Abstract
Streptomyces roseochromogenes NRRL 3504 is best known as a producer of clorobiocin, a DNA replication inhibitor from the aminocoumarin family of antibiotics. This natural product currently draws attention as a promising adjuvant for co-application with other antibiotics against Gram-negative multidrug-resistant pathogens. Herein, we expand the genetic toolkit for NRRL 3504 by showing that a set of integrative and replicative vectors, not tested previously for this strain, could be conjugally transferred at high frequency from Escherichia coli to NRRL 3504. Using this approach, we leverage a cumate-inducible expression of cluster-situated regulatory gene novG to increase clorobiocin titers by 30-fold (up to approximately 200 mg/L). To our best knowledge, this is the highest level of clorobiocin production reported so far. Our findings set a working ground for further improvement of clorobiocin production as well as for the application of genetic methods to illuminate the cryptic secondary metabolome of NRRL 3504.
Key Points • Efficient system for conjugative transfer of plasmids into NRRL 3504 was developed. • Expression of regulatory genes in NRRL 3504 led to increase in clorobiocin titer. • Secondary metabolome of NRRL 3504 becomes an accessible target for genetic manipulations using the expanded vector set and improved intergeneric conjugation protocol. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
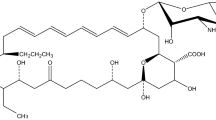
Streptomyces roseochromogenes was first described in the 1970s as a producer of a unique natural product clorobiocin (Clo) (Mancy et al. 1974). Clo belongs to the aminocoumarin family of antibiotics (Fig. 1a) which, via inhibition of DNA gyrases (Flatman et al. 2006), interfere with DNA replication in many Gram-positive bacteria, including multidrug-resistant cocci (Galm et al. 2004). However, clorobiocin has not reached clinical application, unlike its structurally related counterpart novobiocin (Nov). The latter was an FDA-approved drug to treat infections caused by Staphylococus aureus (Walsh et al. 1993; Raad et al. 1995), until the withdrawal in 2011. Clo, however, is a more potent in vitro inhibitor of DNA gyrase than Nov (IC50 against Escherichia coli GyrB are 0.08 and 0.9 µM for Clo and Nov, respectively). Moreover, Clo and Nov (as well as their derivatives) also inhibit topoisomerase IV (Flatman et al. 2006). The clinical relevance of Nov fueled much interest in the family of aminocoumarin antibiotics. Biosynthetic gene clusters (BGCs) for Clo (clo) and Nov (nov) biosynthesis were sequenced and annotated (Fig. 1b) (Steffensky et al. 2000; Pojer et al. 2002; Li and Heide 2006). Later on, the whole genome sequences were reported for NRRL 3504 and novobiocin producer Streptomyces niveus NCIMB 11,891 (Flinspach et al. 2014; Rückert et al. 2014).
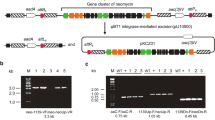
Biosynthetic pathways of clorobiocin (Clo) and novobiocin (Nov). a) Biosynthetic pathways for both Nov and Clo differ by certain tailoring enzymes, encoded within nov and clo BGCs (b). Different portions of clorobiocin and novobiocin and the enzymes responsible for their assembly are color-coded appropriately. Putative regulatory schemes for nov and clo gene expression are shown; green arrows point to the promoters that are/expected to be targeted by the cluster-encoded transcriptional activators NovG and CloG
Early identification of clo and nov BGCs allowed investigating the precise functions of almost all structural genes in a series of genetic and biochemical experiments (see extensive review in Heide 2009 and Pojer et al. 2003a, b). In fact, biosynthetic routes leading to Clo and Nov remain among the most studied secondary metabolic pathways (Fig. 1a). Some investigations were performed in the native producer (Eustáquio et al. 2003; Westrich et al. 2003; Xu et al. 2003), yet the most of our current understanding of aminocoumarin assembly comes from the studies of the heterologous hosts—Streptomyces coelicolor derivatives carrying clo BGC on a cosmid (Freitag et al. 2005a, b, 2006; Wolpert et al. 2007). The native host was manipulated via either protoplast transformation or by E. coli—Streptomyces conjugation to achieve both gene knockouts (Pojer et al. 2002) and knockout complementations (Eustaquio et al. 2003). Plasmids based on φC31 phage integrase or pIJ101 replicon were used for these experiments. Nevertheless, genetic manipulations of NRRL 3504 appeared to be a challenge grave enough to shift most of the work to heterologous hosts (Eustaquio et al. 2005).
More recently, both Nov and Clo were shown to enhance lipopolysaccharide transport in E. coli and now might be considered promising adjuvants for polymyxins and colistins to overcome multiple antibiotic resistance in Gram-negative pathogens (May et al. 2017; Mandler et al. 2018; Mattingly et al. 2020). This makes Clo an interesting target for further investigations, demanding significant amounts of the compound. Purification of Clo from fermentation broths of streptomycetes is currently the only feasible and scalable way to produce this compound. Several strains of S. roseochromogenes have been reported in the literature, some of which resulted from conventional selection for increased Clo accumulation, such as S. roseochromogenes subsp. oscitans DS 12.976, known also as NRRL 3504 (Mancy 1974). The latter was reported to produce 25 mg/L of Clo in an optimized fermentation medium (Eustaquio et al. 2005). Heterologous expression of clo BGC in the engineered S. coelicolor strains is another viable approach (Flinspach et al. 2010). Here, it was possible to reach up to cumulative 160 mg/L of Clo (16%) and its derivatives (Flinspach et al. 2010). While simplifying genetic studies of Clo biosynthesis, heterologous hosts do not offer ample advantages for Clo production as compared to the native strain; moreover, they may suffer from the instability of integrated cosmids. There are no works aimed at improving the titers of Clo in NRRL 3504, which might be naturally best adapted for Clo overproduction. We set out to explore the use of integrative and replicative plasmids for the expression of different regulatory genes in NRRL 3504, which would increase Clo production rates in the native host. An extended toolkit for the manipulation of NRRL 3504 would also facilitate the investigations of the other secondary metabolic pathways in this strain. Here, we show for the first time that one replicative (pSG5-based) and two integrative (actinophage-based) vectors (in addition to φC31-vectors reported previously) can be conjugally transferred and stably maintained in NRRL 3504. We also show that insertional inactivation of genes in NRRL 3504 is possible using non-replicative plasmid pKC1132. The introduction of different regulatory genes into this strain has been achieved, and some of them, particularly novG and adpA, led to quantitative and qualitative changes in aminocoumarin production. This report sets the working ground for further use of genetic tools to manipulate antibiotic production in NRRL 3504.
