Abstract
Streptomycetes are prolific sources of novel biologically active secondary metabolites with pharmaceutical potential. S. collinus Tü 365 is a Streptomyces strain, isolated 1972 from Kouroussa (Guinea). It is best known as producer of the antibiotic kirromycin, an inhibitor of the protein biosynthesis interacting with elongation factor EF-Tu. Genome Mining revealed 32 gene clusters encoding the biosynthesis of diverse secondary metabolites in the genome of Streptomyces collinus Tü 365, indicating an enormous biosynthetic potential of this strain. The structural diversity of secondary metabolisms predicted for S. collinus Tü 365 includes PKS, NRPS, PKS-NRPS hybrids, a lanthipeptide, terpenes and siderophores. While some of these gene clusters were found to contain genes related to known secondary metabolites, which also could be detected in HPLC–MS analyses, most of the uncharacterized gene clusters are not expressed under standard laboratory conditions. With this study we aimed to characterize the genome information of S. collinus Tü 365 to make use of gene clusters, which previously have not been described for this strain. We were able to connect the gene clusters of a lanthipeptide, a carotenoid, five terpenoid compounds, an ectoine, a siderophore and a spore pigment-associated gene cluster to their respective biosynthesis products.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of the genus Streptomyces are ubiquitous gram-positive soil bacteria of the order Actinomycetales. They contain DNA with a high G+C-content (over 70 %) and exhibit a complex life cycle by forming filamentous mycelia and sporulating aerial hyphae [10]. Besides their role in soil ecology, streptomycetes are of particular significance because of their capability to produce natural compounds with a diverse set of bioactivities, including antibiotic, antitumor, antiviral, antifungal, antihypertensive and immunosuppressant properties. About 75 % of all known antibiotics are produced by actinomycetes with the major contribution of the genus Streptomyces [14].
The availability of genome sequences of a large number of microorganisms opened a new area of research by genome mining. Genome mining is defined as the process of technically translating secondary metabolite-encoding gene sequence data into purified molecules in tubes [3]. That Streptomyces are an unexhausted source of natural compounds was highlighted by the identification of 25 and 38 biosynthetic secondary metabolite gene clusters after the complete genome sequences of S. coelicolor and S. avermitilis [4, 19], respectively, became available. Bioinformatics software such as antiSMASH [5, 30, 56], a platform for automated genome mining of secondary metabolite producers, allows the rapid genome-wide identification, analysis and annotation of biosynthesis gene clusters and the identification of similar characterized gene clusters in the MIBiG repository [31]. Another applicable web tool, the Natural Product Domain Seeker (NaPDoS), provides a rapid method to assess secondary metabolite biosynthetic gene diversity, based on the phylogenetic relationships of sequence tags derived from polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) genes [66].
Although biosynthetic genes are encoded in the producers’ genomes, they are often not expressed under laboratory conditions. Yet, there are techniques available in order to successfully activate such silent gene clusters, optimize production yields and manipulate biosynthesis pathways [57]. One approach is to “awaken” pathways in the native host strain by insertion of additional promoters upstream of the biosynthesis genes, which are either independently regulated or constitutively expressed. Otherwise, expression of gene clusters in heterologous hosts can lead to expression and biosynthesis of the products and thus represents a promising alternative for activation of secondary metabolite gene clusters present in less amenable strains.
Streptomyces collinus Tü 365 has been described as the producer of the antibiotic kirromycin, a narrow-spectrum antibiotic which has activity against some bacterial pathogens and the malaria parasite Plasmodium falciparum [60]. Its biosynthesis has been analyzed in great detail in previous studies [35, 58].
Here, we provide an extended analysis of the potential of S. collinus Tü 365 to synthesize further secondary metabolites.
Genome mining of S. collinus Tü 365
Recently, the genome of S. collinus Tü 365 was sequenced using a combination of Sanger, 454 and illumina technology (Genbank ID: CP006259) [43]. S. collinus Tü 365 has a linear chromosome and contains two linear plasmids, SCO1 (85,041 bp, 68.3 % G+C), and SCO2 (19,314 bp, 69.9 % G+C). The total genome size is 8.37 Mb with a G+C-content of 72.6 %. The genome is predicted to encode 7193 proteins. A long terminal inverted repeat was found at both ends of the chromosome. Its size was determined to be 630 kb [43]. Genome sequencing indicated that both ends have identical sequences.
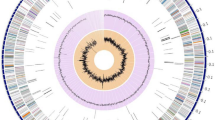
An analysis with antiSMASH using the standard cluster rule based approach resulted in the identification of a range of different natural product classes in S. collinus Tü 365 (Table 1). About 14 % of S. collinus Tü 365 genome is dedicated to secondary metabolism. The secondary metabolite gene clusters are distributed uniformly across the chromosome (Fig. 1). In total, 32 putative biosynthetic gene clusters could be identified (Table 1). Interestingly, three clusters (a lanthipeptide, an atypical NRPS cluster, and the kirromycin biosynthetic gene cluster) are situated on the long terminal inverted repeats of the S. collinus Tü 365 chromosome, and thus are present in two identical copies and gene organizations. Four clusters of S. collinus Tü 365 encode the biosynthesis of polyketide synthase (PKS) compounds (two type I, one type II PKS and one type III PKS cluster) and four clusters encode the synthesis of non-ribosomal peptides. Five clusters code for the biosynthesis of hybrid PKS/NRPS compounds. The modular PKS and NRPS are enzyme assembly lines for the synthesis of chemically diverse natural products. The knowledge on well-characterized PKS and NRPS clusters can be used to predict the encoded secondary metabolites. In S. collinus Tü 365, out of the 13 PKS, NRPS and hybrid clusters, 10 have no significant similarities to characterized pathways and thus either code for known molecules, whose biosynthetic pathways were not yet associated or new compounds that have not been described before. One of the duplicated NRPS/PKS hybrid clusters is known to encode the synthesis of kirromycin. Cluster 16, which codes for a type II PKS, shows strong similarity to the spore pigment cluster of S. coelicolor A3(2) [13].
