Abstract
Genome shuffling is an efficient approach for the rapid engineering of microbial strains with desirable industrial phenotypes. In this study, a strategy of incorporating streptomycin resistance screening into genome shuffling (GS-SR) was applied for rapid improvement of doramectin production by Streptomyces avermitilis NEAU1069. The starting mutant population was generated through treatment of the spores with N-methyl-N’-nitro-N-nitrosoguanidine and ultraviolet (UV) irradiation, respectively, and five mutants with higher productivity of doramectin were selected as starting strains for GS-SR. Finally, a genetically stable strain F4-137 was obtained and characterized to be able to yield 992 ± 4.4 mg/l doramectin in a shake flask, which was 7.3-fold and 11.2-fold higher than that of the starting strain UV-45 and initial strain NEAU1069, respectively. The doramectin yield by F4-137 in a 50-l fermentor reached 930.3 ± 3.8 mg/l. Furthermore, the factors associated with the improved doramectin yield were investigated and the results suggested that mutations in ribosomal protein S12 and the enhanced production of cyclohexanecarboxylic coenzyme A may contribute to the improved performance of the shuffled strains. The random amplified polymorphic DNA analysis showed a genetic diversity among the shuffled strains, which confirmed the occurrence of genome shuffling. In conclusion, our results demonstrated that GS-SR is a powerful method for enhancing the production of secondary metabolites in Streptomyces.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
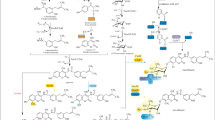
Doramectin (Fig. 1), sold commercially as Dectomax, is a broad-spectrum macrocyclic lactone anthelmintic that can be produced by avermectin-producing strain Streptomyces avermitilis in the presence of cyclohexanecarboxylic acid (CHC) [8, 14, 25, 26, 31]. It belongs to the avermectin class drug family, but possesses better pharmacokinetic characteristics and efficacy than avermectins [14]. Doramectin has already been approved by the Food and Drug Administration (FDA) as a veterinary drug for the treatment of parasites such as gastrointestinal roundworms, lungworms, eyeworms, grubs, sucking lice, and mange mites in cattle, sheep, swine, and others. The properties and applications of doramectin have promoted the commercial production of this compound. Unfortunately, industrialization of this valuable antibiotic is limited by the low production in wild-type strain. Although the biosynthetic pathway of doramectin has been thoroughly elucidated, and several methods such as site-specific mutagenesis, semi-synthetic DNA shuffling, and combinatorial biosynthesis have been employed to improve doramectin productivity, the doramectin yield is still unsatisfactory because that secondary metabolism is a complicated network involved in multiple genes and it is difficult to improve the doramectin yield by modifying single or several genes [25, 26, 31].
Classical strain improvement methods such as mutation and random selection have succeeded in generating strains with industrial potential, but it is a time-consuming and expensive process [7]. The advent of molecular biology led to new strain improvement techniques that depend on the targeted modification, elimination, or over-expression of key genes involved in phenotypes. However, these approaches are only applicable to genes that have been characterized, along with some knowledge of what modifications need to be made to obtain the desired effect [7]. Due to the complexity, the classic or molecular biological approach may be inadequate for improving complex desired phenotypes. Therefore, researchers attempted to manipulate the microbial host to express complex phenotype in industrial settings through integration of classic and modern strain improvement approaches [1, 22]. Genome shuffling is such a tool to address this issue, which is based on the genetic recombination without the need of the knowledge of detailed genetic information [7, 21]. Different genes associated with production can be recombined during several rounds of genome shuffling and desired phenotypes can be obtained. Phenotypic improvement by genome shuffling is undoubtedly an important milestone in both industrial and medical application [24]. So far, genome shuffling has been successfully applied to enhance the yield of secondary metabolites by actinomycetes [6, 11, 17, 42], to improve the performance of Saccharomyces cerevisiae [12, 15, 16, 23, 28, 33, 43], and to increase the synthesis of microbial enzymes [30, 39, 40]. However, to the best of our knowledge, no attempts have been reported to improve doramectin-producing strains by genome shuffling as yet.
In the present study, a strategy designated as GS-SR, which incorporates streptomycin resistance screening into genome shuffling, was employed to increase the doramectin yield in the fermentation with a new doramectin-producing strain S. avermitilis NEAU1069. Furthermore, the mechanism for the improvement of shuffled strain was also investigated.
Materials and methods
Microorganism
The strain S. avermitilis NEAU1069 was used as the initial strain and stored in 20 % glycerol at −80 °C, which has been deposited at the China General Microbiology Culture Collection Center (accession no. CGMCC 2943), Institute of Microbiology, Chinese Academy of Science [31, 33].
Medium and cultivation conditions
Streptomyces avermitilis NEAU1069 was maintained on YMS medium containing 1 % soluble starch, 0.2 % yeast extract, 1 % KNO3, 2 % agar, pH 7.0. The seed medium consists of 2 % corn starch, 0.5 % glucose, 1 % yeast extract, 1 % cotton seed flour, pH 7.2. The producing medium used for fermentation in shake flasks and fermenters contains 0.5 % glucose, 10 % corn starch, 1 % peptone, 1 % cotton seed flour, 0.1 % NaCl, 0.2 % K2HPO4, 0.1 % MgSO4·7H2O and 0.7 % CaCO3, pH 7.0. The solid medium used for CHC tolerance test consists of 0.5 % glucose, 1 % malt extract powder, 0.4 % yeast extract, 0.04 % MgSO4·7H2O, 0.005 % CaCO3, 0.0006 % FeCl3, 2.4 % agar, and CHC with different concentrations (pH 7.0). All of the media were sterilized at 121 °C for 20 min. Slant culture was incubated for 6–8 days at 28 °C and 40 % relative humidity condition. Ten milliliters of sterile water was added to the slant of YMS medium. The spores were scraped and transferred to a sterile screw-cap glass tube and shaken vigorously to break spore clump. The spore suspension was then filtered through six layers of sterile filter cheesecloth and adjusted to 107–108 cfu/ml. Two milliliters of the spore suspension was inoculated into a 250-ml Erlenmeyer flask containing 25 ml of seed medium and incubated at 28 °C for 24 h at 250 rpm. Then 8.0 ml of the culture was transferred into a 250-ml Erlenmeyer flask containing 25 ml of producing medium. Fermentation was carried out at 28 °C for 12 days on a rotary shaker at 250 rpm, with the supplement of CHC at a final concentration of 100 and 60 mg/l at 24 and 168 h, respectively.
