q4) unable to store TAG and to form LBs (Beopoulos et al. 2012). It has been shown that overexpression of DGA2 gene in a Q4 background allows higher TAG accumulation than overexpression of DGA1 gene (Beopoulos et al. 2012). Not only the enzymatic arsenal but also the growth conditions are important for SCO production, in particular, the carbon to nitrogen ratio (C/N) (Beopoulos et al. 2009). In excess of carbon source together with nitrogen limitation, carbon is channeled toward lipid storage in oleaginous microorganisms (Ratledge 2004). SCO has the advantage to be produced in fermentation tanks independently of year seasons. SCO can be produced as “tailor-made” lipids thanks to genetic engineering. This is particularly true for Y. lipolytica for which the entire genome is sequenced and numerous genetic tools are available. Modified strains have been already developed to produce lipids with engineered composition matching the demands for biodiesel properties (Papanikolaou and Aggelis 2010) or novel compounds, for example, polyunsaturated fatty acids, and β-carotene (Chuang et al. 2010; Grenfell-Lee et al. 2014).
Molasses is one of the agro-industrial by-products that can be used for bioconversion and already used for ethanol production by yeasts (Arshad et al. 2014). It is composed predominantly of sucrose but contains also other sugars, proteins, and inorganic compounds. One of drawbacks of Y. lipolytica is the lack of invertase activity, unlike the widely used yeast Saccharomyces cerevisiae. To overcome this handicap, an optimized S. cerevisiae SUC2 expression cassette have been recently developed allowing efficient growth on sucrose (Lazar et al. 2013). In this study, we show that lipid accumulation was improved through multicopy integration of a diacylglycerol acyltransferase DGA2 expression cassette and by blocking β-oxidation through MFE1 gene deletion. We developed new strains expressing S. cerevisiae SUC2 gene able to produce lipid from molasses.
Material and methods
Strains and culture conditions
Strains used in this study were derived from the wild-type strain W29 (ATCC 20460) (Table 1 and Fig. 1). Escherichia coli strains were cultivated in LB medium according to standard protocol (Sambrook and Russell 2001). Rich yeast extract peptone dextrose (YPD) medium was used for yeast inoculum preparation and contained 1 % (w/v) yeast extract, 1 % (w/v) peptone, and 2 % (w/v) glucose. Minimal yeast nitrogen base (YNB), YNBsuc, and YNBleu media were used for transformant selection. Composition of minimal YNB medium was 0.17 % (w/v) yeast nitrogen base (without amino acids and ammonium sulfate; BD, France), 0.5 % (w/v) NH4Cl, 50 mM phosphate buffer (pH 6.8), and 2 % (w/v) glucose. Composition of YNBsuc was similar, but 1 % (w/v) sucrose was used instead of glucose. For the selection of leucine auxotroph upon marker excision, transformants were plated on YNBleu, YNB medium supplemented with 0.1 g/L of leucine. Sucrose-based cultivation medium contained 60 g/L sucrose, 1.5 g/L yeast extract (BD, France), 0.5 g/L NH4Cl, 7 g/L KH2PO4, 5 g/L Na2HPO4·12H2O, 0.1 g/L CaCl2, 1.5 g/L MgSO4·7H2O, 10 mg/L ZnSO4·7H2O, 0.6 mg/L FeCl3·6H2O, 0.07 mg/L MnSO4·H2O, and 0.04 mg/L CuSO4·5H2O. Molasses-based cultivation medium was prepared by addition of 0.5 g/L KH2PO4 to a 8 % (v/v) molasses solution (equivalent to 51.1 g/L of total sugars). In fed-batch experiment, diluted 50 % (v/v) molasses without KH2PO4 was added after 3 days to restore initial concentration of 8 % (v/v) molasses. Raw molasses (Table 2) was obtained from sugar beet processing industry (Slovak sugar producing factory Slovenské cukrovary, s.r.o.). The pH was adjusted to 5 before sterilization. Growth was carried out in 500-mL baffled flasks with 100 mL of sterile media. Flasks were inoculated with cells to an optical density at 600 nm (OD600) of 0.1. Cells were cultivated at 28 °C on orbital shaker at 120 rpm.
