Abstract
(R)-(−)-Mandelic acid (R-MA) is an important intermediate with broad uses. Recently, R-MA production using nitrilase has been gaining more and more attention due to its higher productivity and enantioselectivity. In this work, a new bacterium WT10, which exhibited favorable nitrilase activity and excellent enantioselectivity for production of R-MA by enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile, was isolated and identified as a strain of Alcaligenes faecalis. In order to improve its nitrilase activity for industrial application, the wild-type strain WT10 was further subjected to mutagenesis using a combined LiCl–ultraviolet irradiation and low energy N+ ion beams implantation technique. A valuable mutant strain A. faecalis ZJUTB10 was obtained. The nitrilase specific activity of the mutant strain was greatly improved up to 350.8 U g−1, in comparison with wild-type strain WT10 of 53.09 U g−1. The reaction conditions for R-MA production by mutant strain A. faecalis ZJUTB10 were also optimized. Nitrilase activity in mutant strain showed a broad pH optimum at pH 7.7–8.5. The optimal temperature was 35°C. The highest production rate reached 9.3 mmol h−1 g−1. The results showed that mutant strain A. faecalis ZJUTB10 was a new candidate for efficient R-MA production from (R,S)-mandelonitrile and could potentially be used in industrial production.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
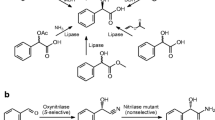
(R)-(−)-Mandelic acid (R-MA) is an important chiral intermediate for the production of pharmaceuticals such as semisynthetic penicillins, cephalosporins, antitumor agents, and antiobesity agents [15]. Currently, R-MA is produced by optical resolution of the racemate with chiral amines, and the maximum theoretical yield of the desired product is only 50% [18]. For the production of the enantiomerically pure R-MA with higher yield, various biological routes have been studied: (i) microbiological hydrolysis of mandelic acid derivatives [19]; (ii) asymmetric reduction of benzoylformic acid with microorganisms [17]; (iii) microbial oxidation of 1-phenyl-1,2-ethanediol [11]; (iv) enzymatic asymmetric synthesis from α-keto aldehyde [12]; (v) microbial oxidation of racemic mandelic acid [10]; (vi) biotransformation of (R,S)-mandelonitrile with nitrilase [20]. Industrial application of the last of these methods is attractive because of its excellent enantioselectivity, cheap starting material which can be synthesized from benzaldehyde by a single step, and above all a possibility of carrying out a dynamic kinetic resolution of substrate which provides theoretically 100% yield of the product (Fig. 1) [13].
Enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile for production of R-MA with nitrilase. The residual (S)-(+)-mandelonitrile which is unreactive to the nitrilase is spontaneously racemized because of chemical equilibrium and then used as the substrate for the nitrilase, thereby being consumed and converted to R-MA
Although a number of microorganisms with nitrilases that can catalyze the formation of carboxylic acids from the corresponding nitriles have been isolated and identified [3], there are only a few reports of microorganisms with nitrilases which can catalyze the formation of R-MA from (R,S)-mandelonitrile [6, 8, 20]. Moreover, the nitrilases from these strains are not satisfactory because of their relatively poor nitrilase activity or stability; thus most of them are not suitable to be applied in industrial process. Therefore, the search for effective biocatalysts has become a hot issue in this field.
In this paper, we describe the screening, identification, and mutagenesis of a newly isolated enantioselective nitrilase-producing strain. This strain was found to be more suitable for the industrial production of R-MA from (R,S)-mandelonitrile as it showed much higher specific activity and stability. The reaction conditions for R-MA production were also investigated by using whole cells.
Materials and methods
Materials
Soil samples used to isolate nitrile-degrading bacteria were collected from different places in Zhejiang Province, China. (R,S)-Mandelonitrile, R-MA, and (S)-(+)-mandelic acid were purchased from J&K Chemical Co., Ltd (Shanghai, China). All the other chemicals were of reagent grade and commercially available.