Materials and methods
Bacterial strains and growth media
S. roseochromogenes NRRL 3504 was used throughout the study. Streptomyces sphaeroides (= niveus) NRRL 2449 served as a source of genomic DNA to clone gene novG. Micrococcus luteus ATCC 4698 was used as a Clo-sensitive test culture in bioassays. E. coli DH5α was used for routine cloning procedures. E. coli ET12567 (pUZ8002) was used as a donor strain for intergeneric conjugations. E. coli strains were grown at 37 °C in liquid or agar LB medium, supplemented with 100 μg/mL of apramycin sulfate, 50 μg/mL of kanamycin sulfate, and 25 μg/mL of chloramphenicol when appropriate (Sambrook and Russell 2001). Streptomyces strains were routinely maintained on SFM agar (Kieser et al. 2000) at 30 °C, while ISP3 (Koshla et al. 2017) was the medium of choice to obtain sporulating lawns of S. roseochromogenes. To reveal endogenous antibiotic activity, S. roseochromogenes strains were grown in liquid and solid GYM (Koshla et al. 2017) medium for up to 120 h at 30 °C. For genomic DNA isolation, S. roseochromogenes and S. sphaeroides, strains were cultivated in TSB for 48–96 h. Recombinant Streptomyces strains were cultivated in presence of 50 μg/mL of apramycin sulfate, when appropriate; cumate (9 μg/mL) and thiostrepton (5 μg/mL) were used to induce novG/cloG expression from plasmids pGCymRP21 and pIJ6902, respectively. See Electronic Supplementary Materials (ESM) for compositions of all media used in this study.
Generation of recombinant plasmids
All vectors and plasmids, used in this study, are listed in Table 1. Oligonucleotide primers used for the recombinant plasmids generation are given in Table 2. Genomic DNA for cloning purposes was isolated according to salting out procedure no.4 or Kirby procedure as described in (Hopwood et al. 1985; Pospiech and Neumann 1995; Kieser et al. 2000). All recombinant vectors were verified with restriction mapping and sequencing.
Recombinant plasmids for novG/cloG expression were generated according to a single scheme. First, coding sequences (along with the putative RBS) of novG (1,012 bp) and cloG (1,022 bp) were amplified from the chromosomal DNA of S. sphaeroides and S. roseochromogenes using novG_XbaI_up/novG_EcoRI_rev and cloG_XbaI_up/cloG_EcoRI_RP primer pairs (Table 2), respectively. The amplicons were digested with EcoRI and XbaI restriction endonucleases, and cloned into the following integrative expression vectors: pmoeE5script (via EcoRI/SpeI sites), to obtain pNOVG101 and pCLOG101 plasmids (the novG and cloG would be under the control of promoter moeEp); pTES (via EcoRI/XbaI sites), yielding pNOVG102 and pCLOG102 (where novG and cloG would be under the control of ermEp); pGCymRP21 (via EcoRI/SpeI sites), to give pNOVG103 and pCLOG103 (the novG and cloG would be under the control of cumate-inducible P21 promoter); pIJ6902 (via EcoRI/XbaI sites) generating pNOVG104 and pCLOG104 plasmids (the novG and cloG would be under the control of thiostrepton-inducible promoter tipAp). Finally, EcoRI/XbaI-treated novG and cloG amplicons were cloned into EcoRI/XbaI-digested replicative vector pOOB83e, to generate pNOVG105 and pCLOG105, where both genes would be under the control of ermEp.
To generate adpA expression plasmid pMoRT4181, pGM4181 (carrying adpA allele from Streptomyces albidoflavus (= albus) J1074 under the control of moeE5p; see Yushchuk et al. 2018) was digested with BamHI/XhoI restriction endonucleases. 3,555-bp fragment, carrying moeE5p-adpA was then ligated with 3,362 bp fragment of pRT801 (Gregory et al. 2003) vector, treated with the same restriction endonucleases. To generate a replicative vector carrying an additional copy of bldA (leucyl(UUA)-tRNA gene from S. albidoflavus J1074), pTOSbldA (Table 1) was digested with HindIII and XbaI. The 777-bp fragment, carrying bldA and its putative promoter region, was subcloned into pKC1139 recognition sites of the same restriction endonucleases. The resulting plasmid was labeled pKCBA.
The plasmid for one-step insertional inactivation of the cloH-I-J-K operon was generated as follows. 2,300-bp fragment of the cloH-I (covering last 1,550 bp of cloH and first 747 bp of cloI) was amplified using nov-clo-inko_F/R primer pair (Table 2). The amplicon was digested with XbaI/EcoRI restriction endonucleases and cloned into XbaI/EcoRI-digested pKC1132 suicide vector, yielding pKOClo.
Preparation of S. roseochromogenes spore suspensions
To obtain spore suspensions, S. roseochromogenes lawns were grown on ISP3 agar for up to 240 h. Each lawn was flooded with 10 mL of 15% (v/v) glycerol and the spores were scraped off the surface of the lawn with a spatula. Obtained suspensions were vigorously vortexed and filtered through sterile cotton wool to eliminate vegetative mycelia and agar debris. Filtered suspensions were examined microscopically to evaluate the homogeneity and the cotton wool filtration step was repeated in case some mycelial fragments were still present in the suspension. Homogeneous spore suspensions were then concentrated by centrifuging for 15 min at 8,000 rcf. Finally, spore titers were estimated by plating serial tenfold dilutions of the spore suspension on TSA plates, incubating for 96 h at 30 °C, and counting the number of colonies. Spore suspensions with defined spore titers were stored at -80 °C in 15% (v/v) glycerol.
