Abstract
The goal of this study was to show that the metabolism of Klebsiella pneumoniae under different aeration strategies could be monitored and predicted by the application of chemometric models and fluorescence spectroscopy. Multi-wavelength fluorescence was applied to the on-line monitoring of process parameters for K. pneumoniae cultivations. Differences observed in spectra collected under aerobiosis and anaerobiosis can be explained by the different metabolic states of the cells. To predict process variables such as biomass, glycerol, and 1,3-propanediol (1,3-PD), chemometric models were developed on the basis of the acquired fluorescence spectra, which were measured continuously. Although glycerol and 1,3-PD are not fluorescent compounds, the results showed that this technique could be successfully applied to the on-line monitoring of variables in order to understand the process and thus improve 1,3-PD production. The root mean square errors of predictions were 0.78 units, 10 g/L, and 2.6 g/L for optical density, glycerol, and 1,3-PD, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Different spectroscopic techniques have been studied for their potential applications in monitoring and controlling various biological and chemical processes [1, 2]. Among these spectroscopy methods, 2D-fluorescence spectroscopy has received considerable attention for the noninvasive monitoring of many biological processes. Using a multi-wavelength excitation (ex)/emission (em) spectrofluorometer, the fluorescence of fluorophors like NADH, amino acids such as tyrosine and tryptophan, and vitamins like pyridoxine and riboflavin can be simultaneously measured [3, 4].
Fluorescence sensors are optical measuring instruments that have been used in the last 15 years for different application fields in biotechnology, e.g., for detection of biomass, characterization of reactors, and particularly for bioprocess monitoring [5–8]. In general, optical sensor measurements are ideal for monitoring bioprocesses because they can be performed on-line and in situ, and do not interfere with cells or media [9]. The potential application of 2D-fluorescence spectroscopy has been investigated in the cultivation of various microorganisms [5, 10]. Marose et al. [5] correlated the off-line cell mass concentration during Saccharomyces cerevisiae cultivation with the culture fluorescence in the different regions of protein, NAD(P)H, riboflavin, and pyridoxine. This method for the on-line detection of biomass is especially suitable if the cells cannot be counted or solid particles in the medium exclude the method of weighing biomass. Even metabolic changes such as anaerobic–aerobic transitions were detected by the 2D-fluorescence spectroscopy. Changes in the relation of oxidized to reduced form of NADH or FADH2 during the aerobic–anaerobic transitions are detected in the region of NADH fluorescence (ex 350/em 450 nm) and FAD/FMN (ex 450/em 530 nm).
Chemometric methods such as the principal component analysis (PCA), the principal components regression (PCR), and the partial least square (PLS) regression are useful for the quantitative analysis of the spectroscopic data [2, 3]. These techniques have been used to analyze metabolic changes in recombinant E. coli [11], study the bioprocess characterization of Claviceps purpurea [1], monitor the state variables in heterologous protein production by Pichia pastoris [12], monitor the on-line process in recombinant E. coli fermentations [2], determine the cell mass and antibiotic polymyxin B in Bacilus polymyxa [13], and to estimate the biomass, glucose, and ethanol in S. cerevisiae cultivation [14].
Klebsiella pneumoniae is a gram-negative facultative bacterium that has been widely investigated owing to its wide substrate tolerance and use, and, concerning this work, its high yields and productivities of 1,3-PD. This chemical has acquired industrial importance in recent years because of its wide range of applications, such as in the synthesis of polyesters, polyethers, and polyurethanes [15, 16]. The production of 1,3-PD by K. pneumoniae has been reported in several works, most of which use pure glycerol as the carbon source for cell growth and product synthesis [17–20]. However, raw glycerol, a by-product from biodiesel synthesis that is becoming an abundant and very cheap raw material, could be used as well in this bioprocess [21–23]. Glycerol metabolism by K. pneumoniae can be followed through the reductive and/or oxidative pathways. In anaerobiosis, cells will take up glycerol, which is first converted to 3-hydroxypropionaldehyde (3-HPA) by glycerol dehydratase (GDHt) and then reduced to 1,3-PD by 1,3-propanediol oxidoreductase (PDOR). Under conditions leading to the oxidative pathway, glycerol is dehydrogenated by NAD+-dependent glycerol dehydrogenase (GDH) to dihydroxyacetone (DHA), which is then phosphorylated by ATP-dependent dihydroxyacetone kinase (DHAK) before entering glycolysis. The main products of these metabolic pathways are ethanol, acetic acid, lactic acid, 2,3-butanediol (2,3-BD), while providing energy and reducing equivalents (NADH2) for biomass and 1,3-PD synthesis [16, 22, 24].