Media
The minimal salts medium consisted of (per liter) KH2PO4 0.2 g, Na2HPO4 0.8 g, MgSO4 0.2 g, CaCl2 0.1 g, and FeCl3 0.005 g, pH 7.0. The rich medium consisted of (per liter) glucose 10 g, yeast extract 5 g, peptone 5 g, K2HPO4 5 g, MgSO4 0.2 g, FeSO4 0.03 g, and NaCl 1 g, pH 7.0. The optimized medium consisted of (per liter) ammonium acetate 12.14 g, yeast extract 7.79 g, K2HPO4 5 g, MgSO4 0.2 g, NaCl 1 g, pH 7.5. All the media were prepared with tap water and autoclaved at 121°C for 20 min. About 1 g l−1 of ε-caprolactam was added to the rich medium to induce the nitrilase activity, whereas 3.29 g l−1 of n-butyronitrile was added to the optimized medium to induce the nitrilase activity.
Screening for enantioselective nitrilase-producing strains
Microorganisms were isolated from soil samples by enrichment culture. Approximately 5.0 g of each soil sample was suspended in 50 ml of distilled water. About 5 ml of the suspension was transferred to 45 ml of the minimal salts medium containing 2 mM phenylacetonitrile in a 250-ml flask and incubated aerobically at 30°C, 150 rpm for 2 or 3 days. Then, 1.0 ml of the culture was transferred to the same fresh medium, and the incubation was carried out under the same conditions for 2 or 3 days. After three-times enrichment, the liquid cultures were diluted tenfold and spread on solid plates of minimal salts medium containing 2.0 mM phenylacetonitrile. Based on colony characteristics, bacteria were selected and picked up. The resulting isolated single colonies were cultivated in rich medium (50 ml in 250-ml flask) and incubated aerobically at 30°C, 150 rpm for 48 h. The cells were harvested by centrifugation at 4°C and 9,000×g for 15 min, then washed with 10 mM phosphate buffer (pH 7.2) and centrifuged at 9,000×g for 15 min. The colorimetric method for detection of nitrilase activity of the isolated strains was performed in a 96-well microplate format according to the method described by Banerjee et al. [2].
Identification
The identification of the strain was based on standard morphological and physiological characteristics and nucleotide sequence analysis of enzymatically amplified 16S rDNA. The morphological characteristics were observed with both an optical microscope (Leica DM4000 B, Germany) and a scanning electron microscope (Hitachi H-7650, Japan). The biochemical characteristics were examined by a standardized method with a VITEK-AMS 60 all-automatic microbiology analysis system (Biomerieux Company, France) according to the manufacturer’s instructions. For the sequence analysis, chromosomal DNA was extracted according to the procedure of bacterial genome extraction kit (Takara, Dalian, China). The polymerase chain reaction (PCR) amplification was carried out with universal primers: p16S-8: 5′-AGA GTT GAT CCT GGC TCA G-3′ and p16S-1541: 5′-AAG GAG GTG ATC CAG CCG CA-3′ in a thermal cycle (Bio-Rad, USA). The standardized PCR conditions were as follows: 5 min at 95°C, 35 cycles of 40 s at 95°C, 60 s at 53°C, 2 min at 72°C, and one final step of 10 min at 72°C. The PCR products were extracted and purified from the agarose gel using a High Pure PCR Product Purification Kit (Roche, Germany). The resulting PCR fragment was ligated with pMD18-T (Takara, Japan) by using the T/A cloning procedure. The constructed vector was transferred to the component cell E. coli JM109 according to the method of Chung et al. [5] and then spread on the LB plate containing X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactoside) (50 mM), IPTG (isopropyl-1-thio-β-d-galactoside) (50 mg l−1), and ampicillin (50 mg l−1). Subsequently a positive clone, designated E. coli JM109/pMD18-T-NIT, was obtained. DNA was sequenced on both strands with an Applied Biosystems Model 377 B automatic DNA sequencer, and a dye-labeled terminator sequencing kit (Applied Biosystems, Foster, CA, USA). The sequences obtained were compiled and compared with sequences in the GenBank databases using the BLAST program. The sequences were aligned by using multiple sequence alignment software, CLUSTAL W ver. 1.81. A phylogenetic tree was constructed with MegAlign software (DNASTAR, Inc., Madison, WI, USA) based on the partial 16S rDNA sequences of 16 strains similar to strain WT10.