Representation of the linear chromosome of S. collinus Tü 365 and predicted secondary metabolite gene clusters. Dark blue and light blue circles represent all genes (reverse and forward strands, respectively). The black arrows symbolize the terminal inverted repeats of the chromosome. Secondary metabolite biosynthetic gene clusters are indicated by red blocks. T1PKS, T2PKS and T3PKS represent type I, II, and III PKS pathways
Among the 32 identified clusters, six biosynthetic gene clusters are predicted to be involved in the biosynthesis of terpene compounds (Table 1) widely distributed among streptomycetes. The terpene biosynthetic gene clusters predicted from S. collinus Tü 365 seem to direct the biosynthesis of the carotenoid isorenieratene (cluster 4), 2-methylisoborneol (cluster 6), albaflavenone (cluster 20), geosmin (cluster 25), pentalenolactone (cluster 27) and hopene/squalene (cluster 28). The production of ectoine, a secondary metabolite playing an important role in protection against osmotic stress is also encoded on the chromosome of S. collinus Tü 365. Furthermore, the genome harbors three clusters involved in biosynthesis of siderophores. One of them, cluster 15, encodes the biosynthesis of desferrioxamine E whose synthesis is based on an NRPS-independent (NIS) mechanism. The remaining gene clusters might encode genes related to bacteriocins (clusters 22 and 24), melanin (cluster 14), and three other compounds (clusters 5, 9 and 29).
In the following sections we present detailed descriptions on the deduced biosynthetic routes of the predicted metabolites as well as results on the induction of the biosynthesis and the chemical characterization of the products.
Kirromycin
Kirromycin production in S. collinus Tü 365 has been described more than 40 years ago [60]. The compound, also referred to under the alternative name mocimycin [53], has been identified in a screening program for narrow-spectrum antibiotics, being the first antibiotic identified targeting the elongation factor EF-Tu. Together with several structurally related compounds, kirromycin belongs to the family of elfamycin antibiotics. Interestingly, so far no kirromycin resistance factor could be unambiguously identified in S. collinus Tü 365. An ABC transporter (FacT) has been reported to be involved in resistance against the related elfamycin antibiotic factumycin [51]. While there is a homologue encoded in the kirromycin biosynthetic gene cluster (KirT, 67 % amino acid identity/80 % similarity), which could have a similar role, we isolated mutations with interrupted kirT gene (unpublished). These strains still are able to produce kirromycin levels comparable to the wild type, indicating additional resistance mechanisms. A different kirromycin producer, S. ramocissimus, codes for 3 EF-Tu genes (tuf1, tuf2 and tuf3) [52], of which the gene product of tuf3 (EF-Tu3) displays a kirromycin-resistant phenotype [38]. The S. collinus Tü 365 genome codes for two elongation factors, EF-Tu1 (WP_020941664.1), and EF-Tu3 (WP_020938678.1) including a homolog to S. ramocissimus EF-Tu3. However, it had been experimentally demonstrated in S. ramocisimus that the EF-Tu3 conferred resistant phenotype is recessive, i.e., the ribosome is still blocked if there are sensitive EF-Tu species present. This is the case during kirromycin production in both, S. ramocissiumus [52], and S. collinus Tü 365 [33]. Thus, kirromycin-resistant EF-Tu seems not to be a main resistance mechanism.
The gene cluster encoding the biosynthesis of kirromycin (MIBiG ID: BGC0001070) was isolated prior to the availability of the S. collinus Tü 365 whole-genome sequence using a S. collinus Tü 365 cosmid library [58, 59]. Kirromycin biosynthesis was the first described example of an alliance between a cis-ATP-type PKS with trans-AT-type PKS in a single pathway [58]. Its biosynthesis, as well as its application for synthetic biological approaches has been reported in recent years [25, 27, 40, 61].
Genome sequencing data now revealed that the gene cluster encoding kirromycin is present in two copies in the terminal inverted repeats of S. collinus Tü 365 genome. This is an explanation for the great difficulty in mutant construction observed in earlier studies. Such duplicated gene clusters, located in the terminal inverted repeats of the chromosome have been identified in different Streptomyces strains, as for example, the type II PKS cluster encoding the biosynthesis of the antibiotic kinamycin in S. amobofaciens [6].
Lanthipeptide
The duplicated cluster situated in the long terminal inverted repeat of S. collinus Tü 365, named stc cluster (MIBiG ID: BGC0001226), codes for a putative lanthipeptide. Lanthipeptides are a prominent group of ribosomally synthesized and post-translationally modified peptides, containing thioether cross-links termed lanthionines and methyllanthionines.
A group of four genes localized on a 6.01-kb cluster shows high homology to biosynthesis genes of the class IV lanthipeptide venezuelin [17] encoded by S. venezuelae. The identified gene cluster contains a short reading frame for the precursor lanthipeptide designated stcA. Genes typically found in lanthipeptide biosynthetic gene clusters are present up- and downstream of stcA, including genes encoding a two component ABC transporter (stcT the ATP-binding element and stcH the permease subunit) and a lanthipeptide synthetase gene stcL. According to their organization and structure, the identified genes encode the biosynthesis of a type IV lanthipeptide (Iftime et al. manuscript submitted).
Terpenoids
Many terpenoid secondary metabolites are known to play significant roles in defense against predators, pathogens, or competitors, or are involved in conveying messages [15]. Terpenes are widespread in Streptomyces. Five clusters of S. collinus Tü 365 encoding terpenes are apparently responsible for the biosynthesis of the carotenoid isorenieratene, of the pentacyclic triterpenoid hopene, three sesquiterpenes such as pentalenolactone, albaflavenone and geosmin and the volatile monoterpene alcohol 2-methylisoborneol.
Isorenieratene
Carotenoids are a class of natural fat-soluble terpenoid pigments, characterized chemically by a long aliphatic polyene chain composed of eight isoprene units. Carotenoids are produced in a wide variety of plants, fungi, algae and bacteria. Besides their role as accessory light harvesting pigments in photosynthetic microorganisms and their potential function as antioxidant in bacteria [2, 11], carotenoids are of high interest in human health as anticancer agents, as immune response stimulants, in prevention of arteriosclerosis or macular degeneration [47]. In this context, the aromatic bicyclic carotenoid isorenieratene is important. 3,3′-Dihydroxyisorenieratene and isorenieratene seem to have multifunctional photoprotective properties and may be suitable natural compounds for the prevention of skin cancer [54]. So far the carotenoid isorenieratene has only been found in green photosynthetic bacteria and in a few actinomycetes [26].