E. coli—S. roseochromogenes conjugation
A standard intergeneric mating protocol (Kieser et al. 2000) was used to transfer vector DNA from E. coli ET12567 (pUZ8002) to germinating spores of S. roseochromogenes. In brief, germination of 108 S. roseochromogenes spores was induced by incubating spores at 50 °C for 10 min. Then, spores were cooled down at room temperature and mixed with 109 cells of an overnight culture of donor E. coli ET12567 (pUB8002), which carried plasmids mentioned in Table 1. Mixed cell suspensions were plated on well-dried SFM or ISP3 plates, optionally supplemented with MgCl2 to 10 mM. After 14–18 h of incubation at 30 °C, plates were overlaid with 1 mL of sterile distilled water containing 1.25 mg of apramycin sulfate and 750 μg of nalidixic acid sodium salt. Emerging transconjugants, selected for apramycin resistance, were analyzed after 120 h. Conjugation efficiency was calculated as a ratio of the total transconjugants number per one plate to the number of spores applied to conjugation. Transconjugants were verified by isolating total DNA with Kirby procedure (Kieser et al. 2000) and amplifying aac(3)IV gene or internal fragment of gusA gene.
Production and extraction of Clo
To initiate the fermentation, 107 spores of S. roseochromogenes strains were inoculated into 50 mL Erlenmeyer flasks with 20 mL of TSB and 6 glass beads (ø5 mm) supplemented with 50 μg/mL apramycin sulfate when appropriate. These pre-cultures were incubated for 48 h at 30 °C on an orbital shaker (220 rpm). Clo-producing cultures were then initiated by inoculating 2.8% (v/v) of pre-culture into a 500 mL Erlenmeyer flask containing 125 mL of GYM medium and 12 glass beads (ø5 mm). Producing cultures were incubated at 30 °C on an orbital shaker (160 rpm) and sampled every 24 h for the downstream determination of biomass accumulation and Clo production. Biomass accumulation was measured as follows. 5 mL of whole cultural broth were collected and the cells were spun down (15 min at 8,000 rcf), the pellet was washed with 15% (v/v) glycerol and left to desiccate for 48 h at 60 °C; finally, the weight of the pellet was determined.
To extract Clo, the pH of the samples, containing 4 mL of whole cultural broth (containing cells and medium, harvested at regular intervals from GYM Clo-producing S. roseochromogenes submerged cultures) was adjusted to 4.0 with concentrated HCl and the samples were vortexed for 30 s. Then, pH was restored to 8.0 by adding NaOH (10 N) and samples were mixed with the equal volume of ethyl acetate. Mixtures were left for 90 min at the orbital shaker (220 rpm) at room temperature, then fractionated by centrifuging 10 min at 8,000 rcf. The organic (upper) phase, containing Clo, was collected. Finally, 2 mL of the organic phase were evaporated in vacuo and the dry pellet was re-suspended in 200 μL of methanol.
Bioassays to determine the antibiotic activity of S. roseochromogenes strains
Bioassays aimed to detect the antimicrobial activity of S. roseochromogenes culture broth samples (well diffusion assay, WDA) or extracts (Kirby–Bauer disk diffusion assay, DDA) were carried out using a similar approach and involved M. luteus ATCC 4698 as a Clo-sensitive test culture. Basic plates for both types of bioassays were made out of two agar layers: the bottom layer was 20 mL of TSA (2% (w/v) agar) and the upper was 10 mL of TSA (0.7% (w/v) agar) where 200 μL of fresh M. luteus cell suspension (OD600 = 0.6) was re-suspended. Then, in the case of DDA, ø5 mm Whatman paper disks soaked with 10 μL of methanol-dissolved ethyl acetate extracts were placed on the surfaces of bioassay plates. In the case of WDA, ø7 mm agar plugs were removed out of the upper agar layer and 10 μL of whole cultural broth were poured into the well. Plates with DDA and WDA were incubated for 24 h at 37 °C, and M. luteus growth inhibition zones were examined and documented.
LC–MS analysis of S. roseochromogenes extracts
Ethyl acetate extracts were obtained as described above, dried in vacuo, and re-dissolved in 200 μL of methanol before analysis. 1 μL of methanol solution was separated on C18 Phenomenex columns (100 × 2.1 mm, 1.7 µm) using Dionex Ultimate 3000 HPLC–DAD system coupled to MaXis Impact HD LC-Q-TOF mass-spectrometer (Bruker Daltonics). The LC runtime was 22 min, at a flow rate of 0.6 mL/min. Solvent system: water + 0.1% formic acid (HFo; solvent A), acetonitrile + 0.1% HFo (solvent B); from 95% A to 0% A in 12 min, then 3 min at 0% A, then reversion to 45% A within the remaining 7 min. Ionization was performed in positive and negative modes. Analysis of LC–MS data was performed using the program Compass Data Analysis 4.2 (Bruker Daltonics). The quantitative analysis of Clo production was done by comparing areas of extracted Clo mass-peak (697.2 Da, [M + H]+) present in NRRL 3504 ethyl acetate extracts to the areas of 697.2-Da mass peak produced by the known amounts of authentic Clo run through the machine. A reference calibration curve was created using the values of areas for 697.2-Da mass peaks obtained when 6 different amounts of Clo were injected and run under conditions mentioned above (see Table S1, Fig. S1). See ESM for the access to raw LC–MS data.
Scanning electron microscopy (SEM)
Before SEM, NRRL 3504 lawns were grown on ISP3 agar for 216 h. Then, thin slices of the lawn surfaces were cut, sprayed with thin layers of copper in vacuo, and imaged with JEOL JSM-T220a SEM (Jeol, Japan) using a 25 kV electron beam.
Qualitative glucuronidase assay
A qualitative glucuronidase assay was utilized to test whether recombinant strains of S. roseochromogenes exhibit gusA-mediated conversion of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) into 5,5'-dibromo-4,4'-dichloro-indigo. For this, 2 μL of X-Gluc (25 mg/mL DMSO solution) was added to the surfaces of the lawns; plates were left for 2 h at 30 °C and then examined for chromogenic conversion of X-Gluc.