Mutagenesis
The wild-type strain WT10 was grown in a 250-ml flask containing 50 ml of rich medium supplemented with 0.5% (w/v) LiCl. After 20 h incubation at 30°C and 150 rpm, cells were harvested by centrifugation, and then washed with 0.9% saline solution. Mutagenesis using 107–108 cells ml−1 with ultraviolet (UV) irradiation was first carried out. The cell suspension was distributed in 9-cm Petri plates (2–3 ml in each plate) and exposed to a UV tube (15 W) for 20 s keeping the distance of the UV source fixed at 25 cm. After exposure, the rich medium supplemented with 0.5% (w/v) LiCl (2.0 ml) was added and the sample was incubated for 2 h in the dark, and then spread on solid plates of rich medium containing 2% (w/v) agar for the growth of mutants. Each mutant was cultivated in rich medium (50 ml in 250-ml flask) and incubated aerobically at 30°C, 150 rpm for 48 h, then centrifuged at 9,000×g for 15 min. The colorimetric method was used for detection of nitrilase activity of the mutants [2]. The mutants with high activity were selected for mutation by low energy N+ ion beams (IB) [9]. The ion implantation system used in the present study has been previously described [22]. Implantation sources were produced by an ion beam bioengineering instrument [21]. The diluted cells suspension (about 109–1010 cells ml−1) was spread on 9-cm Petri plates (0.1 ml evenly spread as a thin layer in each plate). The plates were air-dried at an axenic workbench and then implanted by nitrogen ions. A dose of 7.8 × 1014 ions cm−2 and energy of 10 keV were used for implantation in this experiment. Every sample was kept inside the target chamber for less than 2 min to complete the treatment process, including vacuum pump-down, nitrogen ion beam implantation, and the normal pressure recovery. Each plate was then washed with 1 ml of sterile distilled water. After incubation in rich medium (50 ml in 250-ml flask) and being incubated aerobically at 30°C, 150 rpm for 48 h, the positive mutants with higher nitrilase activity than starting strain were identified and selected. The final mutant capable of converting (R,S)-mandelonitrile to R-MA with highest activity and enantioselectivity was selected for further research.
Analytical methods
The amounts of (R,S)-mandelonitrile and mandelic acid were assayed by HPLC (LC-10AS, Shimadzu, Japan) equipped with an octadecylsilica column (250 × 4.6 mm) (Elite Analytical Instruments Co., Ltd, China) at a flow rate of 1.0 ml min−1 with a solvent system composed of 10 mM NH4H2PO4 and methanol (4:1, v/v). Detection wavelength was set at 228 nm.
The optical purity of mandelic acid was determined by analysis of the enantiomers on a CHIRALCEL-OD-H column (250 × 4.6 mm) (Daicel Chemical Industries, Japan) at a flow rate of 0.8 ml min−1 with a mobile phase containing hexane, isopropanol, and trifluoroacetic acid (90:10:0.1, v/v) at A 228. Before optical purity determination, pretreatment steps were performed on the reaction mixture. The pH of the reaction mixture was adjusted to about 1.5 with 6 M HCl, and then the enantiomers were extracted with equal volume of ethyl acetate. The extract obtained was concentrated and the residual solid was dissolved in the mobile phase for HPLC determination.
