Abstract
Xylanase is an enzyme in high demand for various industrial applications, such as those in the biofuel and pulp and paper fields. In this study, xylanase-producing microbes were isolated from the gut of the wood-feeding termite at 50°C. The isolated microbe produced thermostable xylanase that was active over a broad range of temperatures (40–90°C) and pH (3.5–9.5), with optimum activity (4,170 ± 23.5 U mg−1) at 60°C and pH 4.5. The enzyme was purified using a strong cation exchanger and gel filtration chromatography, revealing that the protein has a molecular mass of 205 kDa and calculated pI of 5.38. The half-life of xylanase was 6 h at 60°C and 2 h at 90°C. The isolated thermostable xylanase differed from other xylanases reported to date in terms of size, structure, and mode of action. The novelty of this enzyme lies in its high specific activity and stability at broad ranges of temperature and pH. These properties suggest that this enzyme could be utilized in bioethanol production as well as in the paper and pulp industry.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Termites are creatures that are notorious for their potential to cause billions of dollars of damage each year by rendering wood to dust. However, termites also harbor more than 200 species of microbes that produce novel cellulosic and hemicellulosic degrading enzymes, which in turn have the potential to be efficiently applied for the production of ethanol from lignocellulosic biomass in the biofuel industries [2].
The gut of the wood-feeding termite is an astonishingly ‘bioreactor’, where diverse array of microbes are utilized to catalyze the conversion of lignified plant cell-wall materials to fermentation products. Without these microbes, termites would be unable to digest cellulose, which is their main food [28, 34]. Molecular phylogenetic data have revealed the presence of hundreds of microbial species in this small environment, but information on their functional diversity is less well known [1, 8]. A meta-genomic analysis of the bacterial community residing in the hindgut of a wood-feeding ‘higher’ Nasutitermes species revealed the presence of a large and diverse set of bacterial genes encoding hydrolytic enzymes involved in cellulose and xylan degradation [36]. Hemicellulose-degrading microorganisms and their enzymes play a vital role in the recycling of carbon [32]. These microorganisms produce the xylanases, which hydrolyze hemicellulose to xylose, arabinose, mannose, glucose, galactose, and acetate [19]. Xylan is a major component of hemicelluloses and a useful biomass for bio-energy and chemical industries. It is composed of a backbone chain that consists of a varying number of β-1,4-d-xylopyranosyl residues (70–130 in softwood xylan and 150–200 in hardwood xylan). Two major kind of enzymes that are responsible for the hydrolysis of the xylan long chain, namely, endo 1,4-β-xylanase (EC 3.2.1.8) and β-xylosidase (EC 3.2.1.37) [24].
Many β-1,4-xylanases have isolated from mesophilic fungi and bacteria, but very few thermostable xylanases have been reported from the thermophiles isolated from the gut of wood-eating termites [7]. In the study reported here, we collected wood-feeding termites from the forest of Rajaji National Park Range, Dehradun (India) and isolated thermophilic bacteria from their gut that produced a thermostable xylanase. We subsequently characterized this novel and thermostable xylanase.
Materials and methods
Termite collection and sample preparation
Approximately 50 wood-feeding higher termites (family Termitidae) were collected from the sissoo (Dalbergia sissoo) plant and washed with sterile saline water. The termites were kept on ice and the guts removed by using sterile tweezers and forceps [27, 32]. The guts were then washed with sterile Milli Q water and crushed gently in a mortar and pestle.
Isolation and screening of thermophiles
A culture enrichment medium [(% w/v): beechwood xylan, 1.0; peptone, 1.0; yeast extract, 1.0; NaCl, 0.5; gelrite, 1.0] was used for the isolation and screening of xylanase-producing thermophiles. Different dilutions of termite gut homogenate were prepared and spread over the beechwood xylan plates and the plates incubated at 50°C for 24 h. The xylanase-producing thermophiles were screened by flooding 0.5% (w/v) congo red dye in 5% (v/v) ethanol on rich medium [(% w/v): beechwood xylan, 0.5; peptone, 1.0; yeast extract, 1.0; NaCl, 0.5; gelrite, 1.0, pH 6.5) plates for 10 min. The plates were repeatedly washed with 1 M NaCl to remove the unbound dye. The bacterial colonies that showed a yellow zone were selected for further study. The replica plate method was used for screening the xylanase-producing strains. The purified strains were maintained on rich medium slants.
Identification of microorganism and culture conditions
The xylanase-producing microorganism was identified on the basis of physiological characteristics and 16S rRNA gene sequence analysis [12]. Using consensus primers, the approximately 1.5-kb 16S rRNA fragment was amplified using PCR. The PCR product was bi-directionally sequenced using the forward and reverse primer as well as an internal primer. Sequence data were aligned and analyzed to identify the closest homologs with a submitted sequence in the NCBI database. The phylogenetic tree was depicted using the TREEVIEW program.