Medium for protoplast formation is YEME medium containing 0.1 % glucose, 0.5 % tryptone, 0.3 % yeast extract, 0.3 % malt extract, 10.3 % sucrose, 0.5 % glycine, 0.1 % MgCl2·6H2O, pH 7.0. YEME culture was incubated at 28 °C and 40 % relative humidity for 20 h.
The regeneration plate medium contains 10.3 % sucrose, 1 % glucose, 0.4 % yeast extract, 0.4 % peptone, 0.1 % casamino acid, 1 % MgCl2·6H2O, 0.025 % K2SO4, 0.735 % CaCl2·2H2O, 2 % trace element solution [10], 20 % TES buffer (5.73 %, adjusted to pH 7.2), and 2 % agar. The pH was adjusted to 7.0 before autoclaving. Soluble fermentation medium (5 % soluble starch, 1.2 % yeast extract, 0.05 % K2HPO4·3H2O, 0.05 % MgSO4·7H2O, 0.4 % KCl, 0.0005 % CoCl2·6H2O) was used to cultivate mycelia for RT-PCR analysis.
Strain mutagenesis and mutant screening
Two approaches were employed for mutagenizing the parent strain S. avermitilis NEAU1069. For N-methyl-N’-nitro-N-nitrosoguanidine (MNNG) mutagenesis, spores of the parent strain were collected and suspended in 50 mM Tris–HCl buffer (pH 6.0). About 5 ml of the spore suspension containing 5 × 107 cells was treated with MNNG (1 mg/ml) for 30 min. Subsequently, the mutant cells were collected by filtration and washed three times with sterile saline. Then, the resulting cells were appropriately diluted and spread on YMS agar plate. Ultraviolet (UV) irradiation was performed by exposing the spore suspension directly to UV light as described as follows. Five milliliters of spore suspension from a slant culture of the parent strain was transferred to an aseptic plate with a rotor. The plate with the cover removed was exposed to UV irradiation for 45 s at a distance of 30 cm from a UV lamp with a wavelength of 254 nm and a power of 30 W. After appropriately diluting, the suspension of survived spores was spread on the YMS agar plate.
All the plates were incubated at 28 °C for 6–7 days. The colonies that appeared to grow the fasted were selected and their doramectin yields were analyzed by fermentation test and high-performance liquid chromatography (HPLC) determination. The mutants with higher production were preserved and taken as starting strains for genome shuffling.
Streptomycin resistance-aided genome shuffling (GS-SR)
The spore suspension (1 ml) of the starting strains was subsequently inoculated into a 250-ml Erlenmeyer flask containing 25 ml of YEME medium. After incubation, mycelia were harvested by centrifugation at 4,000×g for 10 min, and washed twice with 10 ml of P buffer [10]. Lysozyme was then added to a final concentration of 3 mg/ml. After incubation at 30 °C for 30 min, the protoplasts of each strain were prepared. The appearance of spherical cells, as adjusted by phase-contrast microscopy, was used as indicator of protoplast formation. Protoplasts were collected by centrifugation at 1,500×g for 5 min and then suspended in P buffer. Equal numbers of protoplasts from two different populations were mixed and fused by suspension in 2 ml of P buffer containing 40 % PEG4000 for 3 min with gently shaking. After washing twice with 10 ml of P buffer, the fusants was re-suspended in P buffer (5 ml) and transferred to the regeneration plate medium containing 5 μg/ml streptomycin and incubated at 28 °C and 40 % relative humidity for 7–8 days. Then, 75 % of the colonies that appeared well on the regeneration plate medium were selected for fermentation tests in a shake flask. The top five strains with the highest doramectin yields were used as starting strains for subsequent rounds of genome shuffling, in which the concentrations of streptomycin used for resistance screening were 10, 15, and 20 μg/ml, respectively. During the experiments, appropriate dilution should be conducted if the concentration of protoplasts or fusants was too high to isolate individual colonies. On the other hand, a repeated step should be taken to obtain enough colonies if the numbers of colonies were less than 150. To determine the effect of repeated protoplast preparation and regeneration on the improved performances of the recombinant strains [12], one control consisted of protoplasts generated from starting strains that were not fused but regenerated on streptomycin-free medium was conducted. In the first round of experiments, ~50 % of the colonies that appeared well on the regeneration plate medium were randomly selected for fermentation analysis. The top five strains with the highest doramectin yields were subjected to the next protoplast regeneration cycle. After another three rounds, the regenerated strains were analyzed. In order to validate the efficiency of the GS-SR, two additional control experiments designated as the second and third controls were also conducted. The procedure used in the second control was similar to that described in GS-SR except that the protoplasts generated from starting strain were not fused but regenerated by plating on streptomycin-containing medium. The third control designated as GS-WSR was similar to GS-SR except no streptomycin was added into the regeneration plate medium. Additionally, a doramectin resistance-aided genome shuffling was also employed, in which doramectin with different concentrations (200, 300, 400, and 500 mg/l) was used as selective marker and successively added to regeneration plate medium.
Amplification of rpsL gene
Total genomic DNAs of the test strains were extracted and purified according to the standard procedures [13]. The rpsL gene was amplified by PCR from S. avermitilis NEAU1069 genome by using the primers rspL-F (5′-ATTCGGCACAGAAACCGGAGAAG-3′) and rpsL-R (5′-AGAGGAGAACCGTAGACCGGGTCGA-3′) [29]. The amplification protocol consisted of an initial denaturation at 96 °C for 3 min, followed by 30 cycles of denaturation at 98 °C for 10 s and amplification at 60 °C for 1 min, with a final extension at 72 °C for 5 min. The purified PCR products were sequenced by TaKaRa Bio Inc (Dalian, China).