One of five identified terpene clusters of S. collinus Tü 365 codes for putative isorenieratene biosynthesis genes (MIBiG ID: BGC0001227). Seven genes organized in two convergent operons crtVBIE and crtYTU (Fig. 2) could be referred to the isorenieratene cluster of S. collinus Tü 365, which is similar to the isorenieratene gene cluster of S. griseus. Five of these genes, crtBIEYU (Table 2), were demonstrated to be involved in biosynthesis of isorenieratene in S. griseus [29, 46]. The biosynthesis of isorenieratene starts with CrtE, a geranylgeranyl-pyrophosphate synthase. A second enzyme, CrtB, generates phytoene out of two molecules of geranylgeranyl-pyrophosphate. CrtI, a phytoene desaturase, converts phytoene to lycopene, which is the key intermediate for the biosynthesis of most carotenoids. Cyclization of lycopene by CrtY generates β-carotene. In a final step, CrtU, a β-carotene desaturase converts β-carotene to isorenieratene. CrtV and CrtT were proposed to function as methylesterase and methyltransferase, respectively, but they are dispensable for isorenieratene biosynthesis in S. griseus [26].
Compared to other microorganisms with constitutive carotenogenesis, in streptomycetes, carotenogenesis may be induced by light as found in S. coelicolor [49], or by some oxidative stressors as has been described for S. setonii or S. griseus [28].
The crt genes of S. collinus Tü 365 are not expressed under standard laboratory conditions. In this study, we investigated whether the isorenieratene accumulation in S. collinus Tü 365 can be stimulated by treatment with hydrogen peroxide as response to oxidative damage. Therefore, a 3-day culture of S. collinus Tü 365 was supplemented with H2O2 to an end concentration of 100 µM. Extracts prepared from strain S. collinus Tü 365 after oxic shock were analyzed by atmospheric pressure chemical ionization mass spectrum (APCI-MS), revealing traces of isorenieratene in the native strain (Figure S1).
In order to produce greater amounts of isorenieratene a cosmid including the crt genes contained in a region of 25 kb was introduced by conjugation into Amycolatopsis japonica, which does not contain an intrinsic isorenieratene biosynthetic gene cluster [48]. Cultivation of a recombinant A. japonica strain harboring the crt cluster for 5 days on HA agar resulted in orange pigmentation of the mycelium (Fig. 3). The production profile was analyzed by HPLC/APCI-MS (Figure S2). The produced carotenoids were identified as lycopene, β-carotene and isorenieratene by comparing masses and UV–Vis spectra to literature data.
Pentalenolactone
Our screening of the S. collinus Tü 365 genome resulted in the detection of a 11.6-kb spanning region, containing 13 candidate genes that were assigned to the biosynthesis of pentalenolactone (Fig. 2). The closest homologs of these genes were found in the ptl cluster of S. avermitilis (Table 3), which was shown to produce pentalenolactone [50, 62].
Pentalenolactone belongs to the family of sesquiterpene antibiotics that are biosynthesized by a variety of Streptomyces. The compound was first isolated from Streptomyces roseogriseus 1957 and later from over 30 Streptomyces species. This isoprene-derived natural product exhibits various biological activities which include diverse antibacterial and antifungal activity, as well as potent inhibitory activity toward the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase [8]. It was also reported to inhibit the replication of DNA viruses such as HSV-1 and HSV-2 [37].
Based on data available for S. avermitilis, we postulate the following pentalenolactone synthesis pathway in S. collinus Tü 365 (Table 3): The first step in the biosynthesis of pentalenolactone is the cyclization of the precursor farnesyl diphosphate (FPP) to the sesquiterpene hydrocarbon pentalenene [9], catalyzed by a pentalenene synthase, in S. collinus Tü 365 encoded by B446_29695. The oxidative conversion of pentalenene to pentalenolactone requires the presence of four redox enzymes such as cytochrome P450 B446_29700, the non-heme iron, α-ketoglutarate-dependent hydroxylase analog, encoded by gene B446_29665, the 1-deoxy-11-beta-hydroxypentalenate dehydrogenase B446_29675, the flavin-dependent monooxygenase B446_29680 and the dioxygenase B446_29685.
Of the four remaining ORFs, B446_29690 appears to encode a farnesyl diphosphate synthase. Gene B446_29670 was predicted to code for a transmembrane efflux protein, while the B446_29660 gene situated at the 5′-end of the ptl cluster is similar to gap1 of S. avermitilis, which was described as a resistance related gene [50]. Finally, B446_29655 resembles ptlR, a MarR-family transcriptional regulator that was shown to be responsible for the regulation of the pentalenolactone biosynthesis [65] in S. avermitilis.
A group of two genes located at the 3′-end of the ptl cluster, ptlJL, in S. avermitilis that were assigned to encode a lyase and a hypothetical protein are lacking in S. collinus Tü 365. In contrast to the pentalenolactone cluster of S. avermitilis, the cluster in S. collinus Tü 365 contains three additional genes (B446_29705, B446_29710 and B446_29715) related to isoprene biosynthetic genes belonging to the non-mevalonate pathway. These findings indicate that the pentalenolactone building blocks may be synthesized via the alternative non-mevalonate pathway in this strain. In S. avermitilis the gene sav_3006 is located near the ptl cluster. This gene encodes a putative polyprenyl diphosphate synthase, which may be responsible for the biosynthesis of FPP through the mevalonate pathway. Therefore, we speculate that despite the most notable similarity between the pentalenolactone gene clusters of S. collinus Tü 365 and S. avermitilis, the precursors for pentalenolactone are provided by different pathways in S. avermitilis and S. collinus Tü 365.
To prove the functionality of the pentalenolactone biosynthetic pathway in S. collinus Tü 365, we examined different culture extracts of S. collinus Tü 365 by GS-MS.
The production of pentalenene, an intermediate in biosynthesis of pentalenolactone was detected in S. collinus Tü 365 (Figure S3), suggesting that this pathway indeed encodes pentalenolactone biosynthesis.
Hopanoids
Our genome mining revealed a cluster of genes encoding the hopene and aminotrihydroxybacteriohopane (ATBH; also referred to as aminobacteriohopanetriol) biosynthesis in S. collinus Tü 365.
Hopanoids are a class of pentacyclic triterpenoid lipids. They are important components in bacterial membranes being involved in membrane fluidity, stability and permeability, similar to sterols in eukaryotic membranes [22]. They are composed of isoprene units and are present in a wide range of Gram-positive and Gram-negative bacteria. Hopanoids are synthesized by cyclization of the linear precursor squalene to pentacyclic hopene in a reaction catalyzed by the hopene-squalene cyclase enzyme. Also many Streptomyces strains have been described to produce hopanoids [42], including the model organism S. coelicolor A3(2) [41].