Strain construction scheme. Strain Po1d (Leu− Ura−) was derived from wild-type strain W29 (ATCC 20460). Prototrophic JMY2593 was derived from Po1d by insertion of SUC2 gene into zeta platform. JMY1877 (Q4) was prepared by deletion of DGA1, DGA2, LRO1, and ARE1 genes. From JMY1877 strain, JMY1915 was derived by deletion of MFE1 gene. To JMY1877 or JMY1915, pTEF-DGA2-URA3ex and pTEF-DGA2-LEU2ex were inserted to construct mono-, double-, and triple-copy strains. Insertion of pTEF-SUC2-URA3ex or pTEF-SUC2-LEU2ex was performed to ensure invertase activity. Auxotrophic Leu− strains were transformed with LEU2 fragment when necessary to obtain prototrophic strains. Dotted arrows indicate hidden steps, which are described in detail elsewhere (Haddouche et al. (2011), Lazar et al. (2013))
Growth in microtiter plates
Yeast strain growth in 96-well plates was performed in 200 μL of minimal medium containing 0.17 % (w/v) yeast nitrogen base (without amino acids and ammonium sulfate, BD, France), 0.15 % (w/v) NH4Cl, 50 mM phosphate buffer (pH 6.8), and 3 % (w/v) sucrose. Cells were precultivated for 24 h in YPD medium. Cells were washed and inoculated to an OD600 of 0.2 in each well. Triplicate experiments were performed at 28 °C under constant agitation with a Biotek Synergy MX microtiter plate reader (Biotek Instruments, Colmar, France). Growth was monitored measuring optical density at 600 nm every 20 min for 48 h.
Dry cell weight determination
Cells were harvested by centrifugation (2880×g, 5 min) after 3 and 6 days, washed two times with 0.9 % NaCl solution, once with deionized water, and lyophilized. Lyophilized cells were used for further analysis. For gravimetric determination of dry cell weight (DCW), cells from 5 mL of suspension were centrifuged, washed as described, and dried until constant weight at 105 °C.
Measurement of sugars
Sucrose, glucose, and fructose were determined by HPLC (Agilent) using an Aminex HPX-87H column coupled to RI detector as described previously (Lazar et al. 2011).
General genetic techniques
Standard molecular biology techniques were used in this study (Sambrook and Russell 2001). Restriction enzymes were obtained from New England Biolabs (Ipswich, England). Yeast genomic DNA was prepared as described by Querol et al. (1992). PCR amplifications were carried out in an Applied Biosystems 2720 thermal cycler with Taq DNA polymerase (Promega, Madison, WI). DNA fragments were recovered from agarose gels with a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany).
Strain construction
All E. coli and Y. lipolytica strains used in this study are described in Table 1. Plasmid from JME1657 was obtained by replacing the RedStar2 gene from JMP1394 plasmid by the SUC2 gene using BamHI and AvrII restriction enzyme. Plasmid JMP1822 was constructed from JMP1132 plasmid by replacing URA3ex marker for LEU2ex marker from JMP1394 using I-SceI restriction enzyme. Expression vectors were digested with NotI and subjected to electrophoresis. The bands corresponding to the expression cassettes were extracted from the gel and used for transformation. Expression cassettes were integrated at random in Y. lipolytica genome as described previously (Pignède et al. 2000). NotI expression cassette of plasmids from JME1132 (pTEF-DGA2-URA3ex), JME1822 (pTEF-DGA2-LEU2ex), JME1462 (pTEF-SUC2-URA3ex), and JME1657 (pTEF-SUC2-LEU2ex) were used for the following transformations. Yeast cells were transformed by the lithium acetate method (Le Dall et al. 1994). All strains were derived from JMY1877, the Q4 strain deleted for the four acyltransferases (Beopoulos et al. 2012). Strains used as hosts were JMY1884 (Q4-DGA2 x1, Leu−) (Beopoulos et al. 2012) and JMY1915 (Q4-∆mfe, Leu− Ura−) (Haddouche et al. 2011). Schematic representation of successive strain constructions is depicted in Fig. 1. JMY1884 and JMY1915 were transformed with pTEF-DGA2-LEU2ex cassette to construct JMY3357 (Q4-DGA2 x2) and JMY3369 (Q4-∆mfe, DGA2, Ura−). JMY3369 was transformed with pTEF-SUC2-URA3ex cassette to obtain JMY3588 (Q4-∆mfe SUC2 DGA2). JMY1915 was transformed with pTEF-DGA2-LEU2ex and pTEF-DGA2-URA3ex cassettes at once yielding JMY3365 (Q4-∆mfe DGA2 x2). Auxotrophies in JMY3357 and JMY3365 were restored via marker excision using the Cre-lox recombinase system, by transformation with the replicative plasmid pUB4-Cre1 (JME547) (Fickers et al. 2003) yielding JMY3435 and JMY3463, respectively. These strains were transformed with pTEF-SUC2-URA3ex cassette to obtain JMY3532 (Q4-SUC2 DGA2 x2) and JMY3517 (Q4-∆mfe SUC2 DGA2 x2), respectively. These strains were then transformed with pTEF-DGA2-LEU2ex cassette to give rise to JMY3582 (Q4-SUC2 DGA2 x3) and JMY3586 (Q4-∆mfe SUC2 DGA2 x3), respectively. Strain JMY1884 was transformed with pTEF-SUC2-LEU2ex to give rise to JMY3534. Prototrophic strains JMY3400, JMY3592, and JMY3596 were prepared from JMY1884, JMY3532, and JMY3517, respectively, by transformation of cells with SalI-digested LEU2 fragment from plasmid JME1868. For selection of prototrophic strains, YNB plates were used and leucine auxotrophs were selected on YNBleu plates. Insertions of the expression cassettes were confirmed by PCR with primers pairs DGA2in-Leu2in and DGA2in-Ura3in listed in Table 3. SUC2-positive strains were selected on YNBsuc plates.