Semiquantitative RT-PCR analysis of polyketide synthases transcription
Total RNA was prepared from the mycelial grown in YEME liquid medium using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNase-free DNase (Promega, Madison, WI, USA) was used to eliminate all DNA. The quality and quantity of RNA were determined using gel electrophoresis and spectrophotometer assays (Eppendorf, Hamburg, Germany). The transcription profiles of polyketide synthases (PKS) involving the biosynthesis of avermectin/doramectin and oligomycin were conducted by a semiquantitative RT-PCR analysis with Qiagen One-Step RT-PCR kit (Qiagen, Hilden, Germany) and using primers aveA2-F/R (5′-GAGCGAACAGGATTAGATACCC-3′ and 5′-TTGAGGGCTGATGTGTGTATC-3′) and olmA1-F/R (5′-GAACTCGGTACGGTCCAGG-3′ and 5′-CGTCAACACGGCACTCAC-3′), respectively. RT-PCRs without reverse transcription were used as a control for absence of residual DNA. Reaction mixtures contained 10 pmol of each primer and 200 ng RNA in a total volume of 20 μl. RT-PCR conditions were as follows: cDNA synthesis, 50 °C for 30 min and 55 °C for 30 min followed by 95 °C for 15 min; amplification, 30 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 1 min; final extension, 72 °C for 10 min. The transcription of the 16S rRNA gene was used as an internal control and a pair of primers 16S-F/R (5′-GAGCGAACAGGATTAGATACCC-3′ and 5′-AACCCAACATCTCACGACAC-3′) was used. Products were detected by 1.5 % agarose gel electrophoresis and visualized by staining with ethidium bromide. RT-PCR band images were quantified using Quantity One Analysis software 4.6.6 (Bio-Rad, Hercules, CA, USA). The level of transcription in each assay was normalized to the corresponding level of transcription of 16S rRNA.
RAPD analysis
Thirty randomly selected RAPD primers with a 60–70 % G + C content (Shanghai Sangon Co., Shanghai, China) were used for RAPD analysis. The RAPD amplification reaction was conducted in a buffer (25 μl) that contained 1× PCR buffer, 20 pmol of primer, 0.2 mmol/l of dNTP, 100 ng of genomic DNA, and 2.5 U of Taq LA DNA polymerase (TaKaRa Biotechnology Co., Dalian, China). Amplification was performed in Bio-Rad’s imaging system Universal Hood II (Hercules, CA, USA). Thermal cycling parameters consisted of 4-min denaturation (94 °C) followed by 40 cycles of 1-min denaturation (94 °C), 1-min annealing (32 °C), and 1-min extension (72 °C). The cycling was terminated with a final extension at 72 °C for 10 min. PCR products were fractionated by electrophoresis in 1 % (w/v) agarose gels stained with ethidium bromide.
Doramectin fermentation in a 50-l fermentor
Five milliliters of spore suspension of the shuffled strain F4-137 or the initial strain S. avermitilis NEAU1069 was inoculated into a 1-l Erlenmeyer flask containing 200 ml of seed medium. After incubation for 24 h, 1 l of seed broth was inoculated into a 50-l stirred-tank fermentor (FUS-50L (A), Shanghai Guoqiang Bioengineering Equipment Co. Ltd., Shanghai, China) containing 30 l of producing medium and incubated at 28 °C with an agitation speed of 250 rpm and an aeration rate of 0.9 vvm. The CHC was added to the fermentation broth at 24 and 192 h with a final concentration of 100 and 60 mg/l, respectively. During the fermentation, 1 ml of the fermentation broth was sampled every day and doramectin yield was determined by HPLC.
Analytical method
Glucose and amino nitrogen were analyzed by Fehling’s reagent method and formaldehyde titration method, respectively [35]. Mycelial growth was estimated by measuring the packed mycelium volume (PMV). Ten milliliters of fermentation broth was centrifuged at 3,000×g for 15 min, and then the PMV was measured. To analyze the yield of doramectin and oligomycin, 1 ml of the whole fermentation broth was sampled and directly mixed with 2 ml of ethanol and then sonicated at room temperature for 30 min to extract the product. Subsequently, the mixture was centrifuged at 4,000×g for 15 min and the supernatant was analyzed by reversed-phase HPLC (Waters ACQUITY, Waters Ltd, Milford, MA, USA) with a Novapak C18 column (Waters Ltd, Milford, MA, USA, 3.9 × 150 mm, 5 μm). The mobile phase was developed with methanol–water (85:15 v/v). The detection wavelength was 242 nm. The elution was performed at a flow rate of 1.0 ml/min in ambient conditions at 25 °C. Doramectin yield was determined from a standard curve.
Statistical analysis
All of the experiments were conducted in triplicate. Data were expressed as the mean ± standard deviations (SDs). Data were analyzed using one-way analysis of variance (ANOVA) followed by each pair of Student’s t tests for multiple comparison. Differences were considered significant if p < 0.05. All analyses were performed using SPSS for Windows, version 11.5 (SPSS, Chicago, IL, USA).
Results
Selection of starting strains for genome shuffling
Compared to other doramectin-producing strains [25, 26, 31], S. avermitilis NEAU1069 is more apt to biosynthesize doramectin in the presence of CHC. However, the doramectin yield was only 88.7 ± 4.9 mg/l in a shake flask. Generally, it is essential for genome shuffling that the parent strain must exhibit some improvement in the trait of interest [1, 17]. Therefore, UV irradiation and MNNG were used as the mutagenizing agents to generate a parental library of starting strains with high doramectin yield. As shown in Fig. 2, the initial strain NEAU1069 was sensitive to MNNG and UV irradiation. Survival rate of the cells was reduced by increasing exposure to the mutagenizing agents. After the treatment with mutagenizing agents, five mutants designated as UV-45, UV-100, UV-120, MNNG-113, and MNNG-152 with the highest doramectin yield were selected and their doramectin yields were 135.3 ± 5.8, 134 ± 2.6, 126.7 ± 5.9, 113.2 ± 6.1, 116.6 ± 5.2 mg/l, respectively (Fig. 3). Further treatment with mutagenizing agents had no influence on the doramectin yield and streptomycin resistance of NEAU1069 (Table 1).