The hopanoid gene cluster of S. collinus Tü 365 contains 12 genes (Fig. 2) transcribed in one orientation. Based on the 12 identified genes and their similarity to hopanoid biosynthesis genes from S. coelicolor A3(2) (Table 4), a biosynthetic pathway can be proposed: The first step implies the IPP synthesis in a non-mevalonate pathway by involvement of 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (B446_30865) and a 1-deoxy-D-xylulose-5-phosphate synthase (B446_30870) gene. Apparently, genes belonging to the non-mevalonate pathway are present twice in the S. collinus Tü 365 genome, once for pentalenolactone production (see above) and once for hopanoid synthesis. Sequence alignment between these two pair of genes revealed 60 and 88 % gene sequence similarity, respectively.
In analogy to the hopanoid biosynthetic pathway S. coelicolor A3(2), the biosynthesis of hopene in S. collinus Tü 365 probable starts with the polyprenyldiphosphate synthase gene B446_30845 required for the formation of FPP. A second step consists in a head-to-tail elongation of FPP to squalene catalyzed by two phytoene synthases B446_30830 and B446_30835. Finally, the cyclization reaction of squalene to hopene is carried out by B446_30850, a squalene-hopene cyclase in a complex reaction.
Since the genes mentioned above are sufficient for the biosynthesis of hopene, we assumed that the radical SAM enzyme B446_30860 is responsible for the addition of an adenosyl radical to the C=C of hopene, while the remaining enzymes, a putative phosphorylase B446_30855 and an aminotransferase B446_30875 should generate the aminotrihydroxy moiety of the ATBH.
We therefore hypothesized that S. collinus Tü 365 can produce both hopene and ATBH. While the intermediates tetrahydrosqualene, dihydrosqualene, and squalene, as well as hopene were experimentally detected in S. collinus Tü 365 (Figure S4), the experimental confirmation of ATBH production still remains elusive.
Albaflavenone
The S. collinus Tü 365 genome also includes a second sesquiterpene cluster consisting of two genes encoding albaflavenone biosynthesis. The deduced gene products of the genes B446_24610 and B446_24615 have 84 and 82 % identity to SCO5222 and SCO5223 from in S. coelicolor A3(2) encoding a terpene cyclase and a cytochrome P450. In S. coelicolor A3(2) (SCO5222 and SCO5223) and S. avermitilis (SAV_3032 and SAV_3031) these enzymes are involved in albaflavenone biosynthesis. As shown for the homologous enzymes B446_24615 catalyzes the cyclization of farnesyl diphosphate to epi-izozizaene [1], while cytochrome P450 directs the two-step oxidation of epi-izozizaene to albaflavenone via the intermediate albaflavenole [64].
Even though efforts were made to isolate albaflavenone and derivatives, none of these compounds could be identified by GS-MS in cultures of S. collinus Tü 365 wild type. Further studies should support the hypothesis that albaflavenone is the metabolic product of this gene cluster identified in S. collinus Tü 365.
Geosmin
The gene B446_28510 located in cluster 25 revealed 79 % identity to geoA (SAV_2163) from S. avermitilis and 83 % identity to SCO6073 from S. coelicolor A3(2), which was demonstrated to mediate the entire conversion of FPP to geosmin in S coelicolor A3(2) [20]. Thus we assume that B446_28510 is probable responsible for the biosynthesis of geosmin in S. collinus Tü 365. The earthy smell of geosmin was detectable on agar plates cultivated with S. collinus Tü 365.
2-Methylisoborneol
2-Methylisoborneol (2-MIB) is an odorous metabolite that causes the earthy taste in drinking water [21]. Actinobacteria are the main producers of these terpenoid compounds in soil [63], biosynthesis genes for 2-MIB being first identified in these organisms [24]. A gene cluster encoding the biosynthesis of 2-MIB was identified in S. collinus Tü 365. In Streptomyces strains known for its production, 2-MIB is biosynthesized in a two-step reaction mechanism, in which a methyltransferase converts first geranyl diphosphate to methyl-geranyl diphosphate and in a second step 2-MIB is synthesized by a 2-MIB-synthase via methyl-geranyl diphosphate cyclization [24]. B446_04425 and B446_04430 from S. collinus Tü 365 were associated to a 2-MIB-synthase and a methyltransferase, based on protein similarity to SAM23877_0409 (53 %) and SAM23877_0410 (60 %), respectively, from S. amobofaciens.
Siderophore biosynthesis: desferrioxamine E
Iron plays an essential role for metabolic processes in microorganisms. To fulfill their iron needs, many microorganisms acquire iron by biosynthesis of high affinity iron chelators called siderophores [55]. Two pathways have been predominantly described to be involved in siderophore biosynthesis [45], a NRPS dependent route [12], and an NIS-route [39]. An example of NIS-encoded siderophores are the desferrioxamines [34], tris-hydroxamate ferric-iron-chelating metabolites produced by many Streptomyces species [32].
We discovered a desferrioxamine E gene cluster in S. collinus Tü 365 (Fig. 2) similar to that of S. coelicolor A3(2). By mass spectrometry we were able to provide evidence for the presence of desferrioxamine E in culture extracts of S. collinus Tü 365 (Figure S5). Based on the high similarity of the S. collinus Tü 365 genes with that of S. coelicolor A3(2) (Table 5), we propose an analogous pathway: in the first step, l-lysine is decarboxylated by the enzyme B446_14675 to yield cadaverine. In a second step, cadaverine can be oxidized by B446_14680 to produce N-hydroxycadaverine. The acyltransferase enzyme, B446_14685 then adds a succinyl group resulting in formation of the N-hydroxy-N-succinylcadaverine. B446_14690 catalyzes the final NTP-dependent reaction, which converts 3 molecules of N-hydroxy-N-succinylcadaverine into desferrioxamine E.
Spore pigment
Spore pigmentation in many Streptomyces strains is accompanied by the biosynthesis of an aromatic polyketide during the maturation of the spores in the aerial hyphae. The first identified cluster involved in spore pigmentation is the whiE cluster of S. coelicolor A3(2). The Chater group showed in 1990, that the expression of the minimal type II PKS of S. coelicolor A(3)2 resulted in the production of the dodecaketide naphtophenone TW95a [13].
Like many other Streptomyces species, S. collinus Tü 365 has grey-pigmented spores. Sequence analysis of the S. collinus Tü 365 genome revealed the presence of a gene cluster that has notable similarity to the whiE gene cluster of S. coelicolor A3(2). The spore pigment-associated gene cluster of S. collinus Tü 365 also consists of seven unidirectionally transcribed genes (ORF I–ORF VII) and one divergently transcribed gene ORF VIII (Fig. 2).