The isolated bacterial strain was grown in medium containing (g l−1): beechwood xylan, 15; peptone, 4.0; yeast extract, 1.0; K2HPO4, 4.0; MgSO4·7H2O, 1.0; KCl, 0.2; trace elements (CaCl2, 2.2 × 10−3; ZnO, 2.5 × 10−3; FeCl3·6H2O, 2.7 × 10−2; MnCl2·4H2O, 1.0 × 10−2; CuCl2·2H2O, 8.5 × 10−4; CoCl2·H2O, 2.4 × 10−3; NiCl3·6H2O, 2.5 × 10−4; H3BO3, 3.0 × 10−4; Na2MoO4, 1.0 × 10−3), 20 ml; the pH was adjusted to 6.0 using 1 M NaOH. The medium was inoculated with 16-h-old culture (early exponential phase) and incubated at 50°C in an orbital shaker at 150 rpm. A 500-μl sample of inoculum was used in 100 ml of medium in a 500-ml flask.
The bacterial strain was also grown on the different carbon and nitrogen sources. Various carbon sources, such as birchwood xylan, beechwood xylan, oatspelt xylan, carboxymethyl cellulose, cellobiose, glucose, xylose, and raw substrates, such as bagasse (untreated and pre-treated with 0.1% H2SO4 at 121°C for 30 min), and corn cob chips (collected from nearby farms), at concentrations ranging from 0.1 to 2.5% (w/v) were used. The effect of nitrogen on enzyme production was studied using different organic sources,such as yeast extract, beef extract, tryptone, casein hydrolysate, peptone, and soybean meal, and inorganic sources, such as ammonium sulphate, ammonium chloride, ammonium nitrate, ammonium acetate, and urea. The concentration of nitrogen sources was varied from 0.1 to 1.0% (w/v). All experiments were carried out using 100 ml medium in a 500-ml Erlenmeyer shake flask.
Enzyme assay
Thermostable xylanase activity was determined by estimating the amount of reducing sugars released from 1% (w/v) beechwood xylan as a substrate at an optimum temperature of 60°C and optimal pH of 4.5. The amount of released reducing sugars was determined by the dinitrosalicylic acid method [29]. Solutions containing different concentrations of xylose were used as the standards. One unit of xylanase activity was defined as the amount of enzyme that released 1 μmol of reducing sugars per minute at 60°C.
Purification of thermostable xylanase
Salt precipitation of xylanase
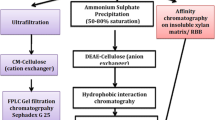
After the culture had been harvested, the broth (1,000 ml) was centrifuged at 34,000 g for 10 min at 4°C in a refrigerated centrifuge (Sorvall Evolution; Thermo Fisher Scientific, Waltham, MA). The cell-free supernatant was passed through an ultra-filtration unit with a 10-kDa membrane. The concentrated enzyme solution (10 ml) was then precipitated by using an ammonium salt precipitation method, with the addition of the ammonium salt in serial increments (0–15, 15–25, 25–35, 35–50, 50–60, 60–70 and 70–80%). The precipitated enzyme solution was dialyzed through a 5-kDa dialysis membrane (HiMedia Laboratories, Mumbai, India) against 20 mM acetate buffer (pH 6.0) with constant stirring under chilled conditions.
Chromatographic separation
An 8-ml of dialyzed enzyme solution was passed through a strong cation exchanger (MacroPrep High S support; Bio-Rad, Hercules, CA) column (1.5 × 30 cm), pre-equilibrated with 20 mM phosphate buffer (pH 6.0). The unbound proteins were eluted first by using same buffer at a flow rate of 1.0 ml min−1, while the bound proteins were eluted later by using linear salt gradient (0–1 M NaCl) in the same buffer at a flow rate of 1.0 ml min−1. One-millititer fractions were collected. Fraction numbers 24–42 showed xylanase activity and were pooled together and concentrated in an Amicon Ultra centrifuge filter device (Ultracel-10K; regenerated cellulose membrane: 10,000 MWCO) (Millipore, Billerica, MA).
The concentrated enzyme solution (500 μl) was applied to a pre-equilibrated P 100 Sephadex gel (1 × 10 cm) (Bio-Rad). The unbound proteins were eluted using 20 mM acetate buffer (pH 5.0) at a flow rate of 1 ml min−1 (1 ml per fraction), and bound proteins were eluted by using a linear salt gradient (0–1 M NaCl) in the same buffer. Among the 50 fractions collected, fraction nos. 4–20 showed xylanase activity and pooled together, concentrated and further analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Electrophoresis and molecular mass determination
Sodium dodecyl sulfate-PAGE was carried out according to Laemmli [20] on a 7.5% SDS-PAGE gel (PROTEAN II xi cell; Bio-Rad). The molecular weight markers (29–205 kDa; Sigma, St. Louis, MO) were run along with the samples. The protein bands were visualized by staining with Coomassie Brilliant Blue G 250, and the molecular mass of the purified enzyme was determined by comparing the band with the markers.