Fatty acid analysis by gas chromatography
Fatty acids (FAs) from lyophilized biomass were converted to their fatty acid methyl esters (FAMEs) by modified method of Čertík and Shimidzu (2000). Biomass (approximately 10–20 mg) was mixed with 1 mL of dichlormethane (containing 0.1 mg of C17:0 as internal standard) and 2 mL of anhydrous methanolic HCl solution, incubated at 50 °C during 3 h to form FAMEs. After transesterification, 1 mL of water was added. FAMEs were extracted with 1 mL of hexane and analyzed by GC-6890 N (Agilent Technologies). FAMEs (1 μL) were automatically injected on DB-23 column (50 % cyanopropyl-methylpolysiloxane, length 60 m, diameter 0.25 mm, film thickness 0.25 μm) and analyzed. Analysis conditions were carrier gas—hydrogen, inlet (230 °C; hydrogen flow: 36.7 mL/min; split—1:20), FID detector (250 °C, hydrogen flow: 40 mL/min, air flow: 450 mL/min), gradient (130 °C—1 min; 130–170 °C—7.0 °C/min; 170–220 °C—6.0 °C/min;. 220 °C—8 min; 220–240 °C—30 °C/min; 240 °C—3.5 min). Chromatograms were analyzed with ChemStation B0103 (Agilent Technologies). Total fatty acid (TFA) content in DCW was calculated from sum of all FA normalized with the internal standard.
Structural analysis of total lipid by TLC
Total cell lipids were extracted by modified procedure of Folch et al. (1957). Approximately 0.5 g of lyophilized biomass was homogenized and extracted by 65 mL of chloroform/methanol (2:1) solution. After 1.5 h., suspension was filtered, 1.2-fold of 0.9 % potassium chloride solution was added to the liquid phase, and solution was mixed. After centrifugation (2880×g, 5 min), upper aqueous methanolic phase was removed, and the lower chloroform phase was dried over anhydrous sodium sulfate. Subsequently, chloroform was evaporated on vacuum evaporator, and lipids were dissolved in hexane/chloroform (1:1) solution. Total cell lipids were loaded on Merck TLC silica gel 60 plates by CAMAG ATS 4 and developed in hexane/ethyl ether/acetic acid 80/20/1 system. Developed plates were dipped in 3.3 % (v/v) sulfuric acid, 50 % (v/v) methanol, and 0.33 % (w/v) MnCl2 water solution and dried at 130 °C for 10 min for visualization. After, visualization plates were scanned by CAMAG TLC Scanner 4 at 400 nm and evaluated by winCATS software ver. 1.4.8 (CAMAG) in order to evaluate the TAG, free fatty acids (FFAs), and DAG proportion in lipid extract.
Results
Engineering yeast cells with improved lipid accumulation
In order to improve lipid accumulation in Y. lipolytica, we evaluated the effect of overexpressing DGA2 under the strong and constitutive promoter pTEF in a Q4 background in multiple copies. Strains with one to three copies of DGA2 were obtained. In addition, in order to broaden the range of utilizable substrates and particularly sucrose, S. cerevisiae SUC2 gene coding for invertase was also introduced. A scheme of the strain constructions is presented in Fig. 1.
First, the capacity of JMY3534, expressing one copy of DGA2 and SUC2 to grow on sucrose, was evaluated. Figure 2 shows that JMY3534 has a similar, even better growth rate compared to JMY2593, parental strain expressing only SUC2, while the control strain, JMY2900, is growing very poorly on sucrose. Then, JMY3534 was evaluated for its capacity to grow directly on industrial raw molasses medium and to produce LBs. JMY3534 (one copy of DGA2) grows well on molasses and produces bigger LBs than the control strain JMY2593 expressing only SUC2 (Fig. 3).