Although some works have reported the K. pneumoniae production of 1,3-PD and tried to elucidate the mechanisms of the enzymatic pathway [24–26], so far none has used the 2D-fluorescence method for following the K. pneumoniae fermentation kinetics. Although not new, 2D-fluorescence could be applied in this process in order to better elucidate the metabolic changes that are involved during K. pneumoniae cultivation. In this context, the aims of this research are to evaluate the use of in situ 2D-fluorescence spectroscopy combined with chemometric modeling for the estimation of metabolic changes of K. pneumonia. Metabolites formed during fermentation using raw glycerol from biodiesel synthesis as the sole carbon source, in particular 1,3-PD, were evaluated.
Materials and methods
Chemicals and substrate
All chemicals used in this research were of analytical grade and purchased from Sigma-Aldrich (St. Louis, USA). The raw glycerol used as the sole carbon source for cell cultivations was supplied by a biodiesel manufacturing plant (Passo Fundo, Brazil) and had a composition of (mass fraction) glycerol, 82.4%; ashes, 5.73%; NaCl, 6.17%; moisture, 11.49%; and pH 5.13, monoacylglycerols, 0.39%.
Microorganism maintenance and inocula preparation
Klebsiella pneumoniae BLh-1 was isolated from the sludge of a soybean processing plant (Esteio, Brazil). This strain was identified by 16S rDNA gene sequencing and identified as K. pneumoniae as reported elsewhere [27]. Stocks of this bacterium are kept at the Microbiology Culture Collection of BiotecLab (UFRGS, Brazil). Working stocks of cultures were maintained frozen at −18°C in (volume fraction) 20% glycerol suspensions, whereas for immediate use, cells were maintained on Luria–Bertani (LB) agar slant at 4°C and subcultured every 2 weeks. Inocula were prepared by transferring one loopful of cells into 500-mL flasks filled with 200 mL of medium and then incubated at 37°C in a rotary shaker at 150 rpm overnight. Inocula were standardized as 1 OD unit at 600 nm. The medium contained (in g/L) 5 yeast extract, 5 peptone, 7 (NH4)2SO4, 7 K2HPO4, 1 mL trace elements solution, and 65 of raw glycerol. The trace elements solution contained (in g/L) 0.1 MnCl2·4H2O, 0.06 H3BO3, 0.0037 CuSO4·5H2O, 0.2 CoCl2·6H2O, 0.025 NiCl2·6H2O, 0.035 Na2MoO4·2H2O, 0.14 ZnSO4·7H2O, and 0.9 mL HCl (volume fraction 37%).
Batch cultivation experiments were performed in a fully equipped 10-L bioreactor (Biostat, B. Braun, Germany). The anaerobic cultivations were run at 37°C and a stirrer speed of 250 rpm. Nitrogen was added to ensure anaerobic conditions. In all cultivations the pH was controlled at 7.0 by automatic addition of either 10 M NaOH or 1 M H3PO4. The aerobic cultivations were carried out with an airflow rate of 0.5 vvm (volumes of air/volume of culture). The bioreactors were inoculated with 10% volume fraction of inoculum. Anaerobic experiments were performed in triplicates, whereas aerobic cultivations were run in duplicates.