Results
Conditions and rates of conjugative transfer of different vectors into S. roseochromogenes using spores as recipients
Integrative, actinophage-based vectors are widely used in Streptomyces genetics because of their efficiency as a tool for stable expression of the cloned genes (Kieser et al. 2000). Currently, vectors based on the int-attP module of actinophages φC31, VWB, and φBT1 are the most popular ones (Bierman et al. 1992; Van Mellaert et al. 1998; Gregory et al. 2003; Ostash et al. 2009). Replicative vectors are a powerful additional tool for gene expression and among them are widely used pSG5 replicon-based moderate copy number (20–50 per genome) plasmids (Bierman et al. 1992; Muth 2018). We, therefore, decided to test the utility of the aforementioned expression systems for Clo producer. Although φC31-based plasmids are known to be transferable into NRRL 3504 (Eustáquio et al. 2003), the other vectors were not tested. NRRL 3504 genome sequence showed that the latter possesses a typical attB site for φC31, as well as intact attB sites for φBT1 and VWB (Fig. S2a, b, and c, respectively, ESM).
We choose the intergeneric conjugation with E. coli ET 12567 (pUZ8002) as a method to deliver recombinant plasmids into cells of NRRL 3504. This method was previously proved efficient for NRRL 3504 (Eustáquio et al. 2003); however, in that case, the recipient was applied to the mating as vegetative mycelium. Such an approach has certain drawbacks, such as low efficiency and reproducibility, as well as a high labor burden. Thus, we decided to test the efficiency of intergeneric conjugation using spores of NRRL 3504 as a recipient material.
First, we tested the growth of NRRL 3504 on a range of common agar media, aiming to detect the optimal sporulation conditions (data not shown). We found out that ISP3 supports abundant sporulation in NRRL 3504 (Fig. S3), allowing to prepare concentrated spore suspensions (see Materials and Methods section).
To test if conjugation with spores works in NRRL 3804 and how efficient it is for different plasmids, we used a previously designed approach (Yushchuk et al. 2020), involving the derivatives of pSET152 (φC31-based integrative vector), pRT801 (φBT1-based integrative vector), pSOK804 (VWB-based integrative vector), and pKC1139 (pSG5-replicon replicative vector)—pSAGA, pRAGA1, pSoAGA2, and pKAGA1, respectively. These plasmids all carry a reporter cassette aac(3)IVp-gusA, which allows to instantly rule out false-positive colonies among transconjugants by adding X-Gluc to the overlay. Thus, we tested the transfer efficiency of four vectors mentioned above from E. coli ET12567 (pUZ8002) to S. roseochromogenes. The pSAGA vector (φC31-based) exhibited the highest transfer efficiency, followed by pRAGA1 (φBT1-based) and pSoAGA2 (VWB-based) (Table 3). The lowest transfer efficiency was shown for a replicative vector pKAGA1 (pSG5-replicon based, Table 3). All transconjugants were able to convert X-Gluc, proving the absence of spontaneous apramycin-resistant colonies (Fig. S4a). Furthermore, we tested three random transconjugants for each platform by PCR and were successfully able to amplify a 1000-bp internal fragment of gusA (Fig. S4b).
We also noticed that conjugative transfer efficiency was medium-dependent: pSAGA- and pRAGA1-carrying transconjugants occurred on ISP3 agar more frequently than on SFM, while for pSoAGA2 and pKAGA SFM turned out to be more favorable (Table 3). Finally, we observed that Mg2+ ions have a positive impact on the conjugative transfer efficiency in S. roseochromogenes, although to a different extent (Table 3). These results are in agreement with many other described cases, where Mg2+ ions improve conjugation efficiencies of different Streptomyces spp.; however, the explanation of this fact remains unknown (Galm et al. 2008; Kim et al. 2008). All platforms mentioned above did not affect the growth, morphology, or antimicrobial activity of NRRL 3504 (data not shown).
Insertional inactivation of cloH-I-J-K operon in S. roseochromogenes
We focused on the combination of well and disk diffusion assays (WDA and DDA, see Materials and Methods section) as surrogates for measuring Clo production. First, WDA would be used to screen the antimicrobial properties of different recombinants at different growth time points and replicates. Then, Clo was extracted (see Materials and Methods section) and tested in DDA. Finally, Clo would be measured quantitatively in these extracts by LC–MS.
We first cultivated NRRL 3504 in GYM medium for 120 h. WDA showed that antimicrobial activity appears in the samples of NRRL 3504 cultural broth starting from 96 h, and reaches the peak at 120 h (Fig. S5a). At the same time, biomass accumulation reaches a peak at 96 h (Fig. S6a). The results of the DDA (Fig. 2a) were in agreement. Finally, we tested the extracts of NRRL 3504 cultural broth samples (collected at 120 h) by LC–MS to detect the presence of Clo (observed m/z 697.2165, calculated Clo [M + H]+ 697.2164 Da, 0.14 ppm difference, Fig. 3).
Results of disk diffusion assays showing antimicrobial activity of ethyl acetate extracts obtained from the cultures of NRRL 3504 and recombinant strains. All strains were cultivated in GYM medium and samples for the extraction were collected at five time points from 24 to 120 h. k− was always a disk soaked with 10 µL of solvent—methanol. Plates here show typical results of three replicates, for the numerical information on growth inhibition zones diameters and statistics, please see Table S4
The total antimicrobial activity revealed by the bioassays could be a net result of the production of Clo as well other uncharacterized compounds. Indeed, streptomycetes are known to produce different antibiotics with similar antimicrobial properties and S. roseochromogenes genome of carries numerous BGCs for specialized metabolic pathways (Rückert et al. 2014). To rule out this possibility, we decided to construct a strain with blocked Clo production. Using the established conjugation protocol, cloH-I-J-K operon (central to Clo production, see Fig. 1b) of NRRL 3504 was insertionaly inactivated with plasmid pKOClo. The bioassays showed the complete cessation of antimicrobial activity of cloH-I::pKOClo mutant against M. luteus (Fig. S5b, Fig. 2b), while its growth rate (measured as dry biomass) was at the slightly lower level than NRRL 3504 (Fig. S6b). Accordingly, LC–MS analysis revealed the absence of Clo mass peak in the extracts of cloH-I::pKOClo strain (Fig. 3). Hence, antimicrobial activity of NRRL 3504 is indeed caused by accumulation of Clo; WDA and DDA are sensitive and reliable methods for the detection of Clo production.