Generation of high yield doramectin-producing strains by GS-SR
In order to obtain recombinants with improved doramectin yield, four successive rounds of GS-SR were carried out with the five mutants including UV-45, UV-100, UV-120, MNNG-113, and MNNG-152 as the starting strains. After the first round of GS-SR, 150 colonies exhibiting 5 μg/ml streptomycin resistance were screened for doramectin productivity, and five strains with the highest doramectin yield ranging from 349 to 370 mg/l in shake flasks were used as starting strains for subsequent rounds of GS-SR (Fig. 3). Among the five starting strains, F1-38 produced the highest doramectin yield with a value of 362.5 ± 5.1 mg/l, which was 4.2-fold higher than that of NEAU1069. It is interesting that F1-38 and F1-140 were derived from the fusion of UV-45 with MNNG-113 and MNNG-152, respectively, and F1-11, F1-104, and F1-112 were derived from the fusion of MNNG-152 with MNNG-113, UV-100, and UV-120, respectively. Therefore, UV-45 and MNNG-152 were used as controls in other experiments. As the concentration of streptomycin increased from 10 to 15 μg/ml, the doramectin production of the top five recombinants reached 617–651 and 813–846 mg/l, respectively. After the fourth round of GS-SR, doramectin yield of the top five recombinants reached 943–992 mg/l (Fig. 3). Among them, strain F4-137 was found to more potential in producing doramectin with a yield of 992 ± 4.4 mg/l, which was approximately 7.3-fold and 11.2-fold higher than that of UV-45 and NEAU1069, respectively. However, the CHC tolerance level of F4-137 (300 mg/l) was only threefold higher than that of NEAU1069 (100 mg/l). As the concentrations of CHC in fermentation medium increased to 150, 200, and 300 mg/l, the doramectin yields by F4-137 were 987 ± 2.1, 993.2 ± 5.7, and 997.5 ± 4.4 mg/l, respectively.
In order to validate the efficiency of the GS-SR, three control experiments were conducted. The first control experiment showed that the doramectin yields of UV-45 and MNNG-152 after repeated protoplast preparation and regeneration procedures were 133.5 ± 1.2 and 107 ± 3.3 mg/l, respectively, suggesting that protoplast preparation and regeneration has minimal effects of on the doramectin yield. The second control experiment, in which the protoplasts of UV-45 or MNNG-152 were regenerated by plating on streptomycin-containing medium, demonstrated that the doramectin yields (356.7 ± 2.2 and 379.1 ± 4.2 mg/l, respectively) of recombinant strains were slightly improved after four rounds of streptomycin resistance screening. Therefore, a certain relationship may exist between streptomycin resistance and doramectin yield. The third control experiment using UV-45 and MNNG-152 as starting strains demonstrated that the highest doramectin yield was 598.7 ± 2.7 mg/l after four rounds of GS-WSR, which was lower than that of GS-SR. The reason may be that more protoplasts including the recombinant and non-recombinant protoplasts could be regenerated on the streptomycin-free medium, leading a lower frequency to obtain the recombinant strains with high doramectin yield. Therefore, the distribution of doramectin yield in 150 randomly selected isolates from the first round of GS-SR or GS-WSR was evaluated, demonstrating that the average doramectin yield of strains obtained from GS-SR was much higher than that from GS-WSR (Fig. 4). However, when 500 strains randomly isolated from the first round of GS-WSR were tested, a strain with streptomycin resistance (5 μg/ml) and a K88R mutation in rpsL was obtained to produce doramectin with a yield of 347.6 ± 3.7 mg/l, which was close to that of high doramectin-producing strains obtained from GS-SR. These results indicated that the selective regeneration of fused protoplasts with streptomycin-resistance contributed to improve the screening efficiency of mutants with improved productivity of doramectin in the genome shuffling process. On the other hand, after the first and second round of doramectin resistance-aided genome shuffling the yields of doramectin produced by the top five strains reached 215–239 and 297–321 mg/l, respectively. However, further treatment had no effect on improving the yield of doramectin.
The histogram representing the classes of doramectin production and the number of producers after the first round of genome shuffling. All the data were obtained by shake-flask fermentation. Classes A–J represent the doramectin yields at the range of <280, 281–290, 291–300, 301–310, 311–320, 321–330, 331–340, 341–350, 351–360 mg/l, and >360 mg/l, respectively. Str− and Str+ represent the strains isolated by GS-WSR and GS-SR, respectively
Genetic instability is a very important issue for high-producing strains originating from various treatments of mutation or recombination [17]. Thus, the genetic stability of F4-137 was evaluated by five successive sub-cultivation tests. The range of doramectin yield among five generations was from 942.3 ± 2.7 to 983.1 ± 4.2 mg/l, indicating that the hereditary characteristic of F4-137 was stable.
RAPD analysis and mutation in rpsL
RAPD analysis [37] was used to distinguish the genomic DNA variation between the shuffled strains and the initial strain NEAU1069. As a result, most of the primers clearly reproduced different RAPD patterns among these strains (data not shown). Especially, primer S113 (5′-GACGCCACAC-3′) was found to produce more polymorphic bands than other primers (Fig. 5). More importantly, 18 bands were obtained from F4-137 using S113 as primer, however, only five bands were obtained from NEAU1069 (Fig. 5). Of these 18 bands, 14 were found to be polymorphic, indicating that genetic variation has occurred in the shuffled strain F4-137.
DNA banding patterns produced by RAPD analysis of the initial strain NEAU1069 and shuffled strains using primer S113. M DNA molecular size marker, lane 1 the shuffled strain F1-38, lane 2 the shuffled strain F2-75, lane 3 the original strain NEAU1069, lane 4 the shuffled strain F3-26, lane 5 the shuffled strain F4-137
The increase of streptomycin resistance is usually believed to be associated with the mutation in rpsL gene encoding the ribosomal protein S12 [2, 3, 19, 27]. Therefore, the rpsL gene in the shuffled strains, the starting strains, and the initial strain was sequenced and analyzed. The results showed that rpsL sequence (NCBI accession no. KC160530) in the initial strain NEAU1069 was identical to that in other doramectin-producing S. avermitilis, however, there are some site-specific substitutions such as S27R, F38I, K43N, N46K, K88E, K88R, Y95H, and A119P in the shuffled strains (Table 1). Of these sites, the mutations in K88 and K43 were previously believed to be associated with high streptomycin resistance [27]. Additionally, other un-described mutations such as S27R, F38I, N46K, Y95H, and A119P were also observed after four rounds of GS-SR.