Off-line analysis
The composition of the cultivation broth was analyzed using a Merck LaChrom 7000 series liquid chromatograph (VWR GmbH, Darmstadt, Germany) equipped with a RID-10A refractive index detector. The stationary and mobile phases were an Aminex HPX-87H column (300 × 7.8 mm, Bio-Rad, USA) and 0.005 mol/L H2SO4 solution at 0.7 mL/min, respectively. The column temperature was controlled at 65°C. The culture samples were prepared by centrifugation at 3,500 g for 10 min to pellet the cell, followed by filtration using cellulose acetate membrane filters (pore size 0.22 μm, Sartorius, Germany). Biomass concentration was estimated by measuring optical density (600 nm) and cell dry weight method, which consisted in filtering (Millipore cellulose nitrate membranes, 0.45 μm pore diameter) 10 mL of sampled culture and drying to constant weight at 80°C in an oven.
Fluorescence spectroscopy
The fluorescence measurements were performed with the BioView® sensor (DELTA Light & Optics, Denmark). Marose et al. [5] presented one of the first and well-detailed works on the use the BioView® sensor for measuring the cell growth and metabolism change of cells of several bacteria and S. cerevisiae. The BioView® sensor is equipped with a xenon flash lamp for the excitation light. The sensor uses two different filter wheels with 16 different filters for excitation and emission and a photomultiplier for detection of the emission light. The liquid light guide is connected to a quartz window of the bioreactor. The measurements were carried out in steps of 20 nm in the excitation wavelength ranging from 270 to 550 nm and the emission wavelength ranging from 310 to 590 nm. During the cultivation 2D-fluorescence spectra were continuously collected.
Chemometric modeling
The chemometric modeling was performed using MATLAB® 7.4.0.287 (R2007a) (The Math Works™). For every process variable, a single partial least square model (PLS-1 model) was calculated on the basis of the data of two anaerobic cultivations. The centered data of the fluorescence spectra were used as independent variables and the values of the process variables determined off-line were the dependent variables. Leave-one-out cross-validation was used for validation of the chemometric models and to determine the number of components for each model, which were used to predict external data and the error of prediction was calculated. The comparison by root mean square error of validation (RMSEV) of predictions of the left out values with the values determined off-line was performed using one to ten principal components. The first principal component is dominated by the fluorescence area of FMN/FAD coenzymes (riboflavin, mainly 450/510, 430/530, and 470/530). The second principal component is also dominated by the fluorescence area of FMN/FAD coenzymes (pyridoxine, mainly 430/490, 410/490, and 390/490). The third principal component is dominated by the fluorescence area of NADH and protein. Finally, the fourth principal component is dominated by the fluorescence area of all principal components mentioned above. The minimal RMSEV decides how many principal components are used for prediction of process variables of a new cultivation run. Here the root of the mean square error of predicted and measured values (RMSEP, Eq. 1) is used to assess the quality of the model.
Results and discussion
Bioreactor cultivations
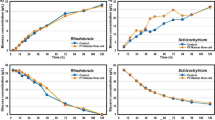
During the cultivations of K. pneumoniae BLh-1, the fluorescence signal was recorded continuously and the process variables (biomass, glycerol consumption, and products formation) were analyzed off-line. The time courses of these process variables during the cultivations are shown in Fig. 1. The anaerobic cultivation of K. pneumoniae (Fig. 1a) showed a quick increase in biomass and product formation up to 10 h. The highest 1,3-PD concentration was 26.84 g/L. Rapid glycerol consumption was observed, being completely depleted by 15 h, showing the exceptional ability of this strain to metabolize raw glycerol. The aerobic kinetics was similar to the anaerobic cultivation concerning biomass and glycerol consumption (Fig. 1b). However, 1,3-PD production was significantly lower, reaching a maximal of 17.67 g/L. Other by-products were produced in both cultivations such as lactic acid, acetic acid, and ethanol, again suggesting the potential use of this strain combined with raw glycerol for other industrial bioprocesses. Table 1 shows the kinetics parameters for K. pneumoniae BLh-1. The yield and productivity were higher for anaerobic cultivation compared with aerobic cultivation. The main by-product was lactic acid with similar yields in both cultivations. These results compare well with other reports found in the literature using glycerol as carbon source. For instance, Zhang et al. [18] studied the rapid conversion of pure glycerol to 1,3-PD by a K. pneumoniae strain isolated from soil. The authors obtained a maximal production of 12.2 g/L of 1,3-PD in batch fermentation with molar yield and productivity of 0.75 and 1.53 g L−1 h−1, respectively. The main by-product was acetate with little accumulations of ethanol and lactate. Cheng et al. [28] studied the 1,3-PD production using K. pneumoniae M5aL using pure glycerol at 50 g/L with different aeration strategies. The authors showed that the higher 1,3-PD yield of 0.54 mol/mol was obtained using N2 in batch fermentation with 0.62 g L−1 h−1 of productivity. With airflow rates of 0.6 vvm, 1,3-PD yield was 0.41 mol/mol with productivity of 0.57 g L−1 h−1. They also observed that the highest concentrations of acetic acid and ethanol were found in the anaerobic batch fermentation. Increasing the airflow rate, acetic acid and ethanol formation decreased while lactic acid formation increased. Using raw glycerol with K. pneumoniae ATCC 700721 to produce 1,3-PD and using experimental design, Oh et al. [23] obtained a final production of 13.74 g/L in batch fermentations and 1,3-PD yield of 0.47 mol/mol.