Consistent with the high level of similarity (Table 6), genes B446_17640, B446_17635 and B446_17630 are predicted to encode the three components of the minimal PKS: ketosynthase (KS), chain length factor (CLF) and acyl carrier protein (ACP). Gene B446_17645 is proposed to encode an aromatase while B446_17625 and B446_17620 are likely responsible for the cyclization of the nascent chain. B446_17655 apparently directs the introduction of a hydroxyl group into the cyclized polyketide. In S. coelicolor A3(2) it was proposed that whiE I encodes a protein needed for targeting the spore pigment within the spores [23]. Moreover, it was speculated that additional genes involved in late tailoring steps may be distributed independently in the linear chromosome [44]. Although the color of S. collinus Tü 365 spores changed to deep grey during the sporulation, the corresponding type II polyketide compound was not isolated so far.
Ectoine
Ectoine biosynthesis is widespread in many bacteria, conferring the ability to microorganisms to survive harsh osmotic stress conditions. In S. collinus Tü 365, a gene cluster resembling the ectABCD operon in S. coelicolor A3(2) [7] was identified (Fig. 2), suggesting that S. collinus Tü 365 is capable to induce the biosynthesis of the water-attracting organic osmolytes ectoine and hydroxyectoine under conditions of high salt concentration.
In concordance to homologous gene function in S. coelicolor A3(2) (Table 7), the first step in hydroxyectoine biosynthesis in S. collinus Tü 365 can be catalyzed by EctB (B446_09705), a diaminobutyric acid aminotransferase, which converts L-aspartate β-semialdehyde to L-diaminobutyrate. The acetyltransferase EctA (B446_09700) is assumed to add an acetyl group to L-diaminobutyrate resulting in acetyl-L-diaminobutyrate, while the ectoine synthase EctC (B446_09710) is proposed to accomplish the formation of ectoine. Finally, EctD (B446_09715) may be responsible for the conversion of ectoine to hydroxyectoine. Until now, the production of ectoine in S. collinus Tü 365 was not experimentally confirmed.
Deoxydehydrochorismic acid: an intermediate in menaquinone biosynthesis
While isolating kirromycin for biochemical studies by preparative chromatography, we observed antibacterial activity against E. coli and B. subtilis in an additional fraction not containing kirromycins. Follow-up of this active fraction yielded deoxydehydrochorismic acid (Figure S6), an intermediate of menaquinone biosynthesis. The single enzyme responsible for biosynthesis of deoxydehydrochorismic acid is MqnA, which converts chorismate to 3-((1-carboxyvinyl)oxy) benzoic acid. Bioinformatical analysis searching for MqnA, revealed the presence of a gene cluster probably encoding menaquinone biosynthesis in S. collinus Tü 365, since it exhibits high similarity to the menaquinone biosynthesis genes of S. coelicolor (Table 8). However, in contrast to S. coelicolor, where the mqn genes were found to be scattered across the genome [16], in S. collinus Tü 365 most genes are clustered with exception of mqnB and mqnD orthologs.
Menaquinone or vitamin K is a lipid-soluble molecule essential for commuting electrons between membrane bound redox enzymes in the electron-transport chain in prokaryotes [18]. In humans, vitamin K plays an important role as cofactor in the biosynthesis of proteins essential for blood clotting and bone health. We detected traces of vitamin K2 in S. collinus Tü 365 extracts, but it might be possible that this compound originates from the media components used for cultivating the strain.
Conclusions
Streptomycetes are a prolific source of secondary metabolites, being an important reservoir for novel bioactive compounds. In this work, we evaluate the capacity of S. collinus Tü 365 to produce secondary metabolites. By in silico analysis of the genome sequence of S. collinus Tü 365 using the antiSMASH genome mining software [5, 30, 56] and a manual curation, a total of 32 putative secondary metabolite biosynthetic gene clusters were identified.
Optimization of the cultivation conditions for the native strain and heterologous expression of two clusters enabled the identification of natural compounds previously not described for this strain.
In addition to the already known kirromycin gene cluster, ten secondary metabolites gene clusters encoded in the S. collinus Tü 365 genome share similarity to well described clusters allowing us to propose a product (Fig. 4). In addition to kirromycin, for five of them, in silico prediction successfully directed the identification of corresponding natural products: a lanthipeptide, the carotenoid isorenieratene, pentalenolactone, hopene and desferrioxamine E. Five gene clusters were putatively associated to the biosynthesis of albaflavenone, geosmin, 2-methylisoborneol, ectoine and a spore pigment. Furthermore, bioactivity-guided fractionation of strain extracts yielded a bioactive small molecule, deoxydehydrochorismic acid, which is an intermediate in menaquinone biosynthesis. Nine of the remaining uncharacterized secondary metabolites clusters in S. collinus Tü 365 are polyketides, non-ribosomal peptides or hybrids of the two structures, confirming that S. collinus Tü 365 is a prolific producer of such molecules. Hence, the activation and enhancement of cryptic pathways in S. collinus Tü 365 is of high interest. Although there are increasing numbers of secondary metabolites being isolated from S. collinus Tü 365, a substantial fraction of the encoded secondary metabolites and their functional roles in this strain remains to be elucidated.
Materials and methods
Cosmid library screening
A cosmid library of S. collinus Tü 365 was constructed using the integrative vector pOJ436 in cooperation with Combinature Biopharm AG, Berlin, Germany. Samples were automatically processed in 384-well plates and transferred to high-density clone arrays. Two identical arrays were used for hybridization experiments to identify cosmids harboring the whole crt gene cluster encoding the biosynthesis of the carotenoid isorenieratene as described earlier [59].
Detection of carotenoids
Extraction of carotenoids was performed using a protocol described for S. albus by Myronovskyi et al. 2014 [36]. The separation and identification of carotenoids was carried out using an Agilent 1200 HPLC System (Agilent Technologies, Waldbronn, Germany) coupled to LC/MSD Ultra Trap System XCT 6330, Agilent Technologies, Waldbronn, Germany) by UV-HPLC/APCI-MS. HPLC separation was carried out at a flow rate of 400 µl/min on a nucleosil column (Nucleosil 100 C18 3 µm, 100 × 2 mm ID fitted with a precolumn 10 × 2 mm, same stationary phase, Dr. Maisch GmbH, Ammerbuch) with the mobile phase composed of formic acid (A = 0.1 %), and formic acid in acetonitrile (B = 0.06 %). A gradient from 70 to 100 % of B in 10 min with a 30-min hold at 100 % for solvent B was used. The column was maintained at 40 °C.