Microscopic observation of control strain JMY2593 (Po1d-SUC2) (a) and overaccumulating strain JMY3534 (Q4-SUC2 DGA2) (b). Cells were cultivated on 8 % (v/v) molasses for 3 days. Q4-SUC2 DGA2 formed large lipid bodies (LBs). White arrows indicate LBs containing storage lipids. White bar indicates 5 μm. Cells were observed under ×100 oil immersion objective
In a second step, effect of the copy number of the pTEF-DGA2 expression cassette on lipid accumulation was evaluated. Strains with two and tree copies were constructed (Fig. 1, Table 1). Strains with three copies of DGA2, cultivated on molasses medium, showed higher lipid accumulation levels compared to the mono- and the double-copy strains. After 5 days of cultivation on 8 % molasses medium, lipid content was 11 % in the control strain JMY2593 (Po1d-SUC2), 24 % in JMY3534 (Q4-SUC2 DGA2), 29 % in JMY3592 (Q4-SUC2 DGA2 x2), and 32 % in JMY3582 (Q4-SUC2 DGA2 x3) strain (Fig. 4). Thus, demonstrating that increasing DGA2 expression allowed improving lipid accumulation.
The next step to increase lipid accumulation was to block FA degradation through deletion of MFE1 gene. Thus, strains Δmfe1, expressing SUC2, with one to three copies of DGA2 were constructed (see Fig. 1). Strains with MFE1 gene deleted accumulated more lipids being 30, 33, and 39 % in the mono, double, and triple copy of DGA2, respectively (Fig. 4).
Growth and FA accumulation on sucrose medium
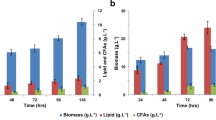
Strains JMY3582 (Q4-SUC2 DGA2 x3) and JMY3586 (Q4-∆mfe SUC2 DGA2 x3) were selected for further analysis as they accumulated the highest percentage of lipids. JMY2593 (Po1d-SUC2) was used as the control strain. First, they were cultivated in minimal sucrose medium with C/N ratio of 80 to investigate their biomass yield and lipid accumulation levels. After 3 days of cultivation, JMY2593 (Po1d-SUC2) yielded 12.8 g/L biomass and lipid content was 20 % of DCW. After 6 days, only a slight increase in lipid content was observed with a lower biomass yield (11.5 g/L) (Fig. 5). However, TFA yield remained at the same level of 2.5 g/L. This indicates that maximum accumulation was reached within 3 days, even though there was still residual substrate in the medium (data not shown). Strains JMY3582 and JMY3586 reached a higher biomass yield compared to control strain, which was connected with higher lipid accumulation. TFA yields were 6.5 and 6.7 g/L in JMY3582 and JMY3586 strains, respectively, which represents 2.5-fold higher yields than in the control strain (Fig. 5a). However, we observed growth differences for these two strains. It was observed that Δmfe1 deletion had negative effect on biomass yield. Indeed, strain JMY3582, similarly to the control strain, reached maximum biomass yield after 3 days (approx. 15 g/L). However, we did not observe a decrease in total biomass as in the control (Fig. 5a) but only small decrease in FA free biomass coupled with slight TFA increase. In contrast, JMY3586 reached maximum DCW yield after 6 days. At this time, TFA content reached 49 % of DCW at the expense of a lower biomass yield than in JMY3582 strain (13.7 vs. 15 g/L) (Fig. 5a, b). It was clearly visible that growth rate of Q4-Δmfe DGA2 x3 strain was reduced compared to JMY3582 due to the redirection of carbon to lipid instead to biomass. Difference in FA composition of all three tested strains was observed. Major FAs in all three strains were oleic acid (55–57 %) and palmitic acid (15–21 %) followed by stearic, palmitoleic, linoleic, and other long-chain FAs (Table 4). However, slight differences could be observed: JMY3582 strain accumulated more palmitic and less linoleic acid, JMY3586 with blocked β-oxidation is characterized by the absence of one of the palmitoleic acids, namely C16:1Δ79, and of the increase of unusual polyunsaturated FAs (Table 4).