Batch fermentation for K. pneumoniae BLh-1. a Anaerobic cultivation; b aerobic cultivation. Raw glycerol (squares), 1,3-PD (circles), acetic acid (up-pointing triangles), lactic acid (inverted triangles), biomass (right-pointing triangles), ethanol (diamonds). Results represent the mean of triplicates (anaerobic) and duplicates (aerobic)
The production of 1,3-PD is clearly associated with cell growth and its physiological role is to regenerate the oxidized form of the reducing equivalents, NAD+, to be used in the energy-producing pathways of glycerol degradation [22]. Therefore, accumulation of lactic acid and ethanol will not only inhibit cell growth, but also consume many more equivalents of NADH, directly competing with the formation of 1,3-PD. On the other hand, the formation of acetic acid by cells is independent of NADH [29, 30], thus not interfering in the conversion of glycerol to 1,3-PD. In the metabolic flux of K. pneumoniae, the pathway of glycerol to acetic acid provides part of the energy (ATP) necessary for growth, but in its undissociated form, acetic acid is considered the main inhibitory metabolite [29, 31, 32]. However, the results obtained in this work show that lactic acid produced during the course of fermentation was the main inhibitor of 1,3-PD production, both under aerobiosis and anaerobiosis (Fig. 1).
Fluorescence data
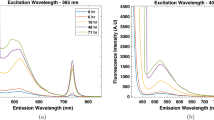
Fluorescence spectra that were gathered during the anaerobic and aerobic cultivations of K. pneumoniae are shown in Fig. 2a, b, respectively, whereas the subtraction of anaerobic minus aerobic spectra showing the differences during the cultivation is shown in Fig. 2c. Figure 2a, b show areas related to fluorescence at ex/em 450/530 nm and ex/em 370/470 nm. These areas are due to the biogenic fluorophors flavine (FAD, FMN) and NADH NADH [1, 5]. As previously reported, fluorescence in the region NADH, riboflavin, and pyridoxine can be used to estimate the cell growth [5, 33]. NADH is involved in redox reactions, carrying electrons from one reaction to another. NADH and FADH2 transfer electrons to O2 during the oxidative phosphorylation, in which ATP is formed. Without oxygen the relation of the oxidized to reduced form is shifted toward the reduced form. NADH can no longer be oxidized via the oxidative phosphorylation and is accumulated. This effect can be visualized in Fig. 2a in a region of NADH fluorescence (ex/em 370/450 nm). When the oxidative phosphorylation is stopped the FMNH2 pool increases. Since the reduced coenzyme does not fluoresce, the FMN fluorescence must decrease under anaerobic conditions.