Properties of recombinant S. roseochromogenes strains carrying extra copies of adpA and bldA genes
Global positive regulator AdpA is known to improve production of different antibiotics in Streptomyces spp. (Ohnishi et al. 2005; Rabyk et al. 2018; Yushchuk et al. 2018). Moreover, AdpA action is tightly intertwined with the expression of bldA—a gene for leu(TTA)-tRNA, involved in the translation of rare TTA codon (Chater and Chandra 2008; Makitrynskyy et al. 2013; Koshla et al. 2017). Within the G-C rich genomes of Streptomyces spp., TTA codon often occurs in genes for morphogenesis and antibiotic biosynthesis, playing a regulatory role (McCormick and Flärdh 2012). The presence of a large number of TTA codons (sixteen, which is four times as many as in nov BGC, see Fig. 1b) in clo genes and multiple presumed operators for pleiotropic regulator AdpA within several intergenic regions of clo BGC (Fig. S7b) prompted us to investigate the effects of additional copies of bldA and adpA genes on Clo production. For this purpose, we have used heterologous genes from S. albidoflavus J1074. This was justified, because AdpA from NRRL 3504 (coded by the M878_RS76560 locus) shares 84% of amino acid sequence identity with its counterpart from J1074 (having identical DNA binding domains), while bldA genes of J1074 and NRRL 3504 (M878_RS74880) share 94% of nucleotide sequence identity. Thus, bldA under the control of native promotor was transferred into NRRL 3504 using pSG5-based low copy number replicative plasmid (pKCBA), while adpA gene under the control of moeE5-promotor was introduced on actinophage φBT1-based integrative plasmid pMoRT4181 (see also Table 1). Both pKCBA+ and pMoRT4181+ recombinants produced more Clo in comparison to the parent strain, as evident from the bioassays and MS measurements (Fig. 2c and d; Fig. S5c and d; Fig. 4). At the same time, biomass accumulation was comparable to the parent strain (Fig. S6c and d). Interestingly, in addition to Clo, pMoRT4181+ accumulated two compounds whose masses fit known Clo intermediates: novclobiocins 101 (observed m/z 661.24, calculated [M—H]− 661.2391, 1.36 ppm difference) and 107 (observed m/z 554.2030, calculated [M—H]− 554.2021, 1.62 ppm difference; see also ESM Fig. S8; see Fig. 1a for chemical structures). We did not observe significant amounts of these intermediates in either NRRL 3504 or pKCBA+ strain, or any other NRRL 3504 derivatives described below (data not shown).
Production of Clo in the recombinant strains carrying additional copies of bldA and adpA genes from S. albidoflavus, as judged from quantitative analysis of MS data. The diagram was derived from MS data as follows. Mass peaks of 697.2 Da, that corresponds to Clo cation ([M + H]+), were extracted from the total ion chromatograms and ratios of the area of pKCBA+ and pMoRT4181+ strains to the wild type (NRRL 3504) were determined and normalized against the same amount of the biomass (dry weight from 5 mL). Results represent mean values (three repeats); error bars represent SD
Overexpression of cluster-situated regulatory genes novG and cloG using different expression platforms
The nov cluster-encoded transcriptional activator NovG is essential for the initiation and sustained production of Nov by S. sphaeroides (Eustáquio et al. 2005a, b). The same functional importance for Clo biosynthesis is suggested for CloG, the NovG ortholog (Heide 2009). Both NovG and CloG belong to the group of StrR-like transcriptional regulators often found to control the biosynthesis of antibiotics (van der Heul et al. 2018). Both proteins share 85% of amino acid sequence identity and so might be functionally interchangeable. Gene cloG contains three TTA codons within the reading frame, while only one TTA is present in novG. We speculated that the presence of three TTA codons might decrease the efficiency of cloG as a tool for Clo overproduction.
We cloned novG and cloG into replicative pKC1139-based vector and a series of actinophage-based vectors so that the expression of these genes would be under the control of different promoters (see Table 1). All novG/cloG-expressing plasmids were transferred into NRRL 3504 and the Clo production by the resulting recombinants and the parental strain was first compared using bioassays. Almost all recombinant strains had better antimicrobial activity as compared to the parental strain (Fig. S5, Fig. 2) while showing no significant decrease in biomass accumulation (Fig. S6). Among them, pIJ6902-derived expression vectors (pCLOG104 and pNOVG104, where regulatory genes are under the control of thiostrepton-inducible promoters) exerted negative effect on antimicrobial activity of recombinants in the absence of inducer, but still increased antimicrobial activity when inducer was added (Fig. S5m-p, Fig. 2m-p). Notably, novG-overexpressing strains had more potent antimicrobial properties than cloG+ strains (Fig. S5, Fig. 2). Then, we have estimated Clo production levels in cultural extracts (120 h time point) by means of LC–MS. Obtained data were again consistent with the WDA and DDA results, showing that novG+ and cloG+ strains specifically overproduced Clo (Fig. 5, Tables S3 and S4). The highest increase in Clo titers (up to 32-fold, Fig. 5, Fig. S9) was observed upon cumate-inducible expression of novG from P21 promoter of vector pGCymRP21 (plasmid pNOVG103, see Table 1); at the same time, expression of cloG gave only 20-fold increase of Clo production under the same conditions (Fig. 5). Such productivity corresponded to 239 and 169 mg/L of Clo, respectively. Although overproducing Clo significantly, pNOVG103+ and pCLOG103+ strains accumulated biomass at the levels comparable with the parent strain (Fig. S6a and j).
Production of Clo in the recombinant strains, carrying additional copies of novG and cloG cluster-situated transcriptional regulatory genes under the control of different promoters. More than 30-fold increase in Clo production was achieved when novG was expressed from pGCymRP21 platform in presence of 9 µg/mL of cumate as an inducer. The diagram was built following the approach explained in the legend for Fig. 4; exact values of Clo peak areas for different measurements are given in Table S5
Discussion
The natural producer of Clo, S. roseochromogenes NRRL 3504, is considered poorly amenable to genetic manipulation and genetic approaches to improve Clo titers in NRRL 3504 were not pursued. Here we report that intergeneric E. coli—Streptomyces conjugation, using spores of NRRL 3504 as a recipient, can be used to transfer integrative and replicative plasmids into S. roseochromogenes and to make gene knockouts in this strain. Overall, we have successfully transferred 12 integrative plasmids (φC31-, φBT1-, and VWB-based) and 4 replicative (pSG5-based) ones, demonstrating high conjugation efficiency. The utility of integrative vectors was demonstrated through the expression of pleiotropic and pathway-specific regulators of secondary metabolism, some of which led to significant enhancements in Clo production levels. This work therefore expands the genetic toolkit for S. roseochromogenes and facilitates the access to large quantities of Clo.