Growth and lipid accumulation on sucrose. Growth was performed in 6 % (w/v) sucrose medium with C/N ratio 80. Samples were taken after 3 and 6 days. DCW yield (a) is depicted as a sum of TFA free biomass (black) and TFA (white). Lipid content (b) expressed as percentage of TFA in DCW of control, Δmfe, and non-Δmfe strains. DCW (a) and TFA/DCW (b) represent mean values of three independent experiments
Growth and FA accumulation on molasses
Next, growth and lipid production were analyzed in 8 % industrial raw molasses medium (Fig. 6). Strains JMY2593 (Po1d-SUC2), JMY3582 (Q4-SUC2 DGA2 x3), and JMY3586 (Q4-Δmfe SUC2 DGA2 x3) were cultivated for 3 days. Biomass yield of control strain JMY2593 was 11.5 g/L, and it accumulated approximately 10 % of TFA in DCW, while strain JMY3582 accumulated 33 % of TFA in DCW. This difference in lipid accumulation resulted in higher biomass yield of JMY3582 strain (16.4 vs. 11.5 g/L). Strain JMY3586 showed lower biomass yield (14.4 g/L) although TFA accumulation was comparable to non-Δmfe strain (31 vs. 33 %) (Fig. 6). The lower growth of JMY3586 compared to JMY3582 is similar to what has been observed during growth on defined sucrose medium. As a result, JMY3586 yielded only 4.4 g/L of TFA compared to 5.4 g/L of JMY3582 (Fig. 6). FA composition of yeast cells was very similar to that of cells cultivated on sucrose medium (Table 5).
Influence of fed-batch culture on growth and lipid accumulation. Cells were grown in 8 % (v/v) molasses for 3 days (3D). After 3 days, fresh molasses was added to restore initial concentration 8 % (v/v) molasses, and cells were grown for three more days (3 + 3D FB) and compared to cells grown without molasses addition (6D). DCW yield (a) is depicted as a sum of TFA free biomass (black) and TFA (white). Lipid content (b) expressed as percentage of TFA in DCW of control, Δmfe, and non-Δmfe strains (b). DCW (a) and TFA/DCW (b) represent mean values of three independent experiments
In attempt to increase biomass and TFA yield, fed-batch was applied. Based on the time course of sugar consumption (Fig. 7), fresh 50 % (v/v) molasses was added on the third day to reach a final concentration of 8 % (v/v). After three more days of incubation, cells were harvested and analyzed. All strains grew well, and no negative effect of fresh molasses was observed. Moreover, addition of fresh molasses improved biomass and TFA yield of all strains (Fig. 6a). JMY3582 (Q4-SUC2 DGA2 x3) strain was the best growing strain with biomass yield of 26.6 g/L and a TFA yield of 8 g/L. TFA content in DCW was not modified by molasses addition and remained approximately at 30 % of DCW, and FA composition was almost unchanged with a small decrease in oleic acid (from 55 to 52 %) and an increase in stearic acid (from 8 to 11–12 %) in JMY3582 and JMY3586 strains (data not shown). TLC analysis showed that major lipidic structure was TAG (Fig. 8). TAG represented about 27 % of all lipidic structures in control strain, 51 % in JMY3582 (Q4-SUC2 DGA2 x3), and 42 % in JMY3586 (Q4-Δmfe DGA2 x3). Strain JMY3586 shows an elevated percentage of FFAs, due to the absence of FA degradation and an increase of DAG (3 % compared to 1.8 % in control strain and JMY3582) (Fig. 8).
Growth and sugar consumption on 8 % (v/v) molasses. Growth of JMY2593 (Po1d-SUC2) (white square), JMY3582 (Q4-SUC2 DGA2 x3) (white triangle), and JMY3586 (Q4-Δmfe SUC2 DGA2 x3) (white circle). Sugar consumption by JMY2593 (Po1d-SUC2) (black square), JMY3582 (Q4-SUC2 DGA2 x3) (black triangle), and JMY3586 (Q4-Δmfe SUC2 DGA2 x3) (black circle)
Lipid separation by TLC for the JMY2593 (Po1d-SUC2), JMY3582 (Q4-SUC2 DGA2 x3), and JMY3586 (Q4-Δmfe SUC2 DGA2 x3). Lipids were extracted by modified procedure of Folch et al. (1957). Lipids were developed in hexane/ether/acetic acid 80/20/1 system. PL polar lipids, DAG diacylglycerols, ERG ergosterol, FFA free fatty acids, TAG triacylglycerols, SE steryl esters, SQ squalene
Discussion
Industry produces many by-products, which are regarded as waste materials with little value. These materials still contain substances which are economically valuable, like complex and simple sugars, nitrogen substances, and inorganic salts. All these compounds are important for growth of different microorganisms, and therefore, there is a strong potential for using these by-products as substrates in biotechnological production. Molasses is regarded as one of the industrial by-products, which is widely used for biotechnological applications (Arshad et al. 2014; Xia et al. 2014; He et al. 2014; Ortiz et al. 2012). Molasses-based cultivation media are undefined media which contain different saccharides, nitrogen compounds, and many other substances impacting cell growth (Olbrich 2006) in various proportions depending on molasses batches. There is a great potential for SCO production using Y. lipolytica with such industrial by-products. However, Y. lipolytica lacks sucrose cleaving enzyme invertase, essential to grow on unpretreated molasses where sucrose is the major sugar. To overcome this