During the first 12 h of cultivation an increase of NADH fluorescence was observed in both cultivations; after that, there was a rapid depletion of glycerol (Fig. 3a). These data show that this compound is related to the metabolic state of K. pneumoniae. The data for riboflavin (Fig. 3b) show the differences in intensity between anaerobic and aerobic cultivations. Under anaerobiosis, a negative intensity was observed in the region of riboflavin (ex/em 450/530 nm), whereas for the aerobic cultivation a positive intensity for this metabolite was observed (Fig. 2a, b). This fact is also related to cell metabolism, since riboflavin is used in a wide variety of redox reactions and is related to cell respiration. Flavins can act as oxidizing agents because of their ability to accept a pair of hydrogen atoms and the reduction of isoalloxazine ring (FAD, FMN oxidized form) yields the reduced forms of the flavoproteins (FMNH2 and FADH2). The study of these parameters is very important in order to increase the production of 1,3-PD, which is associated with growth. The highest values of 1,3-PD were obtained at around 12 h of cultivation when the NADH was at its highest concentration (Fig. 3a). It has been postulated that the synthesis of 1,3-PD requires NADH as coenzyme and the NADH generated from glycerol determines the yields of 1,3-PD [20]. The conversion of the glycerol to 1,3-PD is a chemical reduction, serving as a sink for reducing equivalents, which are generated by oxidation of the second part of glycerol for energy provision and biomass formation [34].
Chemometric evaluations of fluorescence data recorded during cultivation of K. pneumoniae BLh-1 were carried out by PLS modeling of process variables using the fluorescence spectra as well as the off-line measurements from the cultivations. The PLS models for the glycerol, 1,3-PD, and biomass were based on one, two, and four principal components, respectively. The chemometric models were used to perform predictions of the process variables of the second anaerobic batch cultivation of K. pneumoniae BLh-1. Figure 4 shows the predicted courses of the optical density, glycerol, and 1,3-PD in comparison to off-line measurements, and the RMSEP of predicted values with respect to the off-line measurements are presented in Table 2. The obtained results of predictions represent the kinetics of the off-line measurements of all three variables in a satisfactory way.
Although glycerol and 1,3-PD are not fluorescent substances, their concentrations can be predicted from the fluorescence spectra due to their correlation with the fluorophor concentration in the culture broth. Most likely, the prediction error would have been smaller if the database for calibration had been bigger. According to Ödman et al. [14] the batch variation in the data has an important impact on the calibration. Therefore, if more batch data were included in the calibration, the variation in the chosen number of components would be reduced. Lantz et al. [13] studied the determination of cell mass and polymyxin using multi-wavelength fluorescence in cultures of Bacillus polymyxia. The authors conducted batch cultivations varying the medium content of phosphate and nitrogen in order to achieve different biomass/polymyxin ratios under three different growth conditions. PLS regression was used to establish the mathematical relationships between off-line determined cell mass and polymyxin concentrations with on-line fluorescence data. The authors showed that when the same fluorescence data were used to compute a calibration model, the correlation was only 0.55. However, when separate models were performed for each type of cultivation high correlations and low root mean square error of cross validation (RMSECV) (0.4–0.5 g/L) values were found. Ödman et al. [14] studied on-line estimation of biomass, glucose, and ethanol in S. cerevisiae using in situ multi-wavelength fluorescence and standard monitoring sensors. Batch cultivations were performed at high (190–305 g/L) or low (21–25 g/L) glucose gravity conditions. The authors showed that the best models using high gravity cultivation had RMSEPs of 1.0 g/L CDW, 1.8 g/L ethanol, and 5.0 g/L consumed glucose, corresponding to 4, 2, and 2% concentration intervals, respectively. Values obtained for low gravity models were 0.3 g/L CDW, 0.7 g/L ethanol, and 1 g/L consumed glucose, corresponding to 4, 8, and 4% concentration intervals, respectively.
Conclusion
It has been demonstrated that 2D-fluorescence and chemometric models can be used for quantitative prediction of biomass, substrate, and product formation during batch cultures of K. pneumoniae BLh-1. The fluorescence spectra allowed the metabolic state of the cells to be followed. Glycerol and 1,3-PD, which are not fluorescent compounds, could be successfully determined by applying fluorescence in order to monitor the process. The higher than expected predicted error could be a consequence of the exceptionally fast metabolism of K. pneumonia BLh-1 towards glycerol consumption, biomass formation, and 1,3-PD production associated with this metabolism. The 2D-fluorescence measurements produced earlier, more extensive, and equally reliable information about the metabolic changes of cultures when compared with the traditional off-line analytical methods. Another advantage of the 2D-fluorescence method is that it does not introduce sampling effects because it is noninvasive. The combination of fluorescence and chemometric models is suitable for on-line bioprocess monitoring and process prediction thereby creates an effective tool for bioprocess improvement.