Identification of produced terpene products
We identified the production of terpene compounds by cultivation of S. collinus Tü 365 for 6 days on cellophane-covered mannitol-soy agar plates as well as in liquid cultures. For liquid cultures S. collinus Tü 365 was inoculated into 50 ml TSB medium and cultivated for 2 days on a rotary shaker at 27 °C and 180 rpm. Then 200 µl of seed culture were transferred into 200 ml KPM medium (for recipe, see Supporting Information) and incubated for 5 days at 27 °C and 180 rpm for secondary metabolites production. Culture broth was harvested by centrifugation for 20 min at 5000 rpm. Extraction of lipids from S. collinus Tü 365 was carried out using the method described by Bligh and Dyer (1959). The terpene fractions were separated by GS-MS (Shimadzu GC-17A) on a capillary column (Optima 5 MS, 0.25 mm × 15 m, Macherey–Nagel). The temperature program was 45 °C for 5 min isotherm with a temperature ramp of 45–200 °C at 18 °C/min and 200–340 °C at 4 °C/min.
Pentalenene was observed at m/z 204 Da. The observed mass spectrum was compared with those of pentalenene identified from S. avermitilis.
Hopanoids were identified by comparison of their retention times in GS-MS and fragmentation patterns with those of analytical standards. The mass peak at m/z 410, the base peak at m/z 189/191 and the fragmentation pattern are characteristic for hopene.
Identification of desferrioxamine E
Streptomyces collinus Tü 365 was inoculated with a TSB preculture (10 ml) in 500 ml of modified ancovenin production broth (for recipe, see Supporting Information) and incubated for 3 days at 27 °C/180 rpm. The culture broth was harvested by centrifugation for 20 min at 5000 rpm. The supernatant was extracted twice with XAD1180 resin. The XAD1180 resin was then extracted with methanol–acetone (1:1) and the extract was dried in vacuo. The extract was then resolved in 500 µl of MeOH and analyzed using an Agilent 1200 HPLC System (Agilent Technologies, Waldbronn, Germany) coupled to an LC/MSD Ultra Trap System XCT 6330, Agilent Technologies, Waldbronn, Germany).
Chromatographic separation was performed at a flow rate of 400 µl/min using stationary phase C18 column Nucleosil 100 3 µm (100 × 2 mm ID, fitted with a precolumn 10 × 2 mm, same stationary phase, Dr. Maisch GmbH, Ammerbuch) with the mobile phase composed of formic acid (A = 0.1 %), and formic acid in acetonitrile (B = 0.06 %). A gradient from 10 to 100 % of B in 15 min with a 2-min hold at 100 % for solvent B, was used.
Detection of deoxydehydrochorismic acid
Streptomyces collinus Tü 365 was inoculated with a TSB preculture (10 %) in 100 ml of KPM production broth and incubated for 5 days at 27 °C/180 rpm in 500-ml shaking flasks. The culture was extracted twice with 1 volume ethyl acetate at pH 4 and dried in vacuo. The extract was resolved in appropriate volumes of MeOH (200 μl for 50 ml extraction volume).
The samples were analyzed using an Agilent 1200 HPLC System (Agilent Technologies, Waldbronn, Germany) coupled to LC/MSD Ultra Trap System XCT 6330, Agilent Technologies, Waldbronn, Germany). Chromatographic separation was performed at a flow rate of 400 µl/min using stationary phase C18 column Nucleosil 100 3 µm (100 × 2 mm ID, fitted with a precolumn 10 × 2 mm, same stationary phase, Dr. Maisch GmbH, Ammerbuch) with the mobile phase composed of formic acid (A = 0.1 %), and formic acid in acetonitrile (B = 0.06 %). A gradient from 10 to 100 % of B in 15 min with a 2-min hold at 100 % for solvent B, was used.
References
Aaron JA, Lin X, Cane DE, Christianson DW (2010) Structure of epi-isozizaene synthase from Streptomyces coelicolor A3(2), a platform for new terpenoid cyclization templates. Biochemistry 49:1787–1797. doi:10.1021/bi902088z
Avalos J, Carmen Limon M (2015) Biological roles of fungal carotenoids. Curr Genet 61:309–324. doi:10.1007/s00294-014-0454-x
Bachmann BO, Van Lanen SG, Baltz RH (2014) Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol 41:175–184. doi:10.1007/s10295-013-1389-9
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147. doi:10.1038/417141a
Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E, Weber T (2013) antiSMASH 2.0–a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41:W204–W212. doi:10.1093/nar/gkt449
Bunet R, Song L, Mendes MV, Corre C, Hotel L, Rouhier N, Framboisier X, Leblond P, Challis GL, Aigle B (2011) Characterization and manipulation of the pathway-specific late regulator AlpW reveals Streptomyces ambofaciens as a new producer of Kinamycins. J Bacteriol 193:1142–1153. doi:10.1128/JB.01269-10
Bursy J, Kuhlmann AU, Pittelkow M, Hartmann H, Jebbar M, Pierik AJ, Bremer E (2008) Synthesis and uptake of the compatible solutes ectoine and 5-hydroxyectoine by Streptomyces coelicolor A3(2) in response to salt and heat stresses. Appl Environ Microbiol 74:7286–7296. doi:10.1128/AEM.00768-08
Cane DE, Sohng JK (1989) Inhibition of glyceraldehyde-3-phosphate dehydrogenase by pentalenolactone: kinetic and mechanistic studies. Arch Biochem Biophys 270:50–61
Cane DE, Sohng JK (1994) Inhibition of glyceraldehyde-3-phosphate dehydrogenase by pentalenolactone. 2. Identification of the site of alkylation by tetrahydropentalenolactone. Biochemistry 33:6524–6530
Chater K (1999) David Hopwood and the emergence of Streptomyces genetics. Int Microbiol 2:61–68
Clauditz A, Resch A, Wieland KP, Peschel A, Götz F (2006) Staphyloxanthin plays a role in the fitness of Staphylococcus aureus and its ability to cope with oxidative stress. Infect Immun 74:4950–4953. doi:10.1128/IAI.00204-06
Crosa JH, Walsh CT (2002) Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev 66:223–249
Davis NK, Chater KF (1990) Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol 4:1679–1691
Demain AL (2014) Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol 41:185–201. doi:10.1007/s10295-013-1325-z