Additional plasmid-borne copies of bldA and regulatory gene adpA led to increased Clo titers. One of the possible explanations for the case of bldA might be the anomalously high frequency of rare TTA codon in clo genes (16 TTA codons across 28-gene BGC), which may limit the Clo production level. Likewise, the presence of putative AdpA operators within clo BGC motivated us to check the effects of adpA on NRRL 3504. Of note, adpA expression boosted the production of not only Clo but also novclobiocins 101 and 107. We tentatively suggest that this is caused by unbalanced activation of different clo operons by AdpA, although this idea requires further experimental verification. Alternatively, such effect might be due to the exhausted pool of Leu(TTA)-tRNA. Activation of some other biosynthetic pathways in NRRL 3504 by adpA-overexpression is also not excluded; this might explain diffused edges of M. luteus growth inhibition halos caused with pMoRT4181+ culture broth samples (Fig. S5d). In fact, our recent experience in Streptomyces cyanogenus S136 (Yushchuk et al. 2021), which used the same adpA-expression platform, makes this explanation very likely. The work to test these possibilities is underway in our laboratories. While manipulations of global regulatory genes positively influenced Clo production, introducing extra copies of either cloG or novG, which are pathway-specific regulators of biosynthesis of Clo and novobiocin, respectively, into NRRL 3504 led to even greater yields of Clo. The cumate-inducible expression of novG/cloG from a weak P21 promoter yielded the best results. Both the bioassay and MS measurements agree with the estimate that under the described fermentation conditions the parental strain produced Clo at the level of ca. 7 mg/L (Table S5). Consequently, the aforementioned novG-expressing strain would yield around 200 mg/L of Clo, which is the highest titer for this natural product reported so far (Flinspach et al. 2010). Notably, we performed our Clo production experiments in the “generic” liquid medium—GYM—which lacks many properties of industrial production media. Nevertheless, Clo titers obtained here were already significantly higher than in S. coelicolor strains carrying clo gene cluster, which were fermented under highly optimized conditions. Therefore, we believe that recombinant strains overexpressing either novG or cloG might perform even better under more optimal production conditions.
Peculiarly, we noted that novG was more efficient than cloG in terms of increasing Clo production. Perhaps this is caused by the fact that novG is less saturated with TTA codons than cloG is, although we cannot exclude some other reasons. There is ample room for further improvements to Clo production level in NRRL 3504, either via fine-tuning the cloG/novG expression, or a combination of additional copies of pathway-specific and global regulators within one strain. The availability of three different actinophage-based integrative plasmids and a replicative one for manipulation of NRRL 3504 sets a reliable working ground for such endeavors.
Data availability
All data generated or analyzed during this study are included in this published article (and its supplementary information files).
References
Bierman M, Logan R, O’Brien K, Seno ET, Nagaraja Rao R, Schoner BE (1992) Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49. https://doi.org/10.1016/0378-1119(92)90627-2
Chater KF, Chandra G (2008) The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. J Microbiol 46:1–11. https://doi.org/10.1007/s12275-007-0233-1
Eustáquio AS, Gust B, Luft T, Li SM, Chater KF, Heide L (2003) Clorobiocin biosynthesis in Streptomyces: identification of the halogenase and generation of structural analogs. Chem Biol 10:279–288. https://doi.org/10.1016/S1074-5521(03)00051-6
Eustáquio AS, Gust B, Galm U, Li SM, Chater KF, Heide L (2005a) Heterologous expression of novobiocin and clorobiocin biosynthetic gene clusters. Appl Environ Microbiol 71:2452–2459. https://doi.org/10.1128/AEM.71.5.2452-2459.2005
Eustáquio AS, Li S-M, Heide L (2005b) NovG, a DNA-binding protein acting as a positive regulator of novobiocin biosynthesis. Microbiology 151:1949–1961. https://doi.org/10.1099/MIC.0.27669-0
Flatman RH, Eustaquio A, Li SM, Heide L, Maxwell A (2006) Structure-activity relationships of aminocoumarin-type gyrase and topoisomerase IV inhibitors obtained by combinatorial biosynthesis. Antimicrob Agents Chemother 50:1136–1142. https://doi.org/10.1128/AAC.50.4.1136-1142.2006
Flinspach K, Westrich L, Kaysser L, Siebenberg S, Gomez-Escribano JP, Bibb M, Gust B, Heide L (2010) Heterologous expression of the biosynthetic gene clusters of coumermycin A1, clorobiocin and caprazamycins in genetically modified Streptomyces coelicolor strains. Biopolymers 93:823–832. https://doi.org/10.1002/BIP.21493
Flinspach K, Rückert C, Kalinowski J, Heide L, Apel AK (2014) Draft genome sequence of Streptomyces niveus NCIMB 11891, producer of the aminocoumarin antibiotic novobiocin. Genome Announc 2(1):e01146-e1213. https://doi.org/10.1128/GENOMEA.01146-13
Freitag A, Rapp H, Heide L, Li SM (2005a) Metabolic engineering of aminocoumarins: Inactivation of the methyltransferase gene cloP and generation of new clorobiocin derivatives in a heterologous host. ChemBioChem 6:1411–1418. https://doi.org/10.1002/cbic.200500019