Zymogram preparation
Xylanase activity of the purified enzyme was evaluated by PAGE according to Laemmli [20]. After electrophoresis, the gel was first washed with deionized water, then immersed in 20% (v/v) isopropanol at 4°C for 1 h with gentle shaking, rinsed three times for 10 min each time with deionized water, imbibed in 0.2% (w/v) beechwood xylan solution at 4°C for 1 h to assure xylan penetration into the gel, and incubated in 20 mM acetate buffer (pH 5.0) at 60°C for 1 h to allow the enzyme to digest the xylan in the gel. The gel was then stained with 0.1% (w/v) congo red dye for 20 min, washed with 1 M NaCl for 10 min, immersed in 0.5% (w/v) acetic acid to stop the reaction [35], and photographed immediately using an Imager Gel Doc XR system (Bio-Rad).
Protein estimation
Protein content was measured using the Folin-Lowry method with bovine serum albumin as the standard [26].
Characterization of the enzyme
Influence of temperature and pH on enzyme activity
The effect of temperature on the activity of the isolated enzyme was observed by incubating the enzyme for 30 min at temperatures ranging from 30 to 90°C. After each incubation, the enzyme was kept at 4°C. Xylanase activity was determined by using 1% (w/v) beechwood xylan as a substrate and measuring the reducing sugars from the digestion of beechwood xylan at different temperatures. Similarly, the effect of pH on xylanase activity was determined at 60°C at pHs ranging from 3.0 to 10.0 using a glycine–HCl buffer (pH 3.0–4.0), acetate buffer (pH 5.0–6.0), phosphate buffer (pH 6–8.0) and glycine–NaOH buffer (pH 9–10.0). The enzyme was incubated with these buffers at 60°C for 10 min without a substrate, and the activity was determined on 1% (w/v) beechwood xylan by measuring the amount of reducing sugars.
Effects of metal ions and chemicals
The effect of metal ions on enzyme activity was observed by incubating the enzyme in solutions containing 10 mM of MnSO4, CaCl2, NaCl, CoCl2, MgCl2, FeSO4, HgCl2, KCl, ZnSO4, and CuSO4 separately, as well as in solutions containing denaturing agents, such as dithiothreitol (DTT), β-mercaptoethanol (β-ME; 10 mM), SDS (1% w/v), urea (1% w/v), and triton (0.5% v/v) separately, for 30 min without a substrate. The effect of ethylenediaminetetraacetic acid (EDTA) was also observed on enzyme activity. After the incubation, the enzyme activity was determined using 1% (w/v) beechwood xylan at 60°C and pH 4.5. The control was used without any metal ion or denaturing reagents and reported as 100% activity. The relative activity of the enzyme in each of the different metal solutions was determined in reference to that of the control.
Kinetic constants
The initial rate of hydrolysis on beechwood and birchwood xylan by the purified enzyme was calculated by estimating the amount of reducing sugars at different xylan concentrations (range 1–10 mg ml−1). The Michaelis constant (K m) and the rate of reaction (V max) were determined according to Lineweaver–Burk plot [23].
Substrate specificity
Different pure xylans, such as beechwood xylan, birchwood xylan (Sigma), and oatspelt xylan (Hi-Media), and lignocellulosic biomass, such as corn cob chips and dried 40-mesh sieved bagasse both untreated and pre-treated with 0.1% (w/w) H2SO4, were used to determine the substrate specificity of the enzyme. The hydrolysis was carried out at a 1% (w/v) substrate concentration in 20 mM acetate buffer (pH 4.5) at 60°C.
Mode of enzyme action
The enzyme was incubated with 1% (w/v) beechwood xylan at 60°C and pH 4.5. The samples were removed at an interval of 1 h to determine the amount of released reducing sugars. The end products were analyzed by high-performance liquid chromatography using a High Performance Carbohydrate Column (Waters, Milford, MA) at 30°C with an acetonitrile and water mixture (75:25) as a mobile carrier at a flow rate of 1.4 ml min−1. The end products were detected using a Waters 2414 refractive index detector [18].
Effect of protease on xylanase and stability of enzyme
Protease activity was checked during xylanase production in culture broth at 2-h intervals. The effect of protease on purified xylanase was also checked using Proteinase K (Sigma). The purified xylanase was incubated with 10 U of Proteinase K at room temperature for 30 min. Xylanase activity was then determined by analyzing the reducing sugars obtained with 1% (w/v) beechwood xylan at 60°C and pH 4.5. Purified xylanase was kept at 4°C without any preservative in 20 mM acetate buffer (pH 4.5) and 20 mM glycine-NaOH buffer (pH 9.0). After 2 months of incubation, the enzyme was tested for its stability. The assay was carried out at 60°C using both pH 4.5 and 9.0 buffers and 1% (w/v) beechwood xylan.