drawback, Y. lipolytica strains expressing SUC2 gene coding for the S. cerevisiae invertase were constructed. These strains were very successful in utilizing sucrose-based media as it was previously reported by Lazar et al. (2013). In this study, we show that molasses-based cultivation medium is convenient for growth and lipid accumulation by invertase secreting Y. lipolytica recombinant cells. Control strain JMY2593 (Po1d-SUC2) accumulated 20 % of storage lipids when grown on sugar-rich sucrose medium with C/N ratio 80. Accumulation was about one half lower when 8 % (v/v) raw molasses was used. To improve FA accumulation in yeast cells, strategy of multiple gene integration into yeast genome was employed. We overexpressed the DGA2 gene coding Y. lipolytica’s diacylglycerol acyltransferase belonging to DGAT1 family. Overexpression of this gene increased the TFA content in a Q4 background (Beopoulos et al. 2012) with a gene dosage effect. Expressing DGA2 gene under constitutive TEF promoter and introducing several copies of this construct into Q4 strain allowed a 2-fold increase in storage lipid accumulation which confirmed that this step is limiting for TAG synthesis. Similar improvement of lipid accumulation was obtained through overexpression of the DGA1 gene coding Y. lipolytica’s diacylglycerol acyltransferase belonging to DGAT2 family in multicopies. However, we observed an increase in lipid body number, a decrease in size, and differences in distribution and inheritance when DGA1 was expressed (Gajdoš et al. unpublished).
To prevent degradation of storage lipids, deletion of MFE1, a gene encoding an enzyme catalyzing the second step of β-oxidation pathway, was carried out. Indeed, we previously demonstrated a strong improvement of lipid accumulation by blocking the β-oxidation pathway through either deletion of the six acyl-CoA oxidases (Δpox1–pox6) or by deletion of the multifunctional enzyme (Δmfe1) (Dulermo and Nicaud 2011). Surprisingly, deletion of MFE1 gene led to enhanced FA accumulation on sucrose media where lipid content reached nearly 50 % of DCW; however, this had no effect on molasses media. On the contrary, it affected biomass yields, suggesting different adaptation of cells to nitrogen depletion conditions possibly by degradation of intracellular proteins, in line with metabolic activities in oleaginous cells after nitrogen depletion described by Zhu et al. (2012), between sucrose and molasses media. Strain JMY3582 produced more citric acid into medium (data not shown), which was not connected with significant lipid degradation as it was observed by Makri et al. (2010).
After 3 days of cultivation on 8 % (v/v) molasses medium, more than 30 % of TFA in DCW were accumulated by both Δmfe and non-Δmfe strains. TFA yield reached 5.4 g/L which is comparable to the results reported by Tsigie et al. (2012). To increase biomass and overall TFA yield, higher amount of sugars in cultivation medium was needed. One possibility would be increasing molasses concentration over 8 % (v/v). However, it was supposed that increasing percentage of molasses at the beginning of cultivation would lead to higher total nitrogen concentration or higher concentration of growth inhibitory compounds, which could negatively influence TFA yield (Karatay and Dönmez 2010). Indeed, starting at higher molasses concentration had a negative effect on growth (data not shown). However, addition of concentrated molasses medium after 3 days of culture allowed increasing biomass production without affecting lipid content. FA composition was similar to the composition observed on glycerol media (Papanikolaou et al. 2013) with low level of linoleic acid (C18:2) compared to glucose media where linoleic acid could represent more than 20 % and up to about 50 % depending on growth conditions (Beopoulos et al. 2008; Beopoulos et al. 2009; Papanikolaou et al. 2013).
Previous reports showed that wild-type Y. lipolytica strains are naturally not able to use sucrose due to the lack of invertase (Nicaud et al. 1989; Lazar et al. 2013). To our knowledge, this is the first report on lipid production by Y. lipolytica on crude industrial molasses without pretreatment and without addition of neither nitrogen nor vitamin sources. However, molasses composition could vary from batch to batch, and we could not exclude that growth could be affected by some composition or inhibiting compounds. By-passing the hydrolysis of molasses to exploit the total carbon source from molasses using strains expressing heterologous invertase provides a strong technological and economical advantage. Moreover, our genetically modified strains show a strong increase in lipid content compared to control strain and demonstrated that increasing DGA2 gene copy number increase lipid accumulation. Thus, improving fermentation process on molasses will certainly lead to higher lipid content. Improved strains developed in this work has a major application potential as a tool for conversion of industrial residues containing sucrose into value added products based on SCO.