Freitag A, Wemakor E, Li SM, Heide L (2005b) Acyl transfer in clorobiocin biosynthesis: involvement of several proteins in the transfer of the pyrrole-2-carboxyl moiety to the deoxysugar. ChemBioChem 6:2316–2325. https://doi.org/10.1002/cbic.200500252
Freitag A, Li SM, Heide L (2006) Biosynthesis of the unusual 5,5-gem-dimethyl-deoxysugar noviose: Investigation of the C-methyltransferase gene cloU. Microbiology 152:2433–2442. https://doi.org/10.1099/mic.0.28931-0
Galm U, Heller S, Shapiro S, Page M, Li SM, Heide L (2004) Antimicrobial and DNA gyrase-inhibitory activities of novel clorobiocin derivatives produced by mutasynthesis. Antimicrob Agents Chemother 48:1307–1312. https://doi.org/10.1128/AAC.48.4.1307-1312.2004
Galm U, Wang L, Wendt-Pienkowski E, Yang R, Liu W, Tao M, Coughlin JM, Shen B (2008) In vivo manipulation of the bleomycin biosynthetic gene cluster in Streptomyces verticillus ATCC15003 revealing new insights into its biosynthetic pathway. J Biol Chem 283:25236–28245. https://doi.org/10.1074/jbc.M804971200
Gregory MA, Till R, Smith MCM (2003) Integration site for Streptomyces phage φBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323. https://doi.org/10.1128/JB.185.17.5320-5323.2003
Heide L (2009) The aminocoumarins: biosynthesis and biology. Nat Prod Rep 26:1241–1250. https://doi.org/10.1039/b808333a
Herrmann S, Siegl T, Luzhetska M, Jilg LP, Welle E, Erb A, Leadlay PF, Bechthold A, Luzhetskyy A (2012) Site-specific recombination strategies for engineering actinomycete genomes. Appl Environ Microbiol 78:1804–1812. https://doi.org/10.1128/AEM.06054-11
Horbal L, Fedorenko V, Luzhetskyy A (2014) Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other actinobacteria. Appl Microbiol Biotechnol 98:8641–8655. https://doi.org/10.1007/s00253-014-5918-x
Huang J, Shi J, Molle V, Sohlberg B, Weaver D, Bibb MJ, Karoonuthaisiri N, Lih CJ, Kao CM, Buttner MJ, Cohen SN (2005) Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Mol Microbiol 58:1276–1287. https://doi.org/10.1111/j.1365-2958.2005.04879.x
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) Practical Streptomyces genetics. John Innes Foundation, Norwich
Kim M-K, Ha H-S, Choi S-U (2008) Conjugal transfer using the bacteriophage φC31 att/int system and properties of the attB site in Streptomyces ambofaciens. Biotechnol Lett 30:695–699. https://doi.org/10.1007/s10529-007-9586-0
Koshla O, Lopatniuk M, Rokytskyy I, Yushchuk O, Dacyuk Y, Fedorenko V, Luzhetskyy A, Ostash B (2017) Properties of Streptomyces albus J1074 mutant deficient in tRNALeuUAA gene bldA. Arch Microbiol 199:1175–1183. https://doi.org/10.1007/s00203-017-1389-7
Koshla O, Yushchuk O, Ostash I, Dacyuk Y, Myronovskyi M, Jäger G, Süssmuth RD, Luzhetskyy A, Byström A, Kirsebom LA, Ostash B (2019) Gene miaA for post-transcriptional modification of tRNAXXA is important for morphological and metabolic differentiation in Streptomyces. Mol Microbiol 112:249–265. https://doi.org/10.1111/mmi.14266
Li SM, Heide L (2006) The biosynthetic gene clusters of aminocoumarin antibiotics. Planta Med 72:1093–1099. https://doi.org/10.1055/s-2006-946699
Makitrynskyy R, Ostash B, Tsypik O, Rebets Y, Doud E, Meredith T, Luzhetskyy A, Bechthold A, Walker S, Fedorenko V (2013) Pleiotropic regulatory genes bldA, adpA and absB are implicated in production of phosphoglycolipid antibiotic moenomycin. Open Biol 3:130121. https://doi.org/10.1098/rsob.130121
Mancy D, Ninet L, Preud'Homme J (1974) Antibiotic 18631 RP. U.S. patent. 3(793):147
Mandler MD, Baidin V, Lee J, Pahil KS, Owens TW, Kahne D (2018) Novobiocin enhances polymyxin activity by stimulating lipopolysaccharide transport. J Am Chem Soc 140:6749–6753. https://doi.org/10.1021/JACS.8B02283
Mattingly AE, Cox KE, Smith R, Melander RJ, Ernst RK, Melander C (2020) Screening an established natural product library identifies secondary metabolites that potentiate conventional antibiotics. ACS Infect Dis 6:2629–2640. https://doi.org/10.1021/ACSINFECDIS.0C00259
May JM, Owens TW, Mandler MD, Simpson BW, Lazarus MB, Sherman DJ, Davis RM, Okuda S, Massefski W, Ruiz N, Kahne D (2017) The antibiotic novobiocin binds and activates the ATPase that powers lipopolysaccharide transport. J Am Chem Soc 139:17221–17224. https://doi.org/10.1021/jacs.7b07736
McCormick JR, Flärdh K (2012) Signals and regulators that govern Streptomyces development. FEMS Microbiol Rev 36:206–231. https://doi.org/10.1111/j.1574-6976.2011.00317.x
Muth G (2018) The pSG5-based thermosensitive vector family for genome editing and gene expression in actinomycetes. Appl Microbiol Biotechnol 102:9067–9080. https://doi.org/10.1007/s00253-018-9334-5
Myronovskyi M, Welle E, Fedorenko V, Luzhetskyy A (2011) β-Glucuronidase as a sensitive and versatile reporter in actinomycetes. Appl Environ Microbiol 77:5370–5383. https://doi.org/10.1128/AEM.00434-11
Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem 69:431–439. https://doi.org/10.1271/bbb.69.431
Ostash B, Makitrinskyy R, Walker S, Fedorenko V (2009) Identification and characterization of Streptomyces ghanaensis ATCC14672 integration sites for three actinophage-based plasmids. Plasmid 61:171–175. https://doi.org/10.1016/j.plasmid.2008.12.002
Ostash B, Campbell J, Luzhetskyy A, Walker S (2013) MoeH5: a natural glycorandomizer from the moenomycin biosynthetic pathway. Mol Microbiol 90:1324–1338. https://doi.org/10.1111/mmi.12437
Pojer F, Li S-M, Heide L (2002) Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901–3911. https://doi.org/10.1099/00221287-148-12-3901