Gershenzon J, Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3:408–414. doi:10.1038/nchembio.2007.5
Goble AM, Toro R, Li X, Ornelas A, Fan H, Eswaramoorthy S, Patskovsky Y, Hillerich B, Seidel R, Sali A et al (2013) Deamination of 6-aminodeoxyfutalosine in menaquinone biosynthesis by distantly related enzymes. Biochemistry 52:6525–6536. doi:10.1021/bi400750a
Goto Y, Li B, Claesen J, Shi Y, Bibb MJ, van der Donk WA (2010) Discovery of unique lanthionine synthetases reveals new mechanistic and evolutionary insights. PLoS Biol 8:e1000339. doi:10.1371/journal.pbio.1000339
Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T (2008) An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321:1670–1673. doi:10.1126/science.1160446
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531. doi:10.1038/nbt820
Jiang J, He X, Cane DE (2006) Geosmin biosynthesis. Streptomyces coelicolor germacradienol/germacrene D synthase converts farnesyl diphosphate to geosmin. J Am Chem Soc 128:8128–8129. doi:10.1021/ja062669x
Juttner F, Watson SB (2007) Biochemical and ecological control of geosmin and 2-methylisoborneol in source waters. Appl Environ Microbiol 73:4395–4406. doi:10.1128/AEM.02250-06
Kannenberg EL, Poralla K (1999) Hopanoid biosynthesis and function in bacteria. Naturwissenschaften 86:168–176. doi:10.1007/s001140050592
Kelemen GH, Brian P, Flardh K, Chamberlin L, Chater KF, Buttner MJ (1998) Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3 (2). J Bacteriol 180:2515–2521
Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H (2008) Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci USA 105:7422–7427. doi:10.1073/pnas.0802312105
Koryakina I, McArthur J, Randall S, Draelos MM, Musiol EM, Muddiman DC, Weber T, Williams GJ (2013) Poly specific trans-acyltransferase machinery revealed via engineered acyl-CoA synthetases. ACS Chem Biol 8:200–208. doi:10.1021/cb3003489
Krügel H, Krubasik P, Weber K, Saluz HP, Sandmann G (1999) Functional analysis of genes from Streptomyces griseus involved in the synthesis of isorenieratene, a carotenoid with aromatic end groups, revealed a novel type of carotenoid desaturase. Biochim Biophys Acta 1439:57–64
Laiple KJ, Härtner T, Fiedler HP, Wohlleben W, Weber T (2009) The kirromycin gene cluster of Streptomyces collinus Tü 365 codes for an aspartate-alpha-decarboxylase, KirD, which is involved in the biosynthesis of the precursor beta-alanine. J Antibiot (Tokyo) 62:465–468. doi:10.1038/ja.2009.67
Lee HS, Ohnishi Y, Horinouchi S (2001) A sigmaB-like factor responsible for carotenoid biosynthesis in Streptomyces griseus. J Mol Microbiol Biotechnol 3:95–101
Liu R, Wang T, Zhang B, Qin L, Wu C, Li Q, Ma L (2015) Lutein and zeaxanthin supplementation and association with visual function in age-related macular degeneration. Invest Ophthalmol Vis Sci 56:252–258. doi:10.1167/iovs.14-15553
Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, Breitling R (2011) antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39:W339–W346. doi:10.1093/nar/gkr466
Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC et al (2015) Minimum information about a biosynthetic gene cluster. Nat Chem Biol 11:625–631. doi:10.1038/nchembio.1890
Meiwes J, Fiedler HP, Zähner H, Konetschny-Rapp S, Jung G (1990) Production of desferrioxamine E and new analogues by directed fermentation and feeding fermentation. Appl Microbiol Biotechnol 32:505–510
Mikulik K, Zhulanova E (1995) Sequencing of the tuf1 gene and the phosphorylation pattern of EF-Tu1 during development and differentiation in Streptomyces collinus producing kirromycin. Biochem Biophys Res Commun 213:454–461. doi:10.1006/bbrc.1995.2153
Müller A, Zähner H (1968) Metabolic products of microorganisms. 65. Ferrioxamine from Eubacteriales. Arch Mikrobiol 62:257–263
Musiol EM, Härtner T, Kulik A, Moldenhauer J, Piel J, Wohlleben W, Weber T (2011) Supramolecular templating in kirromycin biosynthesis: the acyltransferase KirCII loads ethylmalonyl-CoA extender onto a specific ACP of the trans-AT PKS. Chem Biol 18:438–444. doi:10.1016/j.chembiol.2011.02.007
Myronovskyi M, Tokovenko B, Brötz E, Rückert C, Kalinowski J, Luzhetskyy A (2014) Genome rearrangements of Streptomyces albus J1074 lead to the carotenoid gene cluster activation. Appl Microbiol Biotechnol 98:795–806. doi:10.1007/s00253-013-5440-6
Nakagawa A, Tomoda H, Hao MV, Okano K, Iwai Y, Omura S (1985) Antiviral activities of pentalenolactones. J Antibiot (Tokyo) 38:1114–1115
Olsthoorn-Tieleman LN, Palstra RJ, van Wezel GP, Bibb MJ, Pleij CW (2007) Elongation factor Tu3 (EF-Tu3) from the kirromycin producer Streptomyces ramocissimus is resistant to three classes of EF-Tu-specific inhibitors. J Bacteriol 189:3581–3590
Oves-Costales D, Kadi N, Challis GL (2009) The long-overlooked enzymology of a nonribosomal peptide synthetase-independent pathway for virulence-conferring siderophore biosynthesis. Chem Commun (Camb) 43:6530–6541. doi:10.1039/b913092f
Pavlidou M, Pross EK, Musiol EM, Kulik A, Wohlleben W, Weber T (2011) The phosphopantetheinyl transferase KirP activates the ACP and PCP domains of the kirromycin NRPS/PKS of Streptomyces collinus Tü 365. FEMS Microbiol Lett 319:26–33. doi:10.1111/j.1574-6968.2011.02263.x
Poralla K, Muth G, Härtner T (2000) Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol Lett 189:93–95
Rohmer M, Bouvier-Nave P, Ourisson G (1984) Distribution of hopanoid triterpenes in prokaryotes. J Gen Microbiol 130:1137–1150. doi:10.1099/00221287-130-5-1137
Rückert C, Szczepanowski R, Albersmeier A, Goesmann A, Iftime D, Musiol EM, Blin K, Wohlleben W, Pühler A, Kalinowski J et al (2013) Complete genome sequence of the kirromycin producer Streptomyces collinus Tü 365 consisting of a linear chromosome and two linear plasmids. J Biotechnol 168:739–740. doi:10.1016/j.jbiotec.2013.10.004