Enzyme assays
The nitrilase activities were assayed in a standard reaction mixture (10 ml) containing 1.0 g (dry weight) l−1 whole cells suspended in phosphate buffer (100 mM, pH 8.0) and (R,S)-mandelonitrile (15 mM). The reactions were carried out at 30°C for 30 min and then stopped by centrifugation (9,000×g, 4°C for 10 min). The amount of mandelic acid and the degree of (R,S)-mandelonitrile conversion were determined by HPLC as described above. One unit of enzyme activity was defined as the amount of enzyme that produced 1.0 μmol of mandelic acid per minute under standard assay conditions. The specific activity (U g−1) was then calculated from the reaction rate and the whole cells (dry weight) employed in the assay. However, for optimization of reaction conditions, the pH, temperature, cell concentration, and added substrate were varied in each experiment, depending upon the reaction condition being examined.
Results
Screening for (R,S)-mandelonitrile-converting strains
Various microbial strains were isolated from about 80 soil samples through enrichment culture using phenylacetonitrile as the sole source of carbon and nitrogen. A colorimetric method was first used to select those strains which were capable of converting (R,S)-mandelonitrile to mandelic acid according to the method described by Banerjee et al. [2]. Those with obvious color change from the reaction were marked for further identification by HPLC. Ten microorganisms bearing desired enantioselective nitrilase activity were selected out of 255 strains that were capable of utilizing (R,S)-mandelonitrile to produce R-MA (data not shown). Among these isolates, strain WT10 with the maximum nitrilase activity (53.09 U g−1) and enantioselectivity (ee > 99%), was selected as the best strain for further studies.
Characterization and identification of strain WT10
The morphological characteristics of strain WT10 showed that it was short, rod-like, 1.2–1.5 × 0.6–0.8 μm, and non-spore-forming, with polar or bipolar tufts (Fig. 2). Both catalase and oxidase test were positive. Strain WT10 was gram-negative and an obligately aerobic bacterium. Colonies of strain WT10 grown on the solid plates of rich medium were gray and translucent with smooth surface, and the margins of the colonies were thin and undulate. In the glucose oxidative fermentation test, these colonies showed alkaline reaction. In broth medium fermentation test, ammonia was produced and the pH of the medium reached above 8.6. Then the identification of the strain was conducted with a VITEK-AMS 60 all-automatic microbiology analysis system. The results are shown in Table 1. Strain WT10 did not metabolize carbohydrates. It utilized acetamide, citrate, and malonate. These data indicated that strain WT10 resembled a member of Alcaligenes faecalis. The electrophoretogram showed the PCR product was about 1.5 kb. The 16S rDNA (GenBank accession no. GU181289) of the strain was sequenced and compared with the other sequences deposited in Genebank by using BLAST. It was most similar to that of A. faecalis by examining phylogenetic tree (Fig. 3). According to the physiological and biochemical characterization as well as the comparison of 16S rDNA gene sequence, WT10 was identified as a strain of A. faecalis.
Improvement of nitrilase activity by mutagenesis
Although wild-type strain WT10 exhibited excellent enantioselectivity for the production of R-MA from (R,S)-mandelonitrile, the nitrilase activity was still unsatisfactory for industrial application. To increase nitrilase activity, the strain was subjected to a combined mutagenesis with LiCl–UV and IB. After each mutagenic treatment, the resultant mutants were screened first on 96-well microplate formats. The small colonies with smooth outline and dry appearance usually gave higher nitrilase activity. The activity of putative positive mutants toward (R,S)-mandelonitrile was further confirmed by HPLC. The genetic stability of the selected positive mutants was checked for several generations. One relatively stable mutant with higher activity and enantioselectivity was selected for the next mutagenesis step by IB. The genealogy of the mutant is shown in Table 2. By combined mutagenesis of WT10 with LiCl–UV and IB, the resultant mutant strain ZJUTB10 showed a nitrilase specific activity of 350.8 U g−1 with an ee of greater than 99%. Compared with the wild-type strain, the nitrilase specific activity has been increased greatly with excellent enantioselectivity. The nitrilase activity and enantioselectivity of mutant strain ZJUTB10 were checked for 10 generations. No marked change was observed in each generation (Fig. 4), suggesting that it was a genetically stable mutant. Therefore, mutant strain ZJUTB10 was selected for the biotransformation of (R,S)-mandelonitrile to produce R-MA and was deposited at the China Center for Type Culture Collection as CCTCC M 208168.