Semiquantitative RT-PCR analysis of PKS transcription
To test whether enhanced yields of doramectin/avermectins and oligomycin are associated with the overexpression of PKS, the transcriptions of aveA2 and olm1 involved in the biosynthesis of these secondary metabolites were analyzed by semiquantitative RT-PCR. As shown in Fig. 6, no obvious difference was observed in the transcription of aveA2 and olm1 between the initial strain NEAU1069 and shuffled strain F4-137.
Doramectin fermentation by F4-137 in a 50-l fermentor
Doramectin fermentation experiments were carried out by F4-137 and NEAU1069 in a 50-l fermentor, and the time courses of fermentation are shown in Fig. 7. The doramectin was detected only at the later stages of the exponential growth phase and stationary phase. In the fermentation process, the PMV of F4-137 was higher than that of NEAU1069 (Fig. 7a). However, the doramectin yield of F4-137 could reach 930.3 ± 3.8 mg/l after 14-day fed-batch fermentation (Fig. 7b), which was a 7.5-fold increase compared to NEAU1069 (124.3 ± 12.9 mg/l). The difference between F4-137 and NEAU1069 with regard to consumption of glucose in the first 24 h of fermentation was not obvious, afterwards the consumption rate of F4-137 was accelerated and evidently exceeded that of NEAU1069 (Fig. 7c), while the amino nitrogen consumption of F4-137 was faster than that of NEAU1069 at the beginning of the fermentation (Fig. 7d). Glucose was still consumed rapidly in the stationary phase although the mycelial growth almost stopped in this phase, which indicated that the consumed glucose was primarily used for doramectin biosynthesis and mycelium maintenance. Similarly, the amino nitrogen was consumed vigorously in the first 5 days.
Discussion
Microbial strain improvement for the overproduction of valuable industrial products has been the hallmark of all industrial commercial fermentation processes [17, 41]. However, traditional breeding such as the classic mutation and random selection are very time- and labor-intensive. With the broad application of recombinant DNA technology and X-omics [21], new methods including rational metabolic engineering, cell engineering, and genome shuffling have been employed for the rapid improvement of industrially important microbial phenotypes [6, 7, 11, 12, 15–17, 23, 28, 30, 33, 38–40, 42, 43]. In the present study, genome shuffling was successfully used to achieve apparently improved doramectin yield in S. avermitilis NEAU1069. To minimize the number of screens required for selecting improved strains during the process of genome shuffling, the streptomycin-resistant screening strategy was incorporated.
Product inhibition is a conventional trait for secondary metabolism, and it can lead to the decrease in cell growth as well as the production of antibiotics. Therefore, the tolerance to products has been commonly used as a selective marker to obtain improved strains with desired phenotypes during the process of genome shuffling [4, 9, 36, 38, 44]. Unfortunately, the extensive improvement in doramectin production stopped after the second round of doramectin resistance-aided genome shuffling. Similar to other known doramectin producers [25, 26, 31], doramectin (CHC-B1) and several undesired doramectin analogs (i.e., CHC-A1, CHC-B2) together with avermectins can be produced as co-metabolites by S. avermitilis NEAU1069 in the presence of CHC [32, 34]. These co-metabolites are biosynthesized by the same PKS and the differences in their chemical structures are caused by different start units (Fig. 1) [25, 26, 31]. Therefore, we hypothesized that the method using doramectin resistance as a selective marker could not only obtain the strains with high productivity of doramectin, but also some strains with high productivity of its analogs. This assumption was supported by comparing the HPLC spectra of the initial strain NEAU1069 with those of doramectin-resistant strains Dora-38 and Dora-106 (Fig. 8). Although these two strains exhibiting doramectin resistance of 200 mg/l and the other top five strains with doramectin yields of 215–239 mg/l were all obtained from the first round of doramectin resistance-aided genome shuffling, lower doramectin yields were observed in Dora-38 and Dora-106 (Table 1). Especially, the doramectin yield of Dora-106 was even lower than that of NEAU1069. However, Dora-38 and Dora-106 exhibited increased yields of doramectin analogs (Fig. 8). Therefore, other selective markers should be employed in the genome shuffling.
Recently, a practical method for increasing antibiotic production in bacteria by modulating ribosomal components (ribosomal proteins or rRNA), specifically by generating mutations conferring drug resistance such as streptomycin resistance, has been described [2, 3, 19, 20, 27]. This approach, known as ribosome engineering, has several advantages, including the ability to screen drug-resistant mutations by simple selection on drug-containing plates. Even if the mutation frequency is extremely low, ribosome engineering has proved to be effective for improving the yields of antibiotics in industrial strains [2, 3, 19, 20, 27]. We have previously described the successful application of streptomycin-resistant selection to obtain a mutant of S. bingchenggensis with high yield of milbemycin, which is a macrocyclic lactone with similar structure to avermectin and doramectin [35]. Furthermore, the introduction of rpsL mutations led to high streptomycin resistance and significantly increased the production of oligomycin (~20- to 40-fold) in S. avermitilis [27]. Therefore, a method referred to as GS-SR using streptomycin-resistance as a selective marker was attempted to improve doramectin yield in S. avermitilis NEAU1069. Recently, the same method has successfully been applied to the improvement of avilamycin production in S. viridochromogenes AS 4.126 [18]. As shown in Fig. 4, a higher frequency to obtain strains with enhanced doramectin yield was observed when streptomycin resistance was used as a selective marker. Meanwhile, the relationship between the doramectin yield and the streptomycin-resistant levels in the shuffled strains was evaluated. The results demonstrated that in most cases the doramectin yields were positively correlated with the streptomycin-resistant levels in strains obtained from resistance screening, however, the same relationship was not observed in the strains obtained from resistance-free screening (data not shown). Therefore, the positive correlations between the doramectin yield and streptomycin-resistant levels further confirmed the efficiency of GS-SR to obtain strains with improved doramectin productivity. Furthermore, the control experiments demonstrated that the doramectin yield increased ~2.7- to 3.5-fold and 4.4- to 5.3-fold, respectively, when individual streptomycin resistance screening and genome shuffling was used to treat strains UV-45 and MNNG-152. However, the doramectin yield in the strains obtained from GS-SR increased ~7.3- to 8.9-fold more than that of UV-45 and MNNG-152, suggesting a synergistic effect of streptomycin resistance screening and genome shuffling.