Results and discussion
Isolation, characterization, and taxonomic classification of IIPSP3
Various xylanases have been isolated from bacteria, fungi, and actinomycetes, but only very few thermostable xylanases that are active under both extreme pH (acidic and alkaline) and high temperature conditions have been reported. Termites provide excellent habitats for hemicellulolytic microorganisms, which in turn produce novel hemicellulose-degrading enzymes. In our study, about 80 different types of colonies were isolated from the termite gut extract; of these, three types of colonies developed the zone of yellow color on the beechwood xylan plates following flooding with the congo red solution. Among these three colonies, colony no. 3 was selected based on maximum xylanase production; hence this colony was named IIPSP3. The strain was maintained for routine purpose on rich medium slants, and the master culture was preserved in 40% (v/v) glycerol at −80°C.
The strain was identified as Paenibacillus macerans based on its 16S rRNA gene sequence (1,478 bp), which was aligned with sequences available in the NCBI database using ClustalX software, and further named as P. macerans IIPSP3. The phylogenetic tree clearly showed that the isolated strain has maximum homology (more than 90% match) with the strain P. macerans. The 16S rRNA gene sequence has been deposited in the GenBank under the accession no. HM246634.1, and the pure culture have been deposited in the MTCC, Institute of Microbial Technology, Chandigarh, India (MTCC 5569).
The isolate was observed to be rod shaped, Gram positive, catalase positive, and indole test negative; it tested positive in the fermentation test for glucose, xylose, maltose, lactose, sucrose, galactose, and mannitol. The isolate also has the ability to ferment glycerol, which makes the strain a potent producer of alcohol (data not shown). There have been published reported of various Paenibacillus species producing xylanolytic enzyme complexes [15], but to the best of our knowledge there have been no reports of xylanase enzyme isolated from the P. macerans species.
Production of enzyme
Paenibacillus macerans IIPSP3 was able to grow and produce xylanase at temperatures ranging from 50 to 70°C and a pH ranging from 5.0 to 9.0. The strain showed maximum xylanase production at its optimum growth temperature, i.e., 50°C, and pH 6.0, with a specific activity of 60 ± 2.0 U ml−1. It has been observed that the optimum temperature for bacterial xylanase production varies from 40 to 60°C and a pH of acidic to neutral [17]. Xylanase production started in the log phase, but maximum enzyme production (60 ± 2.0 U ml−1) was observed in the late exponential phase (Fig. 1). No correlation between growth and xylanase production was observed. The increase in xylanase activity during the late exponential phase might be due to the autolysis of aged cells [11]. The xylanase produced by P. macerans IIPSP3 was named IIPSP3 xylanase.
The important factors for efficient xylanase production are carbon and nitrogen sources. Cheap hemicellulosic agro-based feedstocks, such as corn cobs, bagasse, wheat bran, rice bran, rice straw, and corn stalks, are suitable raw materials for the economic production of xylanase. Among the various carbon sources, such as birchwood xylan, beechwood xylan, oat spelt xylan, carboxymethyl cellulose, cellobiose, glucose, and xylose, and raw substrates, such as bagasse both untreated and pre-treated with 0.1% H2SO4 and corn cob chips, used in our study for enzyme production, maximum enzyme activity was observed in the medium containing 1.5% (w/v) corn cob chips (100 ± 2.6 U ml−1) and oat spelt xylan (88 ± 2.7 U ml−1). Xylanase production by P. macerans IIPSP3 seems to be inductive when grown on xylan substrates. Although the strain was able to grow on xylose, carboxy methyl cellulose, and cellobiose, no xylanase production was observed when they were used as substrates. As reported in the literature, most of the bacteria and fungi tested have shown an inducible mode of xylanase production when xylan was used as the carbon source [17]. No xylanase production was observed when xylose was used as a carbon source, possibly due to repression, and similar results of xylanase repression by xylose have been reported with Cryptococcus albidus [5].
Among the organic and inorganic nitrogen sources used, organic nitrogen sources, such as peptone, tryptone, and yeast extract, were able to enhance xylanase yield. P. macerans IIPSP3 secreted high amounts of xylanase (112 ± 2.8 U ml−1) when 0.4% peptone was used as the nitrogen source. Xylanase secretion could be further increased to a specific activity of 120 ± 3.1 U ml−1 when the combination of 0.4% peptone and 0.1% yeast extract was used as the nitrogen source (Table 1). Ninawe and Kuhad [30] reported high xylanase production when peptone was used as the nitrogen source, and Ko et al. [15] reported enhancement of xylanase yield using the combination of 0.5% (w/v) peptone and 0.5% (w/v) yeast extract.