Effect of temperature and pH on activity and enantioselectivity of nitrilase in mutant strain A. faecalis ZJUTB10
The effects of temperature on activity and enantioselectivity of nitrilase in mutant strain A. faecalis ZJUTB10 were investigated at various temperatures (20–50°C, step 5°C). Reactions were carried out for 30 min at specified temperatures in 100 mM phosphate buffer (pH 8.0) with a cell concentration of 1.0 g (dry weight) l−1. The effect of temperature was assayed as a relative activity. The results are shown in Fig. 5. Within the examined temperature between 20 and 35°C, nitrilase activity increased with increasing reaction temperature and reached its maximum at 35°C. Further increase in temperature led to a slight drop in activity, which might be due to the partial inactivation of the whole cells at higher temperatures. The ee value of product was kept above 99% and showed little variation in temperature range from 20 to 50°C.
Effect of temperature on activity and enantioselectivity of nitrilase in A. faecalis ZJUTB10. Reactions were carried out for 30 min at specified temperatures in 100 mM phosphate buffer (pH 8.0) with a cell concentration of 1.0 g (dry weigh) l−1. The substrate concentration was 15 mM. Open circles relative activity; filled squares ee
The effects of pH on nitrilase activity and enantioselectivity were determined by using reaction mixtures adjusted to specified pH values as follows: pH 4.9–8.0, 100 mM phosphate buffer; pH 8.0–9.0, 100 mM Tris-hydrochloride buffer. Reactions were carried out for 30 min at 30°C in the specified buffers (pH from 4.9 to 9.0) with a cell concentration of 1.0 g (dry weight) l−1. The results illustrated that pH had an obvious effect on both enzyme activity and enantioselectivity (Fig. 6). The profiles of activity and ee value were similar over the pH range from 4.9 to 9.0. Nitrilase activity and enantioselectivity in A. faecalis ZJUTB10 showed a broad pH optimum at pH 7.7–8.5. When the pH was lower than 7.7, the activity and ee value sharply decreased. But, when the pH was above 8.5, the activity and ee value only slightly decreased. Therefore, A. faecalis ZJUTB10 exhibited relatively high nitrilase activity and enantioselectivity under alkaline conditions compared with acidic conditions.
Effect of pH on activity and enantioselectivity of nitrilase in A. faecalis ZJUTB10. Reactions were carried out for 30 min at 30°C in specified buffers (pH from 4.9 to 9.0) with a cell concentration of 1.0 g (dry weight) l−1. The substrate concentration was 15 mM. Open circles relative activity; filled squares ee
Thermostability of nitrilase in mutant strain A. faecalis ZJUTB10
Thermostability is an index of great importance for nitrilase application. To determine the thermal stability of the biocatalyst exhibiting nitrilase activity from A. faecalis ZJUTB10, whole cells were suspended in phosphate buffer (100 mM, pH 8.0). The suspensions were pre-incubated at 30, 38, and 45°C, respectively. Samples were withdrawn at different time to determine the nitrilase activity. Residual activity was calculated by comparing with the initial activity of nitrilase, which had not been subjected to thermal pre-incubation. The natural logarithm of residual nitrilase activity (LnRA) was plotted against time and is shown in Fig. 7. The slopes of the individual line correspond to K Deact at the specified temperature. The half-life (t 1/2) of nitrilase activity at 30, 38, and 45°C was 14, 11, and 1.5 days, respectively. Further research indicated that the half-life of nitrilase activity decreased quickly when the temperature was higher than 50°C. The half-life of nitrilase activity at 55 and 63°C was 240 and 55 min, respectively.