References
Arshad M, Ahmed S, Zia MA, Rajoka MI (2014) Kinetics and thermodynamics of ethanol production by Saccharomyces cerevisiae MLD10 using molasses. Appl Biochem Biotechnol 172:2455–2464. doi:10.1007/s12010-013-0689-x
Bankar AV, Kumar AR, Zinjarde SS (2009) Environmental and industrial applications of Yarrowia lipolityca. Appl Microbiol Biotechnol 84:847–865. doi:10.1007/s00253-009-2156-8
Barth G, Gaillardin C (1996) The dimorphic fungus Yarrowia lipolytica. In: Wolf K (ed) Genetics, biochemistry, and molecular biology of non conventional yeasts. Springer, Berlin, pp 313–388
Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, Chardot T, Nicaud JM (2008) Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol 74:7779–7789. doi:10.1128/AEM.01412-08
Beopoulos A, Cescut J, Haddouche R, Uribelarrea JL, Molina-Jouve C, Nicaud JM (2009) Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res 48:375–387. doi:10.1016/j.plipres.2009.08.005
Beopoulos A, Haddouche R, Kabran P, Dulermo T, Chardot T, Nicaud JM (2012) Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl Microbiol Biotechnol 93:1523–1537. doi:10.1007/s00253-011-3506-x
Čertík M, Shimidzu S (2000) Kinetic analysis of biosynthesis by an arachidonic acid-producing fungus, Mortierella alpina 1S-4. Appl Microbiol Biotechnol 54:224–230
Chuang LT, Chen DC, Nicaud JM, Madzak C, Chen YH, Huang YS (2010) Co-expression of heterologous desaturase genes in Yarrowia lipolytica. N Biotechnol 27:277–282. doi:10.1016/j.nbt.2010.02.006
Dulermo T, Nicaud JM (2011) Involvement of G3P shuttle and β-oxidation pathway into the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng 13:482–491. doi:10.1016/j.ymben.2011.05.002
Fickers P, Le Dall MT, Gaillardin C, Thonart P, Nicaud JM (2003) New disruption cassettes for rapid gene disruption and marker rescue in the yeast Yarrowia lipolytica. J Microbiol Methods 55:727–737
Fickers P, Marty A, Nicaud JM (2011) The lipases from Yarrowia lipolytica: genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol Adv 29:632–644. doi:10.1016/j.biotechadv.2011.04.005
Folch J, Lees M, Sloane Stanley GH (1957) A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226:497–509
Gaillardin C, Ribet AM (1987) LEU2 directed expression of beta-galactosidase activity and phleomycin resistance in Yarrowia lipolytica. Curr Genet 11:369–375
Gasmi N, Lassoued R, Ayed A, Tréton B, Chevret D, Nicaud JM, Kallel H (2012) Production and characterization of human granulocyte-macrophage colony-stimulating factor (hGM-CSF) expressed in the oleaginous yeast Yarrowia lipolytica. Appl Microbiol Biotechnol 96:89–101. doi:10.1007/s00253-012-4141-x
Grenfell-Lee D, Zeller S, Cardoso R, Pucaj K (2014) The safety of β-carotene from Yarrowia lipolytica. Food Chem Toxicol 65:1–11. doi:10.1016/j.fct.2013.12.010
Groenewald M, Boekhout T, Neuvéglise C, Gaillardin C, van Dijck PW, Wyss M (2014) Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit Rev Microbiol 40:187–206. doi:10.3109/1040841X.2013.770386
Haddouche R, Poirier Y, Delessert S, Sabirova J, Pagot Y, Neuvéglise C, Nicaud JM (2011) Engineering polyhydroxyalkanoate content and monomer composition in the oleaginous yeast Yarrowia lipolytica by modifying the ß-oxidation multifunctional protein. Appl Microbiol Biotechnol 91:1327–1340. doi:10.1007/s00253-011-3331-2
He J, Wu AM, Chen D, Yu B, Mao X, Zheng P, Yu J, Tian G (2014) Cost-effective lignocellulolytic enzyme production by Trichoderma reesei on a cane molasses medium. Biotechnol Biofuels 7:43. doi:10.1186/1754-6834-7-43
Kabran P, Rossignol T, Gaillardin C, Nicaud JM, Neuvéglise C (2012) Alternative splicing regulates targeting of malate dehydrogenase in Yarrowia lipolytica. DNA Res 19:231–244. doi:10.1093/dnares/dss007
Karatay SE, Dönmez G (2010) Improving the lipid accumulation properties of the yeast cells for biodiesel production using molasses. Bioresour Technol 101:7988–7990. doi:10.1016/j.biortech.2010.05.054