In strains of pentachlorophenol (PCP)-degrading Sphingobium chlorophenolium ATCC 39723 improved by genome shuffling, phenotypes were altered to possess an enhanced growth rate, constitutive expression of the PCP-degradation genes, and enhanced resistance to the toxicity of PCP and its metabolites [5]. In the case of hydroxycitric acid (HCA) producer Streptomyces sp. U121 improved by genome shuffling, the enhanced production of HCA was associated with an increased cell density and enhanced growth rate [9]. In order to further explore the factors influenced on the productivity of doramectin in the shuffled strains, the fermentation properties of F4-137 and NEAU1069 in a 50-l fermentor were investigated. As shown in Fig. 7a and b, the increase in the doramectin yield (~7.5-fold) was much higher than that in the cell density (~1.3-fold), suggesting that the enhanced growth rate only played limited roles in the improved yield of doramectin. Additionally, no significant difference in transcriptions of aveA2 and olm1 was observed between F4-137 and NEAU1069 (Fig. 6), therefore, we speculated that the rpsL mutation may be attributed to the improved yields of oligomycin and avermectin. Substitutions K88E and K88R in the rpsL usually resulted in dramatic improvement of antibiotic productivity [2, 3, 19, 20, 27]. The same substitutions at these two sites have also occurred in shuffled strains (Table 1), suggesting that the substitutions K88E and K88R in the rpsL may play a vital role in the improvement of doramectin productivity. Although F4-28 and F4-137 exhibited similar phenotypes including doramectin yield and streptomycin resistance, an additional mutation A119E was observed in rpsL from F4-137 (Table 1). Thus, the mutation A119E seems to have no effect on doramectin yield. Comparison of doramectin yield and rpsL mutation in F4-57 with those in F4-120 suggested that the mutation S27R might have a negative effect on doramectin yield (Table 1). The yield ratios of avermectins to oligomycin produced by NEAU1069 in the absence and presence of CHC were 1:0.06 and 1:0.09, respectively, implying no obvious effect of CHC on oligomycin production. However, the addition of CHC decreased the yield of avermectins, which confirmed the competition of precursors between the biosynthesis of avermectin and doramectin [25, 26]. Compared to strain NEAU1069, the yields of avermectins and oligomycin produced by strain F4-137 in the presence of CHC increased approximately 1.2 and 11.5-fold, respectively (Table 2). This phenomenon is distinct from the description that there is a balance of avermectin and oligomycin production in S. avermitilis due to the competition of common precursors (malonyl-CoA and methylmalonyl-CoA) between the biosynthesis of avermectin and oligomycin [27]. Because the enhanced transcription of aveA2 and olm1 was not observed in F4-137 (Fig. 6), the increase in the yields of avermectins and oligomycin in F4-137 was speculated to be caused by enhanced production of malonyl-CoA and methylmalonyl-CoA. However, the increase in the yield of avermectins was not significant as that of oligomycin, and this phenomenon may be derived from the competition between the biosynthesis of avermectins and doramectin [25, 26]. Meanwhile, the yield ratios of avermectins to oligomycin produced by F4-137 in the absence and presence of CHC were 1:0.5 and 1:0.96, respectively, implying a small effect of CHC on oligomycin production. Noticeably, besides the enhanced supplement of malonyl-CoA and methylmalonyl-CoA, the production of the start unit CHC-CoA may be another key factor affecting the doramectin yield. It has been reported that the addition of exogenous CHC did not increase the production of doramectin, but the enhanced supplementation of CHC-CoA in vivo by introducing a CHC-CoA biosynthetic gene cassette could lead to a twofold increase in doramectin yield [31]. Therefore, enhanced performance in F4-137 on transforming exogenous CHC into CHC-CoA may contribute to the improved doramectin yield. When CHC was added into the fermentation medium, large amounts of CHC-CoA were recognized by initial acyltransferase domain of PKS responsible for the biosynthesis of avermectin and doramectin, which is the possible reason why the yield of doramectin increased but the yield of avermectins reduced (Table 2). It is known that the addition of CHC had a negative effect on the production of doramectin by inhibiting the cell growth [31]. Thus, the toxic tolerance of the strains to CHC was evaluated, and the results demonstrated that CHC tolerance of the shuffled strain improved from 100 to 300 mg/l. Although high CHC tolerance would be helpful to relieve the strain growth inhibition and improve the yield of doramectin, the effect may be limited because the cell density of F4-137 did not exhibit drastic change with the increasing concentration of CHC during the shake-flask fermentation. Additionally, enhanced doramectin yield was not observed when higher concentrations of CHC were added into the producing medium, which also suggested that the ability of transforming exogenous CHC into intracellular CHC-CoA is an important factor involving the enhanced yield of doramectin. The HPLC spectra of the highest doramectin-producing strains showed that all the yield of avermectins including B2a, A2a, B1a, and A1a increased in F1-38 and F2-75 (Fig. 8d, e), which may be associated with rpsL mutation. However, the yields of these four components, especially the “A” components, decreased in F3-3 and F4-137 (Fig. 8f, g). The reason may be that more precursors (malonyl-CoA and methylmalonyl-CoA) were used for the biosynthesis of doramectin and its analogs due to the competition between the biosynthesis of avermectins and doramectin analogs. This assumption was further confirmed by the fact that a significant increase in doramectin, CHC-B2, and CHC-A1 was observed in F3-3 and F4-137 (Fig. 8f, g). The yield of avermectins in F4-137 was lower than that in F1-38, however, it was still higher than that in NEAU1069, suggesting that rpsL mutation may be the major factor to increase the yield of avermectins. On the other hand, the rpsL mutation associated with enhanced supply of malonyl-CoA and methylmalonyl-CoA may also play important roles in improving the doramectin yield. However, the doramectin yield could not significantly increase in the case of lacking enough amounts of start unit CHC-CoA even though large amounts of extent unit (malonyl-CoA and methylmalonyl-CoA) were produced. Therefore, the enhanced doramectin yield in shuffled strains should be caused by a synergistic effect of rpsL mutation and enhanced production of CHC-CoA, which is consistent with the results obtained from the control experiments mentioned as above. In the RAPD profiles (Fig. 5), the genetic diversity was observed between the initial and shuffled strains, implying that genome shuffling indeed took place in shuffled strains. Because genome shuffling is a whole-genome engineering approach for the rapid improvement of complex cellular phenotypes, the polymorphic bands might be related to the improved productivity of doramectin and the differences in RAPD profiles may be useful to elucidate the molecular mechanisms of doramectin overproduction in S. avermitilis NEAU1069.