Pojer F, Kahlich R, Kammerer B, Li SM, Heide L (2003a) CloR, a bifunctional non-heme iron oxygenase involved in clorobiocin biosynthesis. J Biol Chem 278:30661–30668. https://doi.org/10.1074/JBC.M303190200
Pojer F, Wemakor E, Kammerer B, Chen H, Walsh CT, Li S-M, Heide L (2003b) CloQ, a prenyltransferase involved in clorobiocin biosynthesis. Proc Natl Acad Sci 100:2316–2321. https://doi.org/10.1073/PNAS.0337708100
Raad I, Darouiche R, Hachem R, Sacilowski M, Bodey GP (1995) Antibiotics and prevention of microbial colonization of catheters. Antimicrob Agents Chemother 39:2397. https://doi.org/10.1128/AAC.39.11.2397
Rabyk M, Yushchuk O, Rokytskyy I, Anisimova M, Ostash B (2018) Genomic insights into evolution of AdpA family master regulators of morphological differentiation and secondary metabolism in Streptomyces. J Mol Evol 86:204–215. https://doi.org/10.1007/s00239-018-9834-z
Rückert C, Kalinowski J, Heide L, Apel AK (2014) Draft genome sequence of Streptomyces roseochromogenes subsp oscitans DS 12.976, producer of the aminocoumarin antibiotic clorobiocin. Genome Announc 2(1):e01147-13. https://doi.org/10.1128/GENOMEA.01147-13
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New-York
Sekurova ON, Brautaset T, Sletta H, Borgos SEF, Jakobsen ØM, Ellingsen TE, Strøm AR, Valla S, Zotchev SB (2004) In vivo analysis of the regulatory genes in the nystatin biosynthetic gene cluster of Streptomyces noursei ATCC 11455 reveals their differential control over antibiotic biosynthesis. J Bacteriol 186:1345–1354. https://doi.org/10.1128/JB.186.5.1345-1354.2004
Steffensky M, Mühlenweg A, Wang ZX, Li SM, Heide L (2000) Identification of the novobiocin biosynthetic gene cluster of Streptomyces spheroides NCIB 11891. Antimicrob Agents Chemother 44:1214–1222. https://doi.org/10.1128/AAC.44.5.1214-1222.2000
van der Heul HU, Bilyk BL, McDowall KJ, Seipke RF, van Wezel GP (2018) Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat Prod Rep 35:575–604. https://doi.org/10.1039/C8NP00012C
van Mellaert L, Mei L, Lammertyn E, Schacht S, Anne J (1998) Site-specific integration of bacteriophage VWB genome into Streptomyces venezuelae and construction of a VWB-based integrative vector. Microbiology 144:3351–3358. https://doi.org/10.1099/00221287-144-12-3351
Walsh TJ, Standiford HC, Reboli AC, John JF, Mulligan ME, Ribner BS, Montgomerie JZ, Goetz MB, Mayhall CG, Rimland D, Stevens DA, Hansen SL, Gerard GC, Ragual RJ (1993) Randomized double-blinded trial of rifampin with either novobiocin or trimethoprim-sulfamethoxazole against methicillin-resistant Staphylococcus aureus colonization: prevention of antimicrobial resistance and effect of host factors on outcome. Antimicrob Agents Chemother 37:1334. https://doi.org/10.1128/AAC.37.6.1334
Westrich L, Heide L, Li SM (2003) CloN6, a novel methyltransferase catalysing the methylation of the pyrrole-2-carboxyl moiety of clorobiocin. ChemBioChem 4:768–773. https://doi.org/10.1002/cbic.200300609
Wolpert M, Gust B, Kammerer B, Heide L (2007) Effects of deletions of mbtH-like genes on clorobiocin biosynthesis in Streptomyces coelicolor. Microbiology 153:1413–1423. https://doi.org/10.1099/MIC.0.2006/002998-0
Xu H, Kahlich R, Kammerer B, Heide L, Li SM (2003) CloN2, a novel acyltransferase involved in the attachment of the pyrrole-2-carboxyl moiety to the deoxysugar of clorobiocin. Microbiology 149:2183–2191. https://doi.org/10.1099/mic.0.26314-0
Yushchuk O, Ostash I, Vlasiuk I, Gren T, Luzhetskyy A, Kalinowski J, Fedorenko V, Ostash B (2018) Heterologous AdpA transcription factors enhance landomycin production in Streptomyces cyanogenus S136 under a broad range of growth conditions. Appl Microbiol Biotechnol 102:8419–8428. https://doi.org/10.1007/s00253-018-9249-1
Yushchuk O, Homoniuk V, Datsiuk Y, Ostash B, Marinelli F, Fedorenko V (2020) Development of a gene expression system for the uncommon actinomycete Actinoplanes rectilineatus NRRL B-16090. J Appl Genet 61:141–149. https://doi.org/10.1007/s13353-019-00534-7
Yushchuk O, Ostash I, Mösker E, Vlasiuk I, Deneka M, Rückert C, Busche T, Fedorenko V, Kalinowski J, Süssmuth RD, Ostash B (2021) Eliciting the silent lucensomycin biosynthetic pathway in Streptomyces cyanogenus S136 via manipulation of the global regulatory gene adpA. Sci Rep 11:3507. https://doi.org/10.1038/s41598-021-82934-6
Acknowledgements
Prof. Andriy Luzhetskyy (Saarland University) and Prof. Margaret Smith (University of York) are thanked for kind provision of some vectors used in this study.
Funding
This project has received funding from the Ministry of Education and Science of Ukraine under grants BG-80F, M-26 (to B.O.), and BG-09F (to V.F.).
Author information
Authors and Affiliations
Contributions
BO and OY conceived and designed research, with conceptual input from OK, VF, and DK. SM, AS, and IO performed research and analyzed data. MM provided mass-spectrometry assistance and critical reagents. BO, OY, AS, and DK wrote the paper.
Corresponding author
Ethics declarations
Ethical approval
This article does not contain studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Melnyk, S., Stepanyshyn, A., Yushchuk, O. et al. Genetic approaches to improve clorobiocin production in Streptomyces roseochromogenes NRRL 3504. Appl Microbiol Biotechnol 106, 1543–1556 (2022). https://doi.org/10.1007/s00253-022-11814-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-022-11814-4