Lazar Z, Walczak E, Robak M (2011) Simultaneous production of citric acid and invertase by Yarrowia lipolytica SUC + transformants. Bioresour Technol 102:6982–6989. doi:10.1016/j.biortech.2011.04.032
Lazar Z, Rossignol T, Verbeke J, Crutz-Le Coq AM, Nicaud JM, Robak M (2013) Optimized invertase expression and secretion cassette for improving Yarrowia lipolytica growth on sucrose for industrial applications. J Ind Microbiol Biotechnol 40:1273–1283. doi:10.1007/s10295-013-1323-1
Le Dall MT, Nicaud JM, Gaillardin C (1994) Multi-copy integration in the yeast Yarrowia lipolytica. Curr Genet 26:38–44
Madzak C, Gaillardin C, Beckerich JM (2004) Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol 109:63–81
Makri A, Fakas S, Aggelis G (2010) Metabolic activities of biotechnological interest in Yarrowia lipolytica grown on glycerol in repeated batch cultures. Bioresour Technol 101:2351–2358. doi:10.1016/j.biortech.2009.11.024
Nicaud J-M, Fabre E, Gaillardin C (1989) Expression of invertase activity in Yarrowia lipolytica and its use as a selective marker. Curr Genet 16:253–260
Olbrich H (2006) The molasses. Biotechnologie-Kempe GmbH, Berlin
Ortiz ME, Fornaguera MJ, Raya RR, Mozzi F (2012) Lactobacillus reuteri CRL 1101 highly produces mannitol from sugarcane molasses as carbon source. Appl Microbiol Biotechnol 95:991–999. doi:10.1007/s00253-012-3945-z
Papanikolaou S, Aggelis G (2002) Lipid production by Yarrowia lipolytica growing on industrial glycerol in a single-stage continuous culture. Bioresour Technol 82:43–49
Papanikolaou S, Aggelis G (2010) Yarrowia lipolytica: a model microorganism used for the production of tailor-made lipids. Eur J Lipid Sci Technol 112:639–654. doi:10.1002/ejlt.200900197
Papanikolaou S, Beopoulos A, Koletti A, Thevenieau F, Koutinasa A, Nicaud JM, Aggelis G (2013) Importance of the methyl-citrate cycle on glycerol metabolism in the yeast Yarrowia lipolytica. J Biotechnol 168:303–314. doi:10.1016/j.jbiotec.2013.10.025
Pignède G, Wang H, Fudalej F, Gaillardin C, Seman M, Nicaud J-M (2000) Autocloning vectors for gene expression and amplification for the yeast Y. lipolytica. Appl Environ Microbiol 66:3283–3289
Querol A, Barrio E, Huerta T, Ramon D (1992) Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl Environ Microbiol 58:2948–2953
Ratledge C (2004) Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie 86:807–815
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York
Tsigie YA, Wang CY, Kasim NS, Diem QD, Huynh LH, Ho QP, Truong CT, Ju YH (2012) Oil production from Yarrowia lipolytica Po1g using rice bran hydrolysate. J Biomed Biotechnol 2012:378384. doi:10.1155/2012/378384
Xia J, Xu Z, Xu H, Liang J, Li S, Feng X (2014) Economical production of poly(ε-l-lysine) and poly(l-diaminopropionic acid) using cane molasses and hydrolysate of streptomyces cells by Streptomyces albulus PD-1. Bioresour Technol 164:241–247. doi:10.1016/j.biortech.2014.04.078
Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, Zhou YJ, Jin G, Ye M, Zou H, Zhao ZK (2012) A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun 3:1112. doi:10.1038/ncomms2112
Acknowledgments
The work was supported by grant VEGA 1/0574/15 from the Grant Agency of the Ministry of Education, Slovak Republic and by grant APVV-0662-11 from the Slovak Research and Development Agency, Slovak Republic. This work was funded by the French National Institute for Agricultural Research (Institut National de la Recherche Agronomique).
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gajdoš, P., Nicaud, JM., Rossignol, T. et al. Single cell oil production on molasses by Yarrowia lipolytica strains overexpressing DGA2 in multicopy. Appl Microbiol Biotechnol 99, 8065–8074 (2015). https://doi.org/10.1007/s00253-015-6733-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-015-6733-8