Salerno P, Persson J, Bucca G, Laing E, Ausmees N, Smith CP, Flardh K (2013) Identification of new developmentally regulated genes involved in Streptomyces coelicolor sporulation. BMC Microbiol 13:281. doi:10.1186/1471-2180-13-281
Schmelz S, Botting CH, Song L, Kadi NF, Challis GL, Naismith JH (2011) Structural basis for acyl acceptor specificity in the achromobactin biosynthetic enzyme AcsD. J Mol Biol 412:495–504. doi:10.1016/j.jmb.2011.07.059
Schumann G, Nürnberger H, Sandmann G, Krügel H (1996) Activation and analysis of cryptic crt genes for carotenoid biosynthesis from Streptomyces griseus. Mol Gen Genet 252:658–666
Shaish A, Daugherty A, O’Sullivan F, Schonfeld G, Heinecke JW (1995) Beta-carotene inhibits atherosclerosis in hypercholesterolemic rabbits. J Clin Invest 96:2075–2082. doi:10.1172/JCI118256
Stegmann E, Albersmeier A, Spohn M, Gert H, Weber T, Wohlleben W, Kalinowski J, Rückert C (2014) Complete genome sequence of the actinobacterium Amycolatopsis japonica MG417-CF17(T) (=DSM 44213T) producing (S, S)-N, N’-ethylenediaminedisuccinic acid. J Biotechnol 189:46–47. doi:10.1016/j.jbiotec.2014.08.034
Takano H, Obitsu S, Beppu T, Ueda K (2005) Light-induced carotenogenesis in Streptomyces coelicolor A3(2): identification of an extracytoplasmic function sigma factor that directs photodependent transcription of the carotenoid biosynthesis gene cluster. J Bacteriol 187:1825–1832. doi:10.1128/JB.187.5.1825-1832.2005
Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H (2006) A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry 45:6179–6186. doi:10.1021/bi060419n
Thaker MN, Garcia M, Koteva K, Waglechner N, Sorensen D, Medina R, Wright GD (2012) Biosynthetic gene cluster and antimicrobial activity of the elfamycin antibiotic factumycin. MedChemComm 3:1020–1026. doi:10.1039/C2MD20038D
Vijgenboom E, Woudt LP, Heinstra PW, Rietveld K, van Haarlem J, van Wezel GP, Shochat S, Bosch L (1994) Three tuf-like genes in the kirromycin producer Streptomyces ramocissimus. Microbiology 140(Pt 4):983–998
Vos C, Verwiel PEJ (1973) Total structure of the novel antibiotic mocimycin (MYC 8003). Tetrahedron Lett 52:5173–5176
Wagener S, Völker T, De Spirt S, Ernst H, Stahl W (2012) 3,3′-Dihydroxyisorenieratene and isorenieratene prevent UV-induced DNA damage in human skin fibroblasts. Free Radic Biol Med 53:457–463. doi:10.1016/j.freeradbiomed.2012.05.022
Wandersman C, Delepelaire P (2004) Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–647. doi:10.1146/annurev.micro.58.030603.123811
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W et al (2015) antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243. doi:10.1093/nar/gkv437
Weber T, Charusanti P, Musiol-Kroll EM, Jiang X, Tong Y, Kim HU, Lee SY (2015) Metabolic engineering of antibiotic factories: new tools for antibiotic production in actinomycetes. Trends Biotechnol 33:15–26. doi:10.1016/j.tibtech.2014.10.009
Weber T, Laiple KJ, Pross EK, Textor A, Grond S, Welzel K, Pelzer S, Vente A, Wohlleben W (2008) Molecular analysis of the kirromycin biosynthetic gene cluster revealed beta-alanine as precursor of the pyridone moiety. Chem Biol 15:175–188. doi:10.1016/j.chembiol.2007.12.009
Weber T, Welzel K, Pelzer S, Vente A, Wohlleben W (2003) Exploiting the genetic potential of polyketide producing streptomycetes. J Biotechnol 106:221–232
Wolf H, Zähner H (1972) Stoffwechselprodukte von Mikroorganismen. 99. Mitteilung: Kirromycin. Arch Mikrobiol 83:147–154
Ye Z, Musiol EM, Weber T, Williams GJ (2014) Reprogramming acyl carrier protein interactions of an Acyl-CoA promiscuous trans-acyltransferase. Chem Biol 21:636–646. doi:10.1016/j.chembiol.2014.02.019
You Z, Omura S, Ikeda H, Cane DE (2007) Pentalenolactone biosynthesis: molecular cloning and assignment of biochemical function to PtlF, a short-chain dehydrogenase from Streptomyces avermitilis, and identification of a new biosynthetic intermediate. Arch Biochem Biophys 459:233–240. doi:10.1016/j.abb.2006.11.016
Zaitlin B, Watson SB (2006) Actinomycetes in relation to taste and odour in drinking water: myths, tenets and truths. Water Res 40:1741–1753. doi:10.1016/j.watres.2006.02.024
Zhao B, Lei L, Vassylyev DG, Lin X, Cane DE, Kelly SL, Yuan H, Lamb DC, Waterman MR (2009) Crystal structure of albaflavenone monooxygenase containing a moonlighting terpene synthase active site. J Biol Chem 284:36711–36719. doi:10.1074/jbc.M109.064683
Zhu D, Wang Y, Zhang M, Ikeda H, Deng Z, Cane DE (2013) Product-mediated regulation of pentalenolactone biosynthesis in Streptomyces species by the MarR/SlyA family activators PenR and PntR. J Bacteriol 195:1255–1266. doi:10.1128/JB.02079-12
Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR (2012) The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity. PLoS ONE 7:e34064. doi:10.1371/journal.pone.0034064
Acknowledgments
This work was supported by ERA-IB-GenoDrug (BMBF FKZ 0315930), The German Center for Infection Research (DZIF) (TTU 09.802) and the Graduate College 1708 (Bacterial Survival Strategies). T. Weber is supported by a grant from the Novo Nordisk Foundation. The authors acknowledge P. Schmieder (FMP Berlin-Buch) for recording the NMR spectra for deoxydehydrochorismic acid.
Author information
Authors and Affiliations
Corresponding author
Additional information
Special Issue: Natural Product Discovery and Development in the Genomic Era. Dedicated to Professor Satoshi Ōmura for his numerous contributions to the field of natural products.
We would like to dedicate this publication to Alfred Pühler on the occasion of his 75th birthday.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Iftime, D., Kulik, A., Härtner, T. et al. Identification and activation of novel biosynthetic gene clusters by genome mining in the kirromycin producer Streptomyces collinus Tü 365. J Ind Microbiol Biotechnol 43, 277–291 (2016). https://doi.org/10.1007/s10295-015-1685-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-015-1685-7