References
Boehl D, Solle D, Hitzmann B, Scheper T (2003) Chemometric two-dimensional fluorescence data for Claviceps purpurea bioprocess characterization. J Biotechnol 105:179–188
Rhee J, Kang TH (2007) On-line process monitoring and chemometric modeling with 2D-fluorescence spectra obtained in recombinant E coli fermentations. Process Biochem 42:1124–1134
Solle D, Geissler D, Stärk E, Scheper T, Hitzmann B (2003) Chemometric modeling based on 2D-fluorescence spectra without a calibration measurement. Bioinformatics 19:173–177
Stärk E, Hitzmann B, Schügerl K, Scheper T, Fuchs C, Köster D, Märkl H (2002) In situ-fluorescence-probes: a useful tool for non-invasive bioprocess monitoring. Adv Biochem 74:22–38
Marose S, Lindemann C, Scheper T (1998) Two-dimensional fluorescence spectroscopy: a new tool for on-line bioprocess monitoring. Biotechnol Progr 14:63–74
Haack MB, Eliasson A, Olsson L (2004) On-line cell mass monitoring of Saccharomyces cerevisiae cultivations by multi-wavelength fluorescence. J Biotechnol 114:199–208
Haack MB, Lantz AE, Mortensen PP, Olsson L (2007) Chemometric analysis of in-line multi-wavelength fluorescence measurements obtained during cultivations with a lipase producing Aspergilus oryzae strain. Biotechnol Bioeng 96:904–913
Hagedorn A, Legge RL, Budman H (2003) Evaluation of spectrofluorometry as a tool for estimation in fed-batch fermentations. Biotechnol Bioeng 83:104–111
Lindemann C, Marose S, Nielsen HO, Scheper T (1998) 2-Dimensional fluorescence spectroscopy for on-line bioprocess monitoring. Sens actuators B 51:273–277
Mukherjee J, Lindemann C, Scheper T (1999) Fluorescence monitoring during cultivation of Enterobacter aerogenes at different oxygen levels. Appl Microbiol Biotechnol 52:489–494
Johansson L, Lidén G (2006) A study of long-term effects on plasmid-containing Escherichia coli in carbon-limited chemostat using 2D-fluorescence spectrofluorimetry. Biotechnol Progr 22:1132–1139
Surribas A, Geissler D, Gierse A, Scheper T, Hitzmann B, Montesinos JL, Valero F (2006) State variables monitoring by in situ multi-wavelength fluorescence spectroscopy in heterologous protein production by Pichia Pastoris. J Biotechnol 124:412–419
Lantz AE, Jorgensen P, Poulsen E, Lindemann C, Olsson L (2006) Determination of cell mass and polymyxin using multi-wavelength fluorescence. J Biotechnol 121:544–554
Ödman P, Johansen CL, Olsson L, Gernaey KV, Lantz EA (2009) On-line estimation of biomass, glucose and ethanol in Saccharomyces cerevisiae cultivations using in situ multi-wavelength fluorescence and software sensors. J Biotechnol 144:102–112
Xue X, Li W, Li Z, Xia Y, Ye Q (2010) Enhanced 1,3-propanediol production by supply of organic acids and repeated fed-batch culture. J Ind Microbiol Biotechnol 37:681–687
Saxena RK, Anand P, Saran S, Isar J (2009) Microbial production of 1,3-propanediol: recent developments and emerging opportunities. Biotechnol Adv 27:895–913
Zhang Q (2009) Metabolic pathway analysis of glycerol metabolism in Klebsiella pneumoniae incorporating oxygen regulatory system. Biotechnol Progr 25:103–115
Zhang G, Ma B, Xu X, Li C, Wang L (2007) Fast conversion of glycerol to 1,3-propanediol by a new strain of Klebsiella pneumoniae. Biochem Eng J 37:256–260