Purification of IIPSP3 xylanase
Following the removal of the cells and cell debris from the broth, the broth was concentrated in an ultra-filtration unit and the concentrated enzyme solution then precipitated using ammonium salt. Enzyme activity was found in the precipitate obtained from the 35–50% ammonium salt precipitation enzyme solution. The precipitate was then subjected to dialysis and the enzyme purified by cation-exchange chromatography. Two peaks were observed during elution, one of which was a broad peak that showed xylanase activity. The xylanase activity was found in the unbound protein fractions collected from the ion exchanger. The concentrated active fractions were run on the P 100 gel column, and a single peak of xylanase activity was obtained (Fig. 2). The SDS-PAGE analysis showed a single protein band (Fig. 3). The purification of xylanase has been summarized in Table 2. The yield of the purified xylanase after P 100 gel filtration was found to be about 21%, with a specific activity of 2,514 U mg−1 of protein. The molecular mass of the purified xylanase was 205 kDa on a 7.5% SDS-PAGE gel.
The zymogram analysis of the purified xylanase revealed a band with significant activity corresponding to 205 kDa. Two kinds of xylanases have been reported from bacteria: a high-molecular-mass acidic xylanase and a low-molecular-mass basic xylanase, respectively [17]. P. macerans IIPSP3 produced high-molecular-mass xylanase. Beg et al. [4] reported that xylanases secret in the form of a high-molecular-mass multienzyme complex called a ‘xylanosome’. Lin and Thomson [22] reported an extracellular xylanosome complex B (CB) from Butyvibrio fibrisolvens H17c that had a molecular weight of more than 669 kDa and was composed of 11 protein bands with xylanase activity and three bands showing endoglucanase activity. Two high-molecular-weight multienzyme complexes of 1,450 and 400 kDa, respectively, were also reported by Pason et al. [31] from P. curdlanolyticus B-6.
Internal amino acid sequence of IIPSP3 xylanase by liquid chromatography–tandem mass spectrometry
The internal amino acid sequence from the digested purified enzyme was analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS) to identify the purified enzyme. The enzyme was identified as:
MKKVVNSVLA SALALTVAPM AFAAEEAATT TAPKMDADME KTVKRLEALG LVAGYGNGEY GVDKTITRAE FATLVVRARG LEQGAKLAQF SNTYTDVKST DWFAGFVNVA SGEEIVKGFP DKSFKPQNQV TYAEAVTMIV RALGYEPSVK GVWPNSMISK ASELNIARSI TTPNNAATRG DIFKMLDNAL RVDLMEQVEF.
The BLAST search results showed only 21% homology with xylanase 10A from Bacillus firmus (accession no. gb AAQ83581.1) and xylanase from Bacillus sp. N16-5 (accession no. gb ADI24221.1). A BLAST search of the NCBI revealed that IIPSP3 xylanase is a novel xylanase as it did not show homology with other reported xylanases, even those isolated from other Paenibacillus spp.
Effects of temperature and pH on enzyme activity
The influence of temperature on enzyme activity was observed by incubating the enzyme at different temperatures. The IIPSP3 xylanase was found to be thermostable over a broad temperature range (40–90°C), with an optimum temperature of 60°C. Its activity was observed to increase sharply with gradual increases in temperature up to 60°C (specific activity 2,600 ± 16.6 U mg−1); with further increases in termperature, its activity declined gradually (Fig. 4). When the enzyme was incubated at temperatures ranging from 30 to 100°C, the enzyme was 100% active up to 60°C, but its xylanase activity decreased by 5, 12, 30 and 49% when the temperature was further increased to 70, 80, 90, and 100°C, respectively. Shi et al. [33] reported that the xylanase from Paenibacillus sp. strain E18 was stable at 40°C but that it rapidly lost its activity above 50°C. IIPSP3 xylanase activity decreased progressively over time at 90°C, with complete thermal inactivation after 3 h. The enzyme was found to be 70% active at 90°C. Ko et al. [16] reported that the xylanase from Paenibacillus sp. 2S-6 had an optimum temperature 50°C. Lee et al. [21] reported that the Paenibacillus sp. DG-22 xylanase expressed in Escherichia coli showed a decrease in xylanase activity at temperatures of more than 65°C.
The pI of IIPSP3 xylanase was calculated to be 5.38. The half-life of the enzyme was found to be 6 h at 60°C and 2 h at 90°C. Lee et al. [21] reported a half life of the Paenibacillus sp. 2S-6 xylanase as 35 min at 55°C. The thermostability of this enzyme may be due to the presence of aliphatic residues in place of aromatic ones. When the aliphatic index of the IIPSP3 xylanase was studied from the internal amino acid sequence (aliphatic index is defined as the relative volume occupied by aliphatic side chains alanine, valine, isoleucine, and leucine), the average mole% of the four amino acids was found to be 34.50, compared to 8.00 of aromatic amino acids. This increased presence of alophatic residues is regarded as a positive factor in terms of increasing the thermostability of globular proteins [13].