Thermostability profiles of nitrilase from A. faecalis ZJUTB10. The cells were pre-incubated at 30, 38, and 45°C in 100 mM phosphate buffer (pH 8.0) and then enzyme activity was determined. Residual activity was calculated by comparing with the initial activity of nitrilase which had not been pre-incubated. Ln (RA) was plotted against time. The slops of the individual lines correspond to K Deact at the specified temperature. Filled squares 30°C; filled triangles 38°C; filled circles 45°C
Effect of cell concentration on enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile
The effects of cell concentration ranging from 0.2 to 1.8 g (dry weight) l−1 on R-MA production from (R,S)-mandelonitrile were carried out at 30°C in 20 mM (R,S)-mandelonitrile in 100 mM phosphate buffer (pH 8.0). The results are shown in Fig. 8. When the cell concentration was 0.2 g l−1, after 180 min reaction, the R-MA concentration was only 7.0 mM. Increasing the cell concentration from 0.6 to 1.4 g l−1 led to a decreased reaction time for maximal R-MA concentration (from 180 to 90 min). The highest production rate (micromoles of R-MA produced per hour per gram dry weight cells) resulted from using a cell concentration of 1.0 g l−1. As the cell concentration was increased from 1.4 to 1.8 g l−1, the reaction time (about 90 min) required to reach the maximum R-MA concentration did not decrease any more. The time course curves of 1.4 and 1.8 g l−1 cell concentrations were even overlapped which illustrated that the additional cells did not increase the reaction rate. The reason may be the mass transfer limitations caused by a higher amount of cells [4].
Effect of cell concentration on enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile to produce R-MA by A. faecalis ZJUTB10. Reactions were carried out at 30°C in 100 mM phosphate buffer (pH 8.0) with specified cell concentrations. The substrate concentration was 20 mM. Filled squares 0.2 g l−1; filled circles 0.6 g l−1; filled triangles 1.0 g l−1; open circles 1.4 g l−1; open squares 1.8 g l−1
Effect of substrate concentration on enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile
Higher substrate concentration can accelerate the reaction rate from the perspective of chemical equilibrium. However, it also may cause inhibition of enzyme activity because of the toxicity of (R,S)-mandelonitrile. The effects of (R,S)-mandelonitrile concentration ranging from 10 to 50 mM on bioconversion to R-MA were studied at 30°C in 100 mM phosphate buffer (pH 8.0) with a cell concentration of 1.0 g (dry weight) l−1. The results are shown in Fig. 9. Increasing the (R,S)-mandelonitrile concentration from 10 to 50 mM led to an increased R-MA concentration from 9.5 to 45.4 mM and an extended reaction time (from 60 to 540 min) for maximal R-MA concentration. The ee value of all the products remained above 99%. The highest production rate of 9.3 mmol h−1 g−1 resulted in the concentration of 20 mM after a batch reaction.
Effect of substrate concentration on enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile to produce R-MA by A. faecalis ZJUTB10. Reactions were carried out at 30°C in 100 mM phosphate buffer (pH 8.0) with a cell concentration of 1.0 g (dry weight) l−1 and specified substrate concentration. Filled squares 10 mM; filled circles 20 mM; filled triangles 30 mM; open circles 40 mM; open squares 50 mM
Discussion
Nitrilases (EC 3.5.5.1) have attracted substantial interest as a valuable alternative in organic chemical processes, which can catalyze hydrolysis of nitriles to the corresponding carboxylic acids in a single step, and do not produce large amounts of by-products [14]. Especially advantageous is that the enzyme-catalyzed hydrolysis of a variety of nitriles can be highly stereoselective [16]. Some nitrilases have been investigated for the preparation of single enantiomers of carboxylic acids. One of the most important application appears to be the biotransformation of racemic mandelonitrile to produce chiral R-MA [3]. Several bacteria such as A. faecalis ATCC 8750 [20], Pseudomonas putida MTCC 5110 [8], and Alcaligenes sp. ECU0401 [6] were reported to hydrolyze (R,S)-mandelonitrile to produce R-MA with enantioselective nitrilase.