In conclusion, GS-SR was successfully used to improve the productivity of doramectin in S. avermitilis NEAU1069. Two possible reasons are related to the improved doramectin yield. First, the streptomycin-resistant strains possess enhanced protein biosynthesis, which would improve the utilizing efficiency of doramectin precursor. Secondly, some ambiguous strain performance associated with the overproduction of doramectin has occurred after successive genome shuffling, for example, the improved production of CHC-CoA, malonyl-CoA, and methylmalonyl-CoA could supply more amounts of precursor used for the biosynthesis of doramectin. Although it is still unclear whether other positive factors are responsible for the improved doramectin yield, GS-SR should be an efficient method to enhance the production of secondary metabolites in Streptomyces.
References
Bajwa PK, Pinel D, Martin VJJ, Trevors JT, Lee H (2010) Strain improvement of the pentose-fermenting yeast Pichia stipitis by genome shuffling. J Microbiol Methods 81:179–186
Baltz RH (2011) Strain improvement in actinomycetes in the postgenomic era. J Ind Microbiol Biotechnol 38:657–666
Beltrametti F, Rossi R, Selva E, Marinelli F (2006) Antibiotic production improvement in the rare actinomycete Planobispora rosea by selection of mutants resistant to the aminoglycosides Streptomycin and Gentamycin and to Rifamycin. J Ind Microbiol Biotechnol 33:283–288
Cao X, Song Q, Wang C, Hou L (2012) Genome shuffling of Hansenula anomala to improve flavour formation of soy sauce. World J Microbiol Biotechnol 28:1857–1862
Dai MH, Copley SD (2004) Genome shuffling improves degradation of the anthropogenic pesticide pentachlorophenol by Sphingobium chlorophenolicum ATCC 39723. Appl Environ Microbiol 70:2391–2397
Gendy MMA, EL-Bondkly AMA (2011) Genome shuffling of marine derived bacterium Nocardia sp. ALAA 2000 for improved ayamycin production. Antonie van Leeuwenhoek 99:773–780
Gong J, Zheng H, Wu Z, Chen T, Zhao X (2009) Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv 27:996–1005
Hafner EW, Holley BW, Holdom KS (1991) Branched-chain fatty acid requirement for avermectin production by a mutant of Streptomyces avermitilis lacking branched-chain 2-oxo acid dehydrogenase activity. J Antibiot 44:349–356
Hida H, Yamada T, Yamada Y (2007) Genome shuffling of Streptomyces sp. U121 for improved production of hydroxycitric acid. Appl Microbiol Biotechnol 73:1387–1393
Hopwood DA, Bibb MJ, Chater KF, Kieser T, Bruton CJ, Kieser HM, Lydiate DJ, Smith CP, Ward JM (1985) Genetic manipulation of Streptomyces, a laboratory manual. John Innes Foundation, Norwich, p 356
Jin Q, Jin Z, Zhang L, Yao S, Li F (2012) Probing the molecular mechanisms for pristinamycin yield enhancement in Streptomyces pristinaespiralis. Curr Microbiol 65:792–798
Jingping G, Hongbing S, Gang S, Hongzhi L, Wenxiang P (2012) A genome shuffling-generated Saccharomyces cerevisiae isolate that ferments xylose and glucose to produce high levels of ethanol. J Ind Microbiol Biotechnol 39:777–787
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Preparation and analysis of genomic and plasmid DNA. In: Practical Streptomyces genetics. The John Innes Foundation, Norwich, pp 161–171
Lanusse C, Lifschitz A, Virkel G, Alvarez L, Sánchez S, Sutra JF, Galtier P, Alvinerie M (1997) Comparative plasma disposition kinetic of ivermectin, moxidectin and doramectin in cattle. J Vet Pharmacol Ther 20:91–99
Liu JJ, Ding WT, Zhang GC, Wang JY (2011) Improving ethanol fermentation performance of Saccharomyces cerevisiae in very high-gravity fermentation through chemical mutagensis. Appl Microbiol Biotechnol 91:1239–1246
Lu Y, Cheng YF, He XP, Guo XN, Zhang BR (2012) Improvement of robustness and ethanol production of ethanologenic Saccharomyces cerevisiae under co-stress of heat and inhibitors. J Ind Microbiol Biotechnol 39:73–80
Luo JM, Li JS, Liu D, Liu F, Wang YT, Song XR, Wang M (2012) Genome shuffling of Streptomyces gilvosporeus for improving natamycin production. J Agric Food Chem 60:6026–6036
Lv X-A, Jin Y-Y, Li Y-D, Zhang H, Liang X-L (2013) Genome shuffling of Streptomyces viridochromogenes for improved production of avilamycin. Appl Microbiol Biotechnol 97(2):641–648
Ochi K, Hosaka T (2012) New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol. doi:10.1007/s00253-012-4551-9
Olano C, Lombó F, Méndez C, Salas JA (2008) Improving production of bioactive secondary metabolites in actinomycetes by metabolic engineering. Metab Eng 10:281–292
Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WP, Ryan CM, del Cardayré S (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712
Petri R, Schmidt-Dannert C (2004) Dealing with complexity: evolutionary engineering and genome shuffling. Curr Opin Biotechnol 15:298–304
Pinel D, D’Aoust F, del Cardayre SB, Bajwa PK, Lee H, Martin VJ (2011) Saccharomyces cerevisiae genome shuffling through recursive population mating leads to improved tolerance to spent sulfite liquor. Appl Environ Microbiol 77:4736–4743