The effect of pH on IIPSP3 xylanase activity was found to be a bell-shaped curve, with an optimal specific activity of 3,000 ± 17.0 U mg−1 at pH 4.5 (Fig. 5). The enzyme was active at pH 3.5 and 9.5, with 40 and 67% of maximum activity, respectively, which makes this xylanase perfectly suitable for applications in both the biofuel and pulp and paper industries. The stability of IIPSP3 xylanase over the broad range of pH appears to be characterized by its spatial distribution of charged residues. Similar results were reported from the xylanase produced by A. kawachii XynC, which was acidophilic but also stable within a pH range of 1–9 [10].
Effects of metal ions and chemicals
The effect of metal ions on IIPSP3 xylanase activity was determined at pH 4.5 and 60°C (Table 3). Enhancement of xylanase activity due to the presence of different metal salts (10 mM) in the reaction mixture was found to be in the following order: K+> Na+> Co2+> Ca2+> Mn2+> Mg2+> Zn2+> Fe2+> Ba2+. IIPSP3 xylanase was highly activated by K+, Na+, and Co2+, with a relative activity of 139, 119, and 117%, respectively, of the original activity. IIPSP3 xylanase was also activated by DTT and 0.5% Triton X 100. Enhancement of xylanase activity by reducing agents DTT and β-ME was also reported for the xylanases from Bacillus sp. JB-99 [14]), Bacillus amyloliquefaciens [6], and Bacillus sp. SPS-0 [3]. The increase in enzyme activity by reducing agents indicates that involvement of disulfide bonds in the native conformation of IIPSP3 xylanase is either negligible or the presence of buried (solvent inaccessible) disulfide bonds.
The enzyme activity was completely inhibited by sulphydryl oxidant metals, such as Cu2+ and Hg2+, and moderately inhibited by Ba2+, Fe2+, and Zn2+. Shi et al. [33], Cho and Bai [9], and Yin et al. [37] reported that xylanase activity produced from Bacillus sp. DSNC 101 was inhibited by Cu2+ and Hg2+. The IIPSP3 xylanase retained 71.4% relative activity in the presence of 10 mM EDTA, which suggests that P. macerans IIPSP3 xylanase is not completely a metallozyme. Most native macromolecules are rapidly unfolded by urea, but IIPSP3 xylanase was found to be 89% stable in the presence of 1% (w/v) urea. IIPSP3 xylanase activity was enhanced in the presence 0.5% (v/v) Triton X 100, with a relative activity of 117.4%, while no effect was observed on its activity in the presence of 1% SDS.
Kinetic constants
The xylanase activity of IIPSP3 xylanase (enzyme concentration 1 mg ml−1) was estimated at different concentrations of beechwood and birchwood xylan (Fig. 6). IIPSP3 xylanase had a high hydrolytic activity on xylans from birchwood, beechwood, and oat spelt, indicating that the substrate binding domain of xylanase has a high affinity for xylans from both softwood (birchwood and beechwood) and hardwood (oat spelt). Apparent K m values for IIPSP3 xylanase on beechwood xylan and birchwood xylan were found to be 6.0 and 6.1 mg ml−1, respectively. The V max values for IIPSP3 xylanase on beechwood xylan and birchwood xylan were found to be 7,407 and 7,575 U mg−1, respectively. The K m and V max values of beechwood and birchwood xylan indicate that IIPSP3 xylanase has the same affinity for both xylans. The K m and V max results of IIPSP3 are comparable with those for bacterial xylanase from Thermotoga maritime MSB8 (K m 1.1 mg ml−1 and V max 4,760 μM min−1 mg−1) and fungal xylanase from Thermomyces lanuginosus-SSBP (K m 3.26 mg ml−1 and V max 6,300 μM min−1 mg−1) [4]. The binding of xylanase to various xylans from birchwood and beechwood and to oat spelt xylan might be due to the absence of xylan-specific binding domains. Specific binding to oat spelt xylan was also found in Streptomyces chattanoogensis CECT 3336 [25].