In this work, a new bacterium A. faecalis WT10 which can enantioselectively transform (R,S)-mandelonitrile into R-MA with high ee (>99%) was isolated. By using a combined mutagenesis methods, we have successfully created a mutant strain A. faecalis ZJUTB10 for R-MA production from (R,S)-mandelonitrile. A comparison of the reported nitrilase specific activities, yield, and ee of the microorganisms with those of the strain used here is listed in Table 3; the mutant strain A. faecalis ZJUTB10 showed the highest nitrilase specific activity and yield.
Our investigations found that the optimal temperature for the biotransformation of (R,S)-mandelonitrile to R-MA using A. faecalis ZJUTB10 as biocatalyst was 35°C. Increasing the reaction temperature from 35 to 50°C led to only a slight drop in relatively activity (from 100 to 87.5%). This result was very different compared with other reports [1]. From the results of enzyme thermostability, it was observed that the thermostability of nitrilase in A. faecalis ZJUTB10 was higher than P. putida MTCC 5110 [8] and A. faecalis MTCC 126 [7]. Effect of pH suggested that the nitrilase activity of A. faecalis ZJUTB10 showed a broad pH optimum and better tolerance to alkaline conditions than the nitrilase activity of P. putida 5510. The former could retain above 90% of enzyme activity at pH 9.0, while the latter could retain only about 30% as whole cells were used as biocatalyst [1]. The excellent thermostabilty and pH tolerance of the enantioselective nitrilase in A. faecalis ZJUTB10 made it more suitable for industrial application.
The effects of (R,S)-mandelonitrile concentration (ranging from 10 to 50 mM) on the bioconversion of (R,S)-mandelonitrile by A. faecalis ZJUTB10 showed that the total molar yield of R-MA against racemic substrate reached above 90% with high ee values (>99%). The highest production rate reached 9.3 mol h−1 g−1. Substrate inhibition did not appear in the concentration range examined. However, for whole cells of P. putida 5110, only about 30% conversion for 2 mM of (R,S)-mandelonitrile was achieved in 10 min and ee dropped quickly from 98 to 96% after 40 min when the conversion was 50% [8]. The improvement of conversion rate led to a decline of ee. For whole cells of Alcaligenes sp. ECU0401, the time-dependent change in the substrate and product concentration showed that the R-MA concentration was only 5.2 mM (yield of 26%) after 4 h reaction at (R,S)-mandelonitrile concentration of 20 mM. The R-MA concentration did not change further after 6 h reaction [6]. The substrate toxicity or the product inhibition may be responsible for the unchanged yield. Obviously, mutant strain ZJUTB10 showed the highest production rate.
Comparing the nitrilase specific activity, yield, production rate, and stability, the mutant strain A. faecalis ZJUTB10 was superior to all other nitrilase-producing strains reported [6, 8, 20]. It is one of the best biocatalysts examined and could potentially be used in industrial production of R-MA from (R,S)-mandelonitrile.
References
Banerjee A, Kaul P, Banerjee UC (2006) Enhancing the catalytic potential of nitrilase from Pseudomonas putida for stereoselective nitrile hydrolysis. Appl Microbiol Biotechnol 72:77–87
Banerjee A, Kaul P, Sharma R, Banerjee UC (2003) A high-throughput amenable colorimetric assay for enantioselective screening of nitrilase-producing microorganisms using pH sensitive indicators. J Biomol Screen 8:559–565
Chen J, Zheng RC, Zheng YG, Shen YC (2009) Microbial transformation of nitriles to high-value acids or amides. Adv Biochem Eng Biotechnol 113:33–77
Chen J, Zheng YG, Shen YC (2008) Biotransformation of p-methoxyphenylacetonitrile into p-methoxyphenylacetic acid by resting cells of Bacillus subtilis. Biotechnol Appl Biochem 50:147–153