Stephanopoulos G (2002) Metabolic engineering by genome shuffling-two reports on whole-genome shuffling demonstrate the application of combinatorial methods for phenotypic improvement in bacteria. Nat Biotechnol 20:666–668
Stutzman-Engwall K, Conlon S, Fedechko R, Kaczmarek F, McArthur H, Krebber A, Chen Y, Minshull J, Raillard SA, Gustafsson C (2003) Engineering the aveC gene to enhance the ratio of doramectin to its CHC-B2 analogue produced in Streptomyces avermitilis. Biotechnol Bioeng 82:359–369
Stutzman-Engwall K, Conlon S, Fedechko R, McArthur H, Pekrun K, Chen Y, Jenne S, La C, Trinh N, Kim S, Zhang YX, Fox R, Gustafsson C, Krebber A (2005) Semi-synthetic DNA shuffling of aveC leads to improved industrial scale production of doramectin by Streptomyces avermitilis. Metab Eng 7:27–37
Tanaka Y, Komoru M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K (2009) Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol 75:4919–4922
Tao X, Zheng D, Liu T, Wang P, Zhao W, Zhu M, Jiang X, Zhao Y, Wu X (2012) A novel strategy to construct yeast Saccharomyces cerevisiae strains for very high gravity fermentation. PLoS ONE 7(2):e31235
Wang G, Inaoka T, Okamoto S, Ochi K (2009) A novel insertion mutation in Streptomyces coelicolor ribosomal S12 protein results in paromomycin resistance and antibiotic overproduction. Antimicrob Agents Chemother 53(3):1019–1026
Wang H, Zhang J, Wang XJ, Qi W, Dai YJ (2012) Genome shuffling improves production of the low-temperature alkalophilic lipase by Acinetobacter johnsonii. Biotechnol Lett 34:145–151
Wang J-B, Pan H-X, Tang G-L (2011) Production of doramectin by rational engineering of the avermectin biosynthetic pathway. Bioorg Med Chem Lett 21:3320–3323
Wang M, Yang X-H, Wang J-D, Wang X-J, Chen Z-J, Xiang W-S (2009) New β-class milbemycin compound from Streptomyces avermitilis NEAU1069: fermentation, isolation and structure elucidation. J Antibiot 62:587–591
Wang PM, Zheng DQ, Liu TZ, Tao XL, Feng MG, Min H, Jiang XH, Wu XC (2012) The combination of glycerol metabolic engineering and drug resistance marker-aided genome shuffling to improve very-high-gravity fermentation performances of industrial Saccharomyces cerevisiae. Bioresource Technol 108:203–210
Wang X-J, Wang M, Wang J-D, Jiang L, Wang J-J, Xiang W-S (2010) Isolation and identification of novel macrocyclic lactones from Streptomyces avermitilis NEAU1069 with acaricidal and nematocidal activity. J Agric Food Chem 58:2710–2714
Wang XJ, Wang XC, Xiang WS (2009) Improvement of milbemycin-producing Streptomyces bingchenggensis by rational screening of ultraviolet- and chemically induced mutants. World J Microbiol Biotechnol 25:1051–1056
Wang Y, Li Y, Pei X, Yu L, Feng Y (2007) Genome-shuffling improved acid tolerance and L-lactic acid volumetric productivity in Lactobacillus rhamnosus. J Biotechnol 129:510–515
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Xu B, Jin Z, Wang H, Jin Q, Jin X, Cen P (2008) Evolution of Streptomyces pristinaespiralis for resistance and production of pristinamycin by genome shuffling. Appl Microbiol Biotechnol 80:261–267
Xu D, Pan L, Zhao H, Zhao M, Sun J, Liu D (2011) Breeding and identification of novel koji molds with high activity of acid protease by genome recombination between Aspergillus oryzae and Aspergillus niger. J Ind Microbiol Biotechnol 38:1255–1265
Xu F, Jin H, Li H, Tao L, Wang J, Lv J, Chen S (2012) Genome shuffling of Trichoderma viride for enhanced cellulase production. Ann Microbiol 62:509–515
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayré SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Zhao J, Li Y, Zhang C, Yao Z, Zhang L, Bie X, Lu F, Lu Z (2012) Genome shuffling of Bacillus amyloliquefaciens for improving antimicrobial lipopeptide production and an analysis of relative gene expression using FQ RT-PCR. J Ind Microbiol Biotechnol 39:889–896
Zheng D-Q, Wu X-C, Wang P-M, Chi X-Q, Tao X-L, Li P, Jiang X-H, Zhao Y-H (2011) Drug resistance marker-aided genome shuffling to improve acetic acid tolerance in Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 38:415–422
Zheng H, Gong J, Chen T, Chen X, Zhao X (2010) Strain improvement of Sporolactobacillus inulinus ATCC 15538 for acid tolerance and production of d-lactic acid by genome shuffling. Appl Microbiol Biotechnol 85:1541–1549
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (31071750), the National Key Project for Basic Research (2010CB126102), the China Postdoctoral Science Foundation (20110491023), the Heilongjiang Postdoctoral Fund (LBH-Z11234), and Doctor Start-up Fund of Northeast Agricultural University (2012RCB05), and the Innovative Program for Postgraduates in Heilongjiang Province (YJSCX2011-087HLJ).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, J., Wang, X., Diao, J. et al. Streptomycin resistance-aided genome shuffling to improve doramectin productivity of Streptomyces avermitilis NEAU1069. J Ind Microbiol Biotechnol 40, 877–889 (2013). https://doi.org/10.1007/s10295-013-1280-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1280-8