Hydrolysis of different substrates
IIPSP3 xylanase was able to efficiently hydrolyze a variety of xylans, including those from water-soluble beechwood and birchwood xylan to oatspelt xylan. A higher affinity was observed with oatspelt xylan, for which the specific activity was found to be 4,171 ± 23.7 U mg−1. When agro-based feedstocks, such as corn cobs, bagasse, and wheat bran, were used as substrates, IIPSP3 xylanase efficiently hydrolyzed all of the lignocellulosic substrates with a relative activity (considering 4,171 ± 23.7 U mg−1 as 100% activity) of 87% on corn cobs, 85% on wheat bran, and 65% on bagasse. The relative hydrolytic activity of IIPSP3 on bagasse pre-treated with 0.1% (w/w) H2SO4 was found 85%. A higher specific activity was observed with pre-treated bagasse than with untreated bagasse. Analysis of the hydrolysis products of pure xylan and raw feedstocks revealed the presence of xylose, xylo-oligosaccharides, and glucose (in the case of lignocellulosic feedstock), which suggests the endo mode of action of xylanase (data not shown).
Effect of protease on xylanase and shelf-life stability of enzyme
The activity of IIPSP3 xylanase (4,170 ± 19.2 U mg−1) was not inhibited when treated with proteinase K (Sigma), indicating that IIPSP3 xylanase is a protease-resistant enzyme. Studies on various xylanases have revealed that metabolic enzymes, such as protease, affect the yield of xylanases [17]. We found that both the purified and crude enzyme showed good stability, without any preservative, in the liquid state, when preserved for 6 months at 4°C; there was no loss of enzyme activity. IIPSP3 xylanase was highly stable in both the pH 4.5 and 9.0 buffers even after 60 days of incubation (specific activity 4,168 ± 23.1 U mg−1 at 60°C and at pH 4.5). IIPSP3 xylanase was found to be highly stable under storage conditions at pH 4.5 and 9.0 and at temperatures between 4 and 25°C, without any loss of activity for up to 6 months. Even when xylanase was frozen, its activity was not lost when checked after thawing at 60°C and at pH 4.5 (4,166 ± 22.8 U mg−1).
Conclusions
In this study, the novel strain P. macerans IIPSP3 isolated from wood-feeding termites could be utilized to digest agro-based waste products, such as corn cobs, as a substrate for growth, producing high levels of xylanase. This enzyme therefore is a potential candidate for industrial production and applications. IIPSP3 xylanase is a novel enzyme that can be used in the biofuel industries as it efficiently works at pH 4.5 and in the paper and pulp industry as it can efficiently work at pH 9.0. This ability to be active at both pH extremes and at high temperatures make this xylanase an ideal enzyme for various industries. The isolated xylanase exhibits a high molecular mass (205 kDa), possibly indicating it is a complex of xylanolytic enzymes. Further study is in progress to determine whether this high-molecular-mass xylanase is a complex of multiple xylanases or a multidomain enzyme.
References
Anonymous (2007) Termite guts may yield novel enzymes for better biofuel production. In: ScienceDaily 25 Nov. Available at: http://www.sciencedaily.com-/releases/2007/11/071121145002.htm. Accessed: 21 Apr 2010
Anonymous (2007) Biofuels: bringing biological solutions to energy challenges. US Department of Energy Office of Science, Washington D.C.
Bataillon M, Cardinali APN, Castillon N, Duchiron F (2000) Purification and characterization of a moderately thermostable xylanase from Bacillus sp. strain SPS-0. Enzyme Microb Technol 26:187–192
Beg QK, Kapoor M, Mahajan L, Hoondal GS (2001) Microbial xylanases and their industrial applications: a review. Appl Microbiol Biotechnol 56:326–338
Biely P (1985) Microbial xylanolytic systems. Trends Biotechnol 3:286–290
Breccia JD, Sineriz F, Baigori MD, Castro GR, Hatti-Kaul R (1998) Purification and characterization of a thermostable xylanase from Bacillus amyloliquefaciens. Enzyme Microb Technol 22:42–49
Brennan YL, Callen WN, Christoffersen L, Dupree P, Goubet F, Healey S et al (2004) Unusual microbial xylanases from insect guts. App Environ Microbiol 70:3609–3617
Brune A (2007) Woodworker’s digest. Nature 450:487–488
Cho MC, Bai S (1997) Purification and characterization of xylanase from Bacillus sp. strain DSNC 101. J Microbiol Biotechnol 7:386–390
Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23
Cordeiro CAM, Martins MLL, Luciano AB, da Silva RF (2002) Production and properties of xylanase from thermophilic Bacillus sp. Braz Arch Biol Technol 45:413–418
Dheeran P, Kumar S, Jaiswal YK, Adhikari DK (2010) Characterization of hyperthermostable α-amylase from Geobacillus sp. IIPTN. Appl Microbiol Biotechnol 86:1857–1866