Chung CT, Niemela SL, Miller RH (1989) One-step preparation of competent Escherichia coli—transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA 86:2172–2175
He YC, Xu JH, Xu Y, Ouyang LM, Pan J (2007) Biocatalytic synthesis of (R)-(−)-mandelic acid from racemic mandelonitrile by a newly isolated nitrilase-producer Alcaligenes sp ECU0401. Chin Chem Lett 18:677–680
Kaul P, Banerjee A, Banerjee UC (2006) Stereoselective nitrile hydrolysis by immobilized whole-cell biocatalyst. Biomacromolecules 7:1536–1541
Kaul P, Banerjee A, Mayilraj S, Banerjee UC (2004) Screening for enantioselective nitrilases: kinetic resolution of racemic mandelonitrile to (R)-(−)-mandelic acid by new bacterial isolates. Tetrahedron Asymmetry 15:207–211
Liu ZQ, Zhang JF, Zheng YG, Shen YC (2008) Improvement of astaxanthin production by a newly isolated Phaffia rhodozyma mutant with low-energy ion beam implantation. J Appl Microbiol 104:861–872
Miyamoto K, Ohta H (1992) Enantioselective oxidation of mandelic acid using a phenylmalonate metabolizing pathway of a soil bacterium Alcaligenes bronchisepticus Ku1201. Biotechnol Lett 14:363–366
Oda S, Kikuchi Y, Nanishi Y (1992) Synthesis of optically active mandelic acid via microbial oxidation of racemic 1-phenyl-1, 2-ethanediol. Biosci Biotechnol Biochem 56:1216–1220
Patterson MAK, Szajewski RP, Whitesides GM (1981) Enzymic conversion of alpha-keto aldehydes to optically active alpha-hydroxy acids using glyoxalase I and II. J Org Chem 46:4682–4685
Singh R, Banerjee A, Kaul P, Barse B, Banerjee UC (2005) Release of an enantioselective nitrilase from Alcaligenes faecalis MTCC 126: a comparative study. Bioprocess Biosyst Eng 27:415–424
Singh R, Sharma R, Tewari N, Geetanjali, Rawat DS (2006) Nitrilase and its application as a ‘green’ catalyst. Chem Biodivers 3:1279–1287
Tang KW, Yi JM, Huang KL, Zhang GL (2009) Biphasic recognition chiral extraction: a novel method for separation of mandelic acid enantiomers. Chirality 21:390–395
Wang MX (2005) Enantioselective biotransformations of nitriles in organic synthesis. Top Catal 35:117–130
Xiao MT, Huang YY, Shi XA, Guo YH (2005) Bioreduction of phenylglyoxylic acid to R-(−)-mandelic acid by Saccharomyces cerevisiae FD11b. Enzyme Microb Technol 37:589–596
Xiao MT, Huang YY, Ye J, Guo YH (2008) Study on the kinetic characteristics of the asymmetric production of R-(−)-mandelic acid with immobilized Saccharomyces cerevisiae FD11b. Biochem Eng J 39:311–318
Yadav GD, Sivakumar P (2004) Enzyme-catalysed optical resolution of mandelic acid via RS(∓)-methyl mandelate in non-aqueous media. Biochem Eng J 19:101–107
Yamamoto K, Oishi K, Fujimatsu I, Komatsu KI (1991) Production of R-(−)-mandelic acid from mandelonitrile by Alcaligenes faecalis ATCC 8750. Appl Environ Microbiol 57:3028–3032
Yu Z (2000) Ion beam application in genetic modification. IEEE Trans Plasma Sci 28:128–132
Yu Z, Deng J, He J, Huo Y, Wu Y, Wang X, Lui G (1991) Mutation breeding by ion implantation. Nucl Instrum Methods Phys Res B59(60):705–708
Acknowledgments
This work was supported by the Fund of the National High Technology Research and Development Program of China (863 Program) (No. 2009AA02Z203), the Major Basic Research Development Program of China (973 Project) (No. 2009CB724704), and Natural Science Foundation of Zhejiang Province (No. Z4090612).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Xue, YP., Xu, SZ., Liu, ZQ. et al. Enantioselective biocatalytic hydrolysis of (R,S)-mandelonitrile for production of (R)-(−)-mandelic acid by a newly isolated mutant strain. J Ind Microbiol Biotechnol 38, 337–345 (2011). https://doi.org/10.1007/s10295-010-0778-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-010-0778-6