Chen X, Xiu Z, Wang J, Zhang D, Xu P (2003) Stoichiometric analysis and experimental investigation of glycerol bioconversion to 1,3-propanediol by Klebsiella pneumoniae under microaerobic conditions. Enzym Microbiol Tech 33:386–394
Biebl H, Menzel K, Zeng AP, Deckwer WD (1999) Microbial production of 1,3-propanediol. Appl Microbiol Biotechnol 52:289–297
Moon C, Ahn JH, Kim SW, Sang BI, Um Y (2010) Effect of biodiesel-derived raw glycerol on 1,3-propanediol production by different microorganism. Appl Biochem Biotechnol 161:502–510
Huang H, Gong CS, Tsao GT (2002) Production of 1,3-propanediol by Klebsiella pneumoniae. Appl Biochem Biotech 98:687–689
Oh B, Seo JW, Choi MH, Kim CH (2008) Optimization of culture conditions for 1,3-propanediol production from crude glycerol by Klebsiella pneumoniae using response surface methodology. Biotechnol Bioproc E 13:666–670
Hao J, Lin R, Zheng Z, Liu H, Liu D (2008) Isolation and characterization of microorganisms able to produce 1,3-propanediol under aerobic conditions. World J Microb Biot 24:1731–1740
Ahrens K, Menzel K, Zeng AP, Deckwer WD (1998) Kinetic, dynamic, and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture: III. Enzymes and fluxes of glycerol dissimilation and 1,3-propanediol formation. Biotechnol Bioeng 59:544–552
Celinska E (2010) Debottlenecking the 1,3-propanediol pathway by metabolic engineering. Biotechnol Adv 28:519–530
Rossi DM, Costa JB, Souza EA, Peralba MCR, Samios D, Ayub MAZ (2011) Comparison of different pretreatment methods for hydrogen production using environmental microbial consortia on residual glycerol from biodiesel. I J Hydrogen Energy 36:4814–4819
Cheng KK, Liu DH, Sun Y, Liu WB (2004) 1,3-Propanediol production by Klebsiella pneumoniae under different aeration strategies. Biotechnol Lett 26:911–915
Du C, Yan H, Zhang Y, Li Y, Cao Z (2006) Use of oxidoreduction potential as an indicator to regulate 1,3-propanediol fermentation by Klebsiella pneumoniae. Appl Microbiol Biotechnol 69:554–563
Zeng AP, Biebl H (2002) Bulk chemicals from biotechnology: the case of 1,3-propanediol production and the new trends. Adv Biochem Eng Biotechnol 74:239–259
Cheng KK, Liu HJ, Liu D (2005) Multiple growth inhibition of Klebsiella pneumoniae in 1,3-propanediol fermentation. Biotechnol Lett 27:19–22
Cheng KK, Zhang JA, Liu DH, Sun Y, Yang MD, Xu JM (2006) Production of 1,3-propanediol by Klebsiella pneumoniae from glycerol broth. Biotechnol Lett 28:1817–1821
Horvath JJ, Glazier SA, Sprangler CJ (1993) In situ fluorescence cell mass measurements of Sacharomyces cerevisiae using cellular tryptophan. Biotechnol Progr 9:666–670
Sauer M, Marx H, Mattanovich D (2008) Microbial production of 1,3-propanediol. Recent Pat Biotechnol 2:191–197
Acknowledgments
The authors wish to thank CNPQ, CAPES, and FAPERGS (Brazil), and DAAD (Germany) for the financial support of this research.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Rossi, D.M., Solle, D., Hitzmann, B. et al. Chemometric modeling and two-dimensional fluorescence analysis of bioprocess with a new strain of Klebsiella pneumoniae to convert residual glycerol into 1,3-propanediol. J Ind Microbiol Biotechnol 39, 701–708 (2012). https://doi.org/10.1007/s10295-011-1075-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-011-1075-8