Ikai AJ (1980) Thermostability and aliphatic index of globular proteins. J Biochem 88:1895–1898
Johnvesly B, Virupakshi S, Patil GN, Ramalingam A, Naik GR (2002) Cellulase-free thermostable alkaline xylanase from thermophilic and alkalophilic Bacillus sp. JB-99. J Microbiol Biotechnol 12:153–156
Ko CH, Lin ZP, Tu J, Tsai CH, Liu CC, Chen HT, Wang TP (2010) Xylanase production by Paenibacillus campinasensis BL11 and its pretreatment of hardwood kraft pulp bleaching. Int Biodet Biodeg 64:13–19
Ko CH, Tsai CH, Tu J, Yang BY, Hsieh DL, Jane WN, Shih TL (2011) Identification of Paenibacillus sp. 2S–6 and application of its xylanase on biobleaching. Int Biodet Biodeg 65:334–339
Kulkarni N, Shendye A, Rao M (1999) Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev 23:411–456
Kumar S, Singh SP, Mishra IM, Adhikari DK (2009a) Ethanol and xylitol production from glucose and xylose at high temperature by Kluyveromyces sp. IIPE453. J Ind Microbiol Biotechnol 36:1483–1489
Kumar S, Singh SP, Mishra IM, Adhikari DK (2009b) Recent advances in production of bioethanol from lignocellulosic biomass. Chem Eng Technol 32:517–526
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee TH, Lim PO, Lee YE (2007) Cloning characterization and expression of Xylanase A gene from Paenibacillus sp. DG-22 in Escherichia coli. J Microbiol Biotechnol 17:29–36
Lin LL, Thomson JA (1991) An analysis of the extracellular xylanases and cellulases of Butyrivibrio fibrisolvens H17c. FEMS Microbiol Lett 84:197–204
Lineweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56:658–666
Liu CJ, Suzuki T, Hirata S, Kawai K (2003) The processing of high-molecular-weight Xylanase (XynE, 110 kDa) from Aeromonas caviae ME-l to 60-kDa xylanase (XynE60) in Escherichia coli and purification and characterization of XynE60. J Biosci Bioeng 95:95–101
Lopez-Fernandez CL, Rodriguez J, Ball AS, Copa-Patino JL, Perez-Leblic MI, Arias ME (1998) Application of the affinity binding of xylanases to oat-spelt xylan in the purification of endoxylanase CM-2 from Streptomyces chattanoogensis CECT 3336. Appl Microbiol Biotechnol 50:284–287
Lowry OH, Rosebrough AJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–273
Matson E, Ottesen E, Leadbetter J (2007) Extracting DNA from the gut microbes of the termite (Zootermopsis nevadensis). J Vis Exp 4:195
Matsui T, Tokuda G, Shinzato N (2009) Termites as functional gene resources. Recent Patents Biotechnol 3:10–18
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugars. Anal Chem 31:426–428
Ninawe S, Kuhad RC (2005) Use of xylan-rich cost effective agro-residues in the production of xylanase by Streptomyces cyaneus SN32. J Appl Microbiol 99:1141–1148
Pason P, Kyu KL, Ratanakhanokchai K (2006) Paenibacillus curdlanolyticus strain B-6 xylanolytic—ellulolytic enzyme system that degrades insoluble polysaccharides. Appl Environ Microbiol 72:2483–2490
Schäfer A, Konrad R, Kuhnigk T, Kämpfer P, Herte H, König H (1996) Hemicellulose-degrading bacteria and yeasts from the termite gut. J Appl Microbiol 80:471–478
Shi P, Tian J, Yuan T, Liu X, Huang H, Bai Y, Yang P, Chen X, Wu N, Yao B (2010) Paenibacillus sp. strain E18 bifunctional xylanase-glucanase with a single catalytic domain. Appl Environ Microbiol 76:3620–3624
Varma A, Kollia BK, Paula J, Saxenaa S, König H (1994) Lignocellulose degradation by microorganisms from termite hills and termite guts: a survey on the present state of art. FEMS Microbiol Rev 15:9–28
Wang CY, Chan H, Lin HT, Shyu YT (2010) Production, purification and characterization of a novel halostable xylanase from Bacillus sp. NTU-06. Ann Appl Biol 156:187–197
Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT et al (2007) Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450:560–565
Yin LJ, Lin HH, Chiang YI, Jiang ST (2010) Bioproperties and purification of xylanase from Bacillus sp. YJ6. J Agric Food Chem 58:557–562
Acknowledgments
All of the authors thank Dr M.O. Garg, Director IIP, Dehradun for his valuable suggestions and encouragement to carry out this research and also acknowledge their thanks to TCGA, New Delhi for the internal amino acid sequencing using LC–MS/MS. One of the authors gratefully acknowledges a Senior Research Fellowship awarded by the Council of Scientific and Industrial Research (CSIR), India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dheeran, P., Nandhagopal, N., Kumar, S. et al. A novel thermostable xylanase of Paenibacillus macerans IIPSP3 isolated from the termite gut. J Ind Microbiol Biotechnol 39, 851–860 (2012). https://doi.org/10.1007/s10295-012-1093-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